Process optimization of cellulase production from alkali-treated coffee pulp and pineapple waste using Acinetobacter sp. TSK-MASC
Kandasamy Selvam†
a,
Muthusamy Govarthanan†bc,
Seralathan Kamala-Kannanc,
Munisamy Govindharajud,
Balakrishnan Senthilkumar*a,
Thangasamy Selvankumar*bd and
Arumugam Sengottaiyanb
aCentre for Biotechnology, Muthayammal College of Arts and Science, Rasipuram, Namakkal – 637 408, Tamil Nadu, India. E-mail: nbsenthilkumar@gmail.com
bPG & Research Department of Biotechnology, Mahendra Arts and Science College, Kalippatti, Namakkal – 637501, Tamil Nadu, India. E-mail: t_selvankumar@yahoo.com; Tel: +91-9443470394
cDivision of Biotechnology, College of Environmental and Bioresource Sciences, Chonbuk National University, Iksan – 570752, South Korea
dDepartment of Environmental Biotechnology, Bharathidasan University, Tiruchirappalli – 620 024, Tamilnadu, India
First published on 30th January 2014
Abstract
The aim of this study was to assess the mixed combination of coffee pulp waste (CPW) and pineapple waste (PW) residues for cellulase production using newly isolated Acinetobacter sp. TSK-MASC in solid state fermentation. Response surface methodology based Box–Behnken design (BBD) was employed to optimize variables such as pH, incubation time, concentrations of CPW and PW. The BBD design investigation showed a sound adjustment of the quadratic model with the experimental statistics. Statistics based 3-D plots were generated to evaluate the changes in the response surface and to understand the relationship between the enzyme yield and culture parameters. The higher production (888 U mL−1) was achieved after 60 h of incubation with 3.0 g L−1 of CPW and PW at pH 7.0.
1. Introduction
Cellulases are enzymes which hydrolyse the β-1,4-glucosidic linkages of cellulose. The enzyme is mostly involved in degradation and recycling of abundant cellulosic biomass present in the environment. Cellulases play a major role in industries as well as generation of sustainable energy sources like, glucose, ethanol, hydrogen and methanol.1,2 At present cellulases are the third largest enzymes in the industrial enzyme market worldwide. They have a wide range of applications in cotton processing, paper recycling, the detergent industry, and food industries.3Wide variety of microorganisms such as aerobic and anaerobic bacteria4,5 white rot soft fungi6,7 and anaerobic fungi produce cellulase enzyme.8 In addition to that filamentous fungi, actinomycetes, and aerobic bacteria, cellulases are mostly secreted as free molecules. Most of the cellulases exploited for industrial applications are from filamentous fungi such as Trichoderma, Penicillium, Fusarium, Humicola, Phanerochaete, etc., where a large number of cellulases are encountered.7,9 In recent years bacterial enzymes have received much attention as potential enzymes in the industries. The rapid growth rate, ability to grow in different environmental conditions, ability to utilize wide substrates as carbon and nitrogen source, and secretion of different types of extracellular enzymes enhanced the application of bacteria in fermentation studies.10
Solid state fermentation (SSF) is a process in which fermentation is carried out in solid matrix with limited water; however, the substrate must possess enough moisture for microbial growth and metabolic activity. The minimal amount of water used in SSF allows the maximum production of metabolites and reduces the time required for downstream processing. Compared to submerged fermentation processes, SSF is less expensive, small scale operating vessels, less water utilization, lower energy consumption.11,12 Production of industrially important biomolecules by SSF has been greatly influenced by the optimization of physicochemical properties of the medium.10,13 Optimization of fermentation conditions may enhance the production of the desirable products. Conventional optimization methods are involves changing one independent variable at a time, while other variables remain fixed. However the optimization of variables by using statistical methods are quite interesting because of its simplicity, time consume, less man power.14–16
Response surface methodology (RSM), a statistical tool is universally practiced methodology for design the models and analyzes the most significant problems, in which a response is highly influenced by several variables for the production of industrially important biomolecules and secondary metabolites. It helps to identify the successful factors, study interactions, most favorable conditions, calculate the optimum level of the variables, and ensure the maximum production in a fixed number of experiments.10,17,18
Industrial biotechnology offers budding opportunities for cost-effective consumption of agro-industrial residues such as coffee pulp and coffee husk for the production of enzymes and organic acids.19 Coffee pulp wastes (CPW) are generated during the industrial processing of coffee cherries by wet and/or dry process. These wastes are generated by coffee producing countries (India, Brazil, Vietnam etc.) in large amount throughout the year and are the most abundant renewable resources.19 Pineapple wastes (PW) are found to have potential uses as raw materials that can be converted into value-added products. The peel is a rich source of cellulose, hemicelluloses and other carbohydrates. Pineapple wastes are also proportionally increasing. CPW have not as much of sugar content and sufficient protein but PW holds high amounts of sugars and proteins. The sufficient sugars and proteins from the substrates which promote the growth of microorganisms lead to the production of significant yield of enzymes. So this bi-substrate combination contribute the synergistic effect should be useful technology for production of high quantity of cellulase. Waste disposal represents a growing problem since it is usually prone to microbial spoilage and it causes serious environmental problems. The utilization of waste would be an innovation to handle the great deal of waste from processing.20,21 Because of the large availability and sugar composition it is widely used as a substrate in SSF. So far, most of the studies have been reported for the production of enzymes by using single substrate, while there is no report for the production of enzymes from the combination of bi-substrates. Such type of the combination may enhance the bacterial growth in SSF and thereby the product of interest.
2. Materials and methods
2.1. Isolation and screening of cellulase producing bacteria
Soil samples were collected from coffee pulp disposing site at Yercaud, Tamil Nadu, India. The isolation of bacteria from soil was carried out the procedure described by Kamala-Kannan and Krishnamoorthy.22 The serially diluted soil suspension (0.1 mL) was plated using the spread-plate technique onto Nutrient agar (Hi-media Pvt Ltd, India). Plates were incubated at 25 ± 2 °C for 48 h and observed for the bacterial growth. Morphologically distinct colonies were identified, purified, and stored at 4 °C for further study. The isolated bacterial colonies were screened for cellulase production on agar medium composed of 0.5% yeast extract, 0.5% casamino acid, 0.5% MgSO4·7H2O, 0.3% tri-sodium citrate, 0.2% KCl and 5% sodium chloride, supplemented with 0.5% (w/v) carboxy methyl-cellulose (CMC) (pH 9). All the isolates were inoculated into the agar plates, incubated at 30 °C and observed for the clear zone around the colonies after 24 h in the plates.23 Isolation and identification of the isolate were carried out at the PG and Research Department of Biotechnology, Mahendra Arts and Science College, Kalippatti, Tamil Nadu, India.2.2. DNA extraction and identification of the isolate TSK-MASC
Genomic DNA was extracted according to Maniatis et al., Sambrook and Russell.24,25 The partial 16S rRNA gene was amplified using universal primers 27f (5′AGAGTTTGATCCTGGCTCAG3′) and 907r (5′CCCCGTCAATTCATTTGAGTTT3′). The amplicons was purified (QIAGEN, CA, USA) and sequenced using ABI PRISM (Model 3700, CA, and USA). The sequences were compared using BLAST (NCBI) for the identification of isolate TSK-MASC. Phylogenetic analysis was performed using Neighbor-Joining method in CLC WORKBENCH 5.2 software (CLC bio, MA, USA).2.3. Substrate
CPW was procured from coffee processing industry at Yercaud, Salem, Tamil Nadu, India. PW was collected from juice industries at Dharmapuri, Tamil Nadu, India. The substrates were dried at 50 °C for 12 h to decrease the moisture content. The dried particles were sieved (mean size of 1.0 to 2.0 mm) and used for fermentation studies. The substrates were pretreated individually with 1% (w/v) NaOH. Briefly, ten grams of each substrate was dispensed in 500 ml Erlenmeyer flasks containing 100 ml of alkali and left at room temperature for 2 h. The alkali treated substrate was then washed thoroughly with distilled water to remove traces of base followed by drying.262.4. Inoculum
The substrates were vigoursly mixed, and the flasks were autoclaved at 121 °C for 15 min. After sterilization the flasks were cooled to 50 °C and inoculated with 5 mL of TSK-MASC isolate carrying 108 cells mL−1 (0.8 OD at 600 nm) as a seed culture under aseptic condition.2.5. Scanning electron microscopy (SEM)
Scanning Electron Microscopy was used to examine morphological modifications of CPW and PW before and after the alkaline pretreatment according to Díaz-Malváez et al.27 Samples were dehydrated and mounted on stubs and sputter-coated with gold for 300 s using high vacuum and a voltage acceleration of 10 kV. SEM was performed in a Jeol JSM 6390 model.2.6. Statistical optimization of cellulase production
