Pyrene-labeled pyrrolidinyl peptide nucleic acid as a hybridization-responsive DNA probe: comparison between internal and terminal labeling†
Chalothorn Boonluaa,
Boonsong Ditmangkloa,
Nisanath Reenabthueb,
Chaturong Suparppromb,
Nattawee Poomsukc,
Khatcharin Siriwongc and
Tirayut Vilaivan*a
aOrganic Synthesis Research Unit, Department of Chemistry, Faculty of Science, Chulalongkorn University, Phayathai Road, Patumwan, Bangkok 10330, Thailand. E-mail: vtirayut@chula.ac.th
bDepartment of Chemistry, Faculty of Science, Naresuan University, Ta-Po District, Muang, Phitsanulok 65000, Thailand
cMaterials Chemistry Research Unit, Department of Chemistry, Faculty of Science, Khon Kaen University, Khon Kaen 40002, Thailand
First published on 16th January 2014
Abstract
Nucleic acids or nucleic acid analogues carrying an environment sensitive fluorophore that can change optical properties in response to hybridization with complementary DNA or RNA targets are potentially useful as self-reporting hybridization probes that do not require a quencher. In this study, pyrrolidinyl peptide nucleic acids (acpcPNA) carrying a pyrene label at terminal, or internal positions were synthesized and their fluorescence properties investigated. Significant fluorescence increases, between 2.9 and 73 fold, were observed only with internally pyrene-labeled acpcPNA upon hybridization with complementary DNA under a variety of sequence contexts. Hybridization with mismatched DNA targets yielded mostly unchanged or decreased fluorescence, depending on the type and position of the mismatch base. Non-specific fluorescence increase due to the presence of a mismatch located away from the pyrene label could be eliminated by digestion with S1 nuclease, which allows unambiguous discrimination between complementary and all single mismatched DNA targets. The terminally pyrene-labeled acpcPNA showed a much smaller fluorescence change upon hybridization with both complementary and mismatched DNA targets, and the change was in the opposite direction to the internally pyrene-labeled acpcPNA. These results were rationalized by the use of molecular dynamics simulations, which suggested that the pyrene label adopts a different conformation when located at different positions of the acpcPNA probes.
Introduction
Fluorophore-labeled oligonucleotides and analogues are useful as probes for the determination of DNA or RNA sequences. A representative example of such probes is a molecular beacon, specifically a doubly end-labeled oligodeoxynucleotide with a partial self-complementary sequence that can change its conformation significantly upon hybridization with the correct DNA target.1 The conformation change alters the distance between the two labels, which in turn gives rise to a change in optical properties.2 Limitations in the design of these probes are the requirements of a stem-loop or other types of secondary structures, and the presence of two labels on the same probe. As a consequence, several designs of molecular beacons have been reported in the literature to overcome these limitations.3 One of the more recent developments is the discovery that singly labeled oligonucleotides carrying an appropriate fluorophore can yield different fluorescence responses in relation to a hybridization event.4 Combining this concept with a neutral analogue of an oligonucleotide called peptide nucleic acid (PNA),5 which can adopt a compact structure in the single stranded state and extended structure upon hybridization with DNA,6 should result in singly labeled PNA beacons that are simple to design and synthesize, and which exhibit excellent binding specificities. Examples of such singly-labeled, stemless PNA probes include a fluorene-labeled PNA,7 light up probes8 and FIT probes.9In recent years, we have designed several new conformationally restricted pyrrolidinyl PNAs.10 The most successful system was the one carrying an α/β-peptide backbone derived from D-prolyl-(1S,2S)-2-aminocyclopentanecarboxylic acid (acpcPNA), which showed excellent binding affinity and specificity towards DNA.11 Several applications of acpcPNA as probes for DNA sequence determination have been reported.12 Techniques for site-specific labeling of acpcPNA at the backbone13 or the nucleobase14 have also been established. Preliminary studies in a limited sequence context revealed that acpcPNAs having a single pyrene label attached onto the backbone generated significant fluorescence enhancements upon binding to DNA in a sequence specific fashion.13a,b However, it was not clear from our previous studies about the generality of the design, and the structural basis of this fluorescence change. In this report, we: (1) demonstrate the generality of the design by extending the study to several other sequences having different neighboring nucleobases; (2) compare the performance of internally, and terminally pyrene-labeled acpcPNA probes and (3) rationalize the structural basis of the fluorescence change using molecular dynamics (MD) simulations.