Response surface methodology combined with BBD was established using Design Expert software (8.0 trial version). Statistical based BBD with four factorial and three levels was developed to optimize the cultural conditions.28–30 The factors, namely, pH, incubation time, CPW and PW were optimized for enhanced cellulase production using the isolate TSK-MASC under SSF. The temperature was kept constant at 37 °C throughout the experiments. A total of 29 experiments were performed to optimize the process parameters, and experiments were performed according to the experimental design matrix. The results were evaluated by applying the coefficient of determination (R2), analysis of variance (ANOVA) and response plots. Employing RSM, the most widely used second-order polynomial equation developed to fit the experimental results and identify the relevant model terms| Y = β0 + ∑βiXi + ∑βiiXi2 + ∑βijXiXj | (1) |
2.7. Enzyme recovery
The fermented solution from each flask was extracted and mixed with 0.05 M sodium acetate buffer (pH 4.8) and subjected to shaking at constant speed of 150 rpm for 30 min. Later, the mixture was filtered initially through Whatman No. 1 filter paper followed by 0.2 μm membrane filter and again centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min at 4 °C. The supernatant was used as crude enzyme source for further assay.30
000 rpm for 10 min at 4 °C. The supernatant was used as crude enzyme source for further assay.30
2.8. Enzyme assay
The cellulase activity was measured by mixing 100 μl of enzyme solution with 100 μl of 1% (w/v) CMC in 50 mM Tris–HCl buffer (pH 9) at 50 °C for 20 min. The reaction was stopped by adding the 3,5-dinitrosalicylic acid (DNS) reagent. The mixture was boiled for 10 min, cooled in ice and amount of reducing sugars liberated was measured at 550 nm.31 One unit of CMCase was defined as the amount of enzyme required to liberate 1 μmol of glucose per min.232.9. Effect of temperature on activity of purified Cellulase
The optimum temperature for enzyme activity was determined by incubating the reaction mixture (200 μl diluted enzyme solution + 300 μl of 1% CMC) for 1 h at various temperatures ranged between 30 and 80 °C and the residual activity was assayed by standard DNS method.2.10. Effect of pH on activity and stability of purified cellulase
The optimal pH of the purified cellulase activity was evaluated by incubating the reaction mixture at 50 °C for 1 h with different pH buffers. Buffers used were; 50 mM sodium citrate (pH 3–6), 50 mM sodium phosphate (pH 6–8), 50 mM glycine–NaOH (9–11), and 50 mM dilute NaOH for pH 12–13 and the residual activity were assayed by standard DNS method.3. Results and discussion
3.1. Isolation, identification, screening and characterization of cellulase producing bacteria
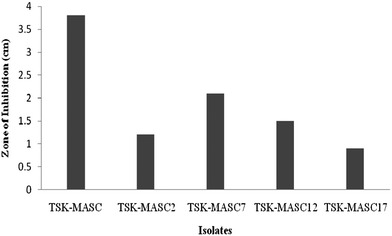
The present study represents an attempt to evaluate the potential of two substrates for the production of industrially important cellulases by SSF. Five morphologically different bacterial colonies were isolated from the coffee pulp disposing site and were screened on CMC agar plates for cellulolytic activity. The results showed that the isolate, designated TSK-MASC, exhibited maximum cellulolytic activity. The isolate showed the clearance zone of 3.8 cm in diameter (Fig. 1). Accordingly, the isolate TSK-MASC was selected for SSF studies. Polymerase chain reaction amplification of the partial 16S rDNA resulted in the predicted 900-bp amplicon in the isolate TSK-MASC. The amplified product was sequenced and compared with the available sequences in NCBI database. The isolate TSK-MASC exhibited 99% identity with Acinetobacter sp. The partial 16s rRNA of the isolate TSK-MASC was submitted in GenBank (Accession No. KC309425). A phylogenetic tree was derived from the partial 16S rDNA sequences of the isolate with existing sequences in the database, and the results are shown in Fig. 2. Expectedly, the isolate TSK-MASC and Acinetobactor sp. GA56 were in the same clusters and which further confirms the identity of the 16S rDNA sequence with Acinetobacter sp.CPW and PW were chosen as substrates for effective cellulases production. The pineapple peel contains high amounts of cellulose, hemicelluloses, and other carbohydrates.21,33 It is also used as a culture broth for the cultivation of microorganisms and cellulase production.34 Coffee pulp is composed of carbohydrates, proteins and fibers.19 India is one of the major coffee producing countries in the world and coffee is one of the major crops in India. Large number of coffee processing industries released wastes such as, coffee pulp and husk. Consumption of these wastes for the production of industrially important biomolecules is very interesting in the field of bioprocess technology and waste management.
3.2. SEM analysis of CPW and PW
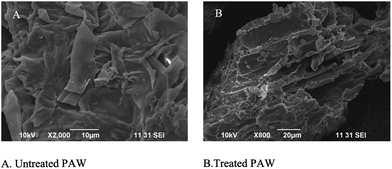
The SEM analysis clearly showed that the pretreatment modified the outer layer of the substrates CPW and PW. The pretreated samples showed that the degradation of external micro fibers (Fig. 3 and 4). Diaz-Malvaez et al.27 reported that the NaOH treated Corn pericarp fibers represented more available desired product for hemicellulolytic activity production during fermentation. Wang et al.35 stated that the pretreatment can alter the structure as well as increased amounts of product of interest by microbial fermentation.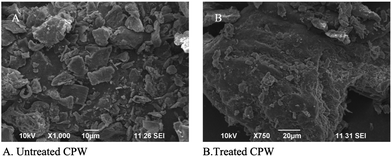 | ||
| Fig. 3 Scanning Electron Microscopy (SEM) of the Coffee Pulp Waste (CPW) surface. (A) Untreated; (B) CPW pretreated with NaOH. | ||
3.3. Optimization of cellulase production by using BBD
The BBD was applied to identify the optimal conditions for the enhanced production of cellulase enzyme. The experimental design is presented in Table 1. ANOVA of the quadratic regression model (Table 2) exhibits that it was a highly significant model, as was evident from the Fisher's F-test with a very low probability value (F value = 16.25). Values of ‘Prob > F’ (0.0500) indicate that the term of the model was significant. The Model F-value of 16.25 implies that the model was significant. There was only a 0.01% chance that a model F-value could occur due to noise. The predicted R2 (0.6663) and adjusted R2 (0.8841) values for cellulase production were in reasonable agreement with the value of R2 (0.9420), which is closer to 1.0, indicating the better fitness of the model in the experimental data. The model for cellulase production by SSF, three different tests, namely, sequential model sum of squares, lack of fit tests and model summary statistics were carried out in the present study.| Experiment No | pH | Incubation time (hrs) | Coffee pulp (%) | Pine apple waste (%) | Cellulase activity (U ml−1) |
|---|---|---|---|---|---|
| 1 | 7.0 | 24.0 | 5.0 | 3.0 | 612 |
| 2 | 7.0 | 60.0 | 3.0 | 3.0 | 885 |
| 3 | 9.0 | 60.0 | 1.0 | 3.0 | 436 |
| 4 | 5.0 | 60.0 | 1.0 | 3.0 | 512 |
| 5 | 7.0 | 96.0 | 3.0 | 1.0 | 612 |
| 6 | 9.0 | 24.0 | 3.0 | 3.0 | 467 |
| 7 | 7.0 | 60.0 | 3.0 | 3.0 | 880 |
| 8 | 5.0 | 60.0 | 5.0 | 3.0 | 561 |
| 9 | 7.0 | 24.0 | 1.0 | 3.0 | 564 |
| 10 | 7.0 | 60.0 | 5.0 | 5.0 | 671 |
| 11 | 9.0 | 60.0 | 5.0 | 3.0 | 567 |
| 12 | 7.0 | 60.0 | 5.0 | 1.0 | 612 |
| 13 | 7.0 | 96.0 | 5.0 | 3.0 | 478 |
| 14 | 5.0 | 96.0 | 3.0 | 3.0 | 450 |
| 15 | 7.0 | 96.0 | 3.0 | 5.0 | 482 |
| 16 | 5.0 | 24.0 | 3.0 | 3.0 | 389 |
| 17 | 7.0 | 60.0 | 1.0 | 1.0 | 456 |
| 18 | 7.0 | 60.0 | 3.0 | 3.0 | 886 |
| 19 | 9.0 | 60.0 | 3.0 | 1.0 | 442 |
| 20 | 5.0 | 60.0 | 3.0 | 5.0 | 565 |
| 21 | 7.0 | 60.0 | 1.0 | 5.0 | 615 |
| 22 | 7.0 | 24.0 | 3.0 | 5.0 | 660 |
| 23 | 7.0 | 60.0 | 3.0 | 3.0 | 883 |
| 24 | 7.0 | 96.0 | 1.0 | 3.0 | 616 |
| 25 | 5.0 | 60.0 | 3.0 | 1.0 | 515 |
| 26 | 9.0 | 60.0 | 3.0 | 5.0 | 565 |
| 27 | 9.0 | 96.0 | 3.0 | 3.0 | 459 |
| 28 | 7.0 | 24.0 | 3.0 | 1.0 | 565 |
| 29 | 7.0 | 60.0 | 3.0 | 3.0 | 888 |
| Source | Sum of Squares | Df | Mean square | F value | p-value |
|---|---|---|---|---|---|
| Model | 6.049 × 105 | 14 | 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 209.25 209.25 |
16.25 | <0.0001a |
| A | 261.33 | 1 | 261.33 | 0.098 | 0.7585 |
| B | 2133.33 | 1 | 2133.33 | 0.80 | 0.3856 |
| C | 7600.33 | 1 | 7600.33 | 2.86 | 0.1130 |
| D | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561.33 561.33 |
1 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 561.33 561.33 |
3.97 | 0.0661 |
| AB | 1190.25 | 1 | 1190.25 | 0.45 | 0.5144 |
| AC | 1681.00 | 1 | 1681.00 | 0.63 | 0.4398 |
| AD | 1332.25 | 1 | 1332.25 | 0.50 | 0.4907 |
| BC | 8649.00 | 1 | 8649.00 | 3.25 | 0.0929 |
| BD | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656.25 656.25 |
1 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 656.25 656.25 |
4.76 | 0.0467 |
| CD | 2500.00 | 1 | 2500.00 | 0.94 | 0.3487 |
| A2 | 3.659 × 105 | 1 | 3.659 × 105 | 137.58 | <0.0001 |
| B2 | 2.202 × 105 | 1 | 2.202 × 105 | 82.80 | <0.0001 |
| C2 | 1.289 × 105 | 1 | 1.289 × 105 | 48.49 | <0.0001 |
| D2 | 1.156 × 105 | 1 | 1.156 × 105 | 43.47 | <0.0001 |
| Residual | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 228.70 228.70 |
14 | 2659.19 | — | — |
| Lack of fit | 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 191.50 191.50 |
10 | 3719.15 | 399.91 | <0.0001a |
| Pure error | 37.20 | 4 | 9.30 | — | — |
| Core total | 6.422 × 105 | 28 | — | — | — |
The 3-D plots were graphical representations was generated (Fig. 5). The results demonstrate that there was significant relation of pH, incubation time, with CPW and PW concentrations for cellulase production. The optimum levels of the variables were obtained by using BBD. The model predicted a maximum cellulase activity of 888 U mL−1 appearing after 60 h cultivation with 3.0 g L−1 of CPW and PW at pH 7.0. Predicted model was validated and experiments were conducted using these optimal conditions. The predicted model values were in good agreement with the values measured in these experiments, thus mitigating the validity of the response model and the necessity for optimal conditions. The graphs highlighted the roles played by the variables for the production of cellulase.
 | ||
| Fig. 5 3-D plots of the combined effects of two variables on cellulase production by Acinetobacter sp. TSK-MASC. | ||
The coefficients of the regression equation were calculated and the following regression equation was obtained.
| Y = 884.40 − 4.67A − 13.33B + 25.17C + 29.67D − 17.25AB + 20.50AC + 18.25AD − 46.50BC − 56.25BD − 25.00CD − 237.49A2 − 184.24B2 − 140.99C2 − 133.49D2 | (2) |
3.4. Effect of temperature and pH on enzyme activity
The effect of temperature on activity of cellulase was determined at various temperatures ranged between 30 °C and 80 °C. The optimum temperature for cellulase activity was found to be 50 °C at pH 7.0 and decreased rapidly as the temperature increased above 60 °C. The optimal temperature of cellulase produced by the bacteria, Acinetobacter sp. TSK-MASC was similar to those produced by B. amyloliquefaciens DL- 3,37 and B. subtilis subsp. subtilis A-53 (ref. 38) (Fig. 6 and 7).4. Conclusion
In this present study we investigated that the mixed substrates produced higher amount of cellulase enzyme. The isolate Acinetobacter sp. TSK-MASC effectively utilizes the mixed substrates combinations as carbon source. Higher cellulase production was obtained when coffee pulp waste supplemented with pineapple waste in the ratio 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 was used as substrates. The mixed substrates with the isolate TSK-MASC can be potentially exploited for the cellulase production. The enzyme production was further enhanced the application of BBD statistical optimization method. The higher production (888 U mL−1) was achieved pH 7.0 at 60 h cultivation with 3.0 g L−1 of CPW and 3.0 g L−1 of PW.
3 was used as substrates. The mixed substrates with the isolate TSK-MASC can be potentially exploited for the cellulase production. The enzyme production was further enhanced the application of BBD statistical optimization method. The higher production (888 U mL−1) was achieved pH 7.0 at 60 h cultivation with 3.0 g L−1 of CPW and 3.0 g L−1 of PW.
References
- B. Kamm and M. Kamm, Chem. Biochem. Eng. Q., 2004, 18, 1–6 CrossRef CAS.
- Y. H. P. Lynd, Biomacromolecules, 2005, 6, 1510–1515 CrossRef PubMed.
- R. R. Singhania, R. K. Sukumaran, A. K. Patel, C. Larroche and A. Pandey, Enzyme Microb. Technol., 2010, 46, 541–549 CrossRef CAS PubMed.
- B. Kumar, P. Trivedi, A. K. Mishra, A. Pandey and L. M. S. Palni, Microbiol. Res., 2004, 159, 141–146 CrossRef CAS PubMed.
- S. Thirumale, D. Swaroopa Rani and K. Nand, Process Biochem., 2001, 37, 241–245 CrossRef CAS.
- P. Shrestha, S. K. Khanal, A. L. Pomettoiii and J. Van Leeuwen, J. Agric. Food Chem., 2009, 57, 4156–4161 CrossRef CAS PubMed.
- C. M. Lo, Q. Zhang, N. V. Callow and L. K. Ju, Bioresour. Technol., 2010, 101, 717–723 CrossRef CAS PubMed.
- M. Dashtban, H. Schraft and W. Qin, Int. J. Biol. Sci., 2009, 5, 578–595 CrossRef CAS.
- J. S. Bak, J. K. Ko, I. G. Choi, Y. C. Park, J. H. Seo and K. H. Kim, Biotechnol. Bioeng., 2009, 104, 471–482 CrossRef CAS PubMed.
- M. Govarthanan, S. H. Park, J. W. Kim, K. J. Lee, M. Cho, S. Kamala-Kannan and B. T. Oh, Prep. Biochem. Biotechnol., 2014, 44, 119–131 CrossRef CAS PubMed.
- R. P. Tengerdy, J. Sci. Ind. Res., 1996, 55, 313–316 CAS.
- A. Pandey, Biochem. Eng. J., 2003, 13, 81–84 CrossRef CAS.
- S. Liu, Y. Fang, L. V. Mingsheng, S. Wang and L. Chen, Bioresour. Technol., 2010, 101, 7924–7929 CrossRef CAS PubMed.
- O. Prakash, M. Talat, S. H. Hasan and R. K. Pandey, Bioresour. Technol., 2008, 99, 7565–7572 CrossRef CAS PubMed.
- H. L. Liu, Y. W. Lan and Y. C. Cheng, Process Biochem., 2004, 39, 1953–1961 CrossRef CAS PubMed.
- F. Amiri, S. M. Mousavi and S. Yaghmaei, Sep. Purif. Technol., 2011, 80, 566–576 CrossRef CAS PubMed.
- S. Mohana, A. Shah, J. Divecha and D. Madamwar, Bioresour. Technol., 2005, 99, 7553–7564 CrossRef PubMed.
- Z. Zhu, G. Zhang, Y. Luo, W. Ran and Q. Shen, Bioresour. Technol., 2012, 112, 254–260 CrossRef CAS PubMed.
- A. Pandey, C. R. Soccol, P. Nigam, D. Brand, R. Mohan and S. Roussos, Biochem. Eng. J., 2000, 6, 153–162 CrossRef CAS.
- A. P. Bartolome, P. Rupbrez and F. Carmen, Food Chem., 1995, 53, 75–79 CrossRef CAS.
- S. Ketnawa, P. Chaiwut and S. Rawdkuen, Food Bioprod. Process., 2012, 90, 385–391 CrossRef CAS PubMed.
- S. Kamala-Kannan and R. Krishnamoorthy, Sci. Total Environ., 2006, 367, 341–353 CrossRef PubMed.
- N. Annamalai, M. V. Rajeswari, S. Elayarajab and T. Balasubramanian, Carbohydr. Polym., 2013, 94, 409–415 CrossRef CAS PubMed.
- T. Maniatis, E. F. Fritsch and J. Sambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, 2nd edn, 1989 Search PubMed.
- J. Sambrook and D. Russell, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbour, NY, 3rd edn, 2001 Search PubMed.
- N. Bansal, R. Tewari, R. Soni and S. K. Soni, Waste Manag., 2012, 32, 1341–1346 CrossRef CAS PubMed.
- F. I. Díaz-Malváez, B. E. García-Almendárez, A. Hernández-Arana, A. Amaro-Reyes and C. Regalado-González, Process Biochem., 2013, 48, 1018–1024 CrossRef PubMed.
- G. E. P. Box and D. W. Behnken, Technometrics., 1960, 2, 455–475 CrossRef.
- R. Soni, A. Nazir and B. S. Chadha, Ind. Crops Prod., 2010, 31, 277–283 CrossRef CAS PubMed.
- N. M. Narra, G. Dixit, J. Divecha, D. Madamwar and R. Amita Shah, Bioresour. Technol., 2012, 121, 355–361 CrossRef PubMed.
- G. L. Miller, Anal. Chem., 1959, 31, 426–428 CrossRef CAS.
- M. M. A. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS.
- D. P. Bartholomew, R. E. Paull and K. G. Rohrbach, Pineapple Botany, Production and Uses, CABI publishing, London, 2003, vol. 1, pp. 1–29, 281–288 Search PubMed.
- P. Omojasola, J. Folakemi, P. Omowumi and S. A. Ibiyemi, Nat. Sci., 2008, 6, 64–81 Search PubMed.
- B. Wang, T. Ezeji, Z. Shi, H. Feng and H. P. Blaschek, Trans. ASABE, 2009, 52, 885–892 CrossRef CAS.
- R. R. Singhania, A. K. Patel, C. R. Soccol and A. Pandey, Biochem. Eng. J., 2009, 44, 13–18 CrossRef CAS PubMed.
- Y. J. Lee, B. K. Kim, B. H. Lee, K. I. Jo, N. K. Lee and C. H. Chung, Bioresour. Technol., 2008, 99, 378–386 CrossRef CAS PubMed.
- K. Kim, B. H. Lee, Y. J. Lee, I. H. Jin, C. H. Chung and J. W. Lee, Enzyme Microb. Technol., 2009, 44, 411–416 CrossRef PubMed.
Footnote |
| † The first two authors equally contributed this work. |
| This journal is © The Royal Society of Chemistry 2014 |