Results and discussion
Synthesis of pyrene-labeled acpcPNA
For terminally-labeled acpcPNA, the pyrenebutyryl (PyBtr) label was attached via an amide bond to the amino group of the N-terminal 2-aminocyclopentanecarboxylic acid (ACPC) spacer. For internally-labeled acpcPNA, the ACPC spacer at the position to be modified was replaced with a trifluoroacetyl (Tfa)-protected (3R,4S)-3-aminopyrrolidine-4-carboxylic acid (APC) spacer.13a After removal of the Tfa and nucleobase protecting groups by treatment with aqueous ammonia–dioxane at 60 °C, the pyrene label was attached to the acpcPNA backbone either by acylation (PyBtr),13a or reductive alkylation (pyrenebutyl label – PyBtl).13b These approaches allow site-specific placement of the pyrene label anywhere in the acpcPNA backbone without the need to pre-synthesize pyrene-labeled monomers. The structure of acpcPNA, position of label attachment and type of pyrene label are shown in Scheme 1. All pyrene-labeled acpcPNAs were purified by reverse phase HPLC to >90% purity and gave the expected mass (MALDI-TOF). Sequence and characterization data of all pyrene-modified acpcPNAs can be found in Table 1 and Fig. S1–S20.†| PNA | Sequence [N → C] | tRa [min] | Yield [%] | m/z (calcd) | m/zb (found) |
|---|---|---|---|---|---|
| a HPLC conditions: C18 column 4.6 × 50 mm, particle size 3 μ; mobile phase A = 0.1% trifluoroacetic acid (TFA) in water; B = 0.1% TFA in MeOH; gradient = 10–90% B over 60 min with 5 min equilibration time at a flow rate of 0.5 mL min−1.b Linear positive ion mode, α-cyano-4-hydroxycinnamic acid (CCA) matrix. | |||||
| T9 | PyBtrTTTTTTTTT | 33.8 | 9.7 | 3407.7 | 3409.1 |
| M10 | PyBtrGTAGATCACT | 22.9 | 8.6 | 3787.1 | 3788.2 |
| M11 | PyBtrCTAAATTCAGA | 23.1 | 8.2 | 4112.5 | 4110.8 |
| M12 | PyBtrAGTTATCCCTGC | 23.1 | 7.4 | 4412.8 | 4410.8 |
| TT | 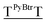 |
24.7 | 31.7 | 1013.2 | 1012.5 |
| AA | 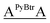 |
24.9 | 32.6 | 1031.2 | 1030.6 |
| CC | 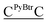 |
24.8 | 37.1 | 983.2 | 982.4 |
| GG |  |
24.1 | 28.2 | 1063.2 | 1062.5 |
| T9_TT | 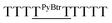 |
23.8 | 6.5 | 3450.8 | 3452.0 |
| M10_AT | 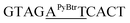 |
23.0 | 4.6 | 3892.2 | 3893.8 |
| M11_TT | 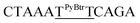 |
28.8 | 3.7 | 4283.7 | 4283.5 |
| M11_TC | 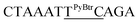 |
23.9 | 3.3 | 4283.7 | 4283.9 |
| M11_AAAA | 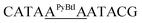 |
21.7 | 5.2 | 4150.4 | 4149.8 |
| M11_ATTA | 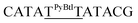 |
21.4 | 8.7 | 4132.4 | 4133.5 |
| M11_ACCA |  |
21.1 | 9.5 | 4102.4 | 4103.1 |
| M11_AGGA |  |
21.2 | 2.6 | 4182.4 | 4182.3 |
| M11_TAAT |  |
21.7 | 5.9 | 4132.4 | 4134.8 |
| M11_TTTT |  |
27.4 | 7.3 | 4114.5 | 4116.9 |
| M12_AT(Btr) | 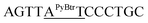 |
22.1 | 4.1 | 4584.0 | 4583.5 |
| M12_AT(Btl) |  |
23.9 | 7.3 | 4569.0 | 4570.7 |
DNA binding and optical properties of internally pyrene-labeled acpcPNA
Melting temperatures, and fluorescence data of internally pyrene-labeled acpcPNAs as well as their hybrids with complementary and non-complementary DNA are shown in Tables 2 and 3. The corresponding data for the terminally pyrene-labeled acpcPNAs are shown in Table 4. In all cases, the pyrene-modified acpcPNA retained strong binding affinity and sequence specificity to complementary DNA as shown by the high Tm values for the hybrids with complementary DNA and significantly lowered Tm values for the hybrids with non-complementary DNA. The Tm data also revealed that the internal labeling resulted in a larger destabilization of both complementary and non-complementary DNA hybrids compared to the terminal labeling, as shown by a relatively large decrease in Tm values from unmodified acpcPNA (T9 = 81.8, M10 = 62.4 and M12 = 75.0 °C under identical conditions). This could be explained by the perturbation of the acpcPNA·DNA binding by the relatively bulky pyrene label, which should be more significant in the case of the internally-modified acpcPNA than the terminally-modified acpcPNA. Nevertheless, CD spectra confirmed that the internal pyrene label does not change the overall structure of the duplexes formed between acpcPNA and DNA (Fig. S21†). In contrast, for an identical sequence and labeling position, the PyBtl label having a CH2–N linker is less destabilizing compared to the PyBtr label attached via a CH2–CO–N linker (compare entries 24 and 28 in Table 2).| Entry | PNA | Label | DNA (3′ → 5′) | Tm (°C) | ΦF(ss) | ΦF(ds)/ΦF(ss) | F(ds)/F(ss) |
|---|---|---|---|---|---|---|---|
| 1 | T9_TT | PyBtr | AAA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) AAAA (comp) AAAA (comp) |
60.3 | 0.0030 | 106 | 72.8 |
| 2 | PyBtr | AAA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) AAAA (mm) AAAA (mm) |
25.4 | 13.2 | 10.8 | ||
| 3 | M10_AT | PyBtr | CATC![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTGA (comp) GTGA (comp) |
45.6 | 0.0440 | 4.41 | 5.20 |
| 4 | PyBtr | CATC![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTGA (mm) GTGA (mm) |
25.4 | 1.00 | 0.87 | ||
| 5 | M11_TC | PyBtr | GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCT (comp) TCT (comp) |
60.2 | 0.1401 | 2.73 | 2.91 |
| 6 | PyBtr | GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) TCT (mm) TCT (mm) |
54.8 | 0.27 | 0.23 | ||
| 7 | PyBtr | GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) TCT (mm) TCT (mm) |
56.7 | 0.31 | 0.22 | ||
| 8 | PyBtr | GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) TCT (mm) TCT (mm) |
43.7 | 0.53 | 0.33 | ||
| 9 | PyBtr | GATTGA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCT (mm) TCT (mm) |
29.8 | 1.42 | 1.04 | ||
| 10 | PyBtr | GATTCA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCT (mm) TCT (mm) |
33.0 | 1.95 | 1.50 | ||
| 11 | PyBtr | GATTAA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCT (mm) TCT (mm) |
34.2 | 1.99 | 1.55 | ||
| 12 | M11_TT | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (comp) GTCT (comp) |
65.0 | 0.0512 | 5.79 | 6.82 |
| 13 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) GTCT (mm) GTCT (mm) |
42.6 | 0.80 | 0.64 | ||
| 14 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) GTCT (mm) GTCT (mm) |
39.6 | 3.05 | 2.36 | ||
| 15 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) GTCT (mm) GTCT (mm) |
37.6 | 1.09 | 0.67 | ||
| 16 | PyBtr | GATTT![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (mm) GTCT (mm) |
43.6 | 1.42 | 1.01 | ||
| 17 | PyBtr | GATTT![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (mm) GTCT (mm) |
45.9 | 7.02 | 4.10 | ||
| 18 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ATCT (mm) ATCT (mm) |
38.9 | 3.33 | 2.72 | ||
| 19 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CTCT (mm) CTCT (mm) |
38.2 | 2.21 | 1.69 | ||
| 20 | PyBtr | GATTT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TTCT (mm) TTCT (mm) |
37.4 | 3.25 | 2.92 | ||
| 21 | PyBtr | GATTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (mm) GTCT (mm) |
39.2 | 5.42 | 5.98 | ||
| 22 | PyBtr | GATTC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (mm) GTCT (mm) |
40.4 | 5.26 | 4.36 | ||
| 23 | PyBtr | GATTG![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GTCT (mm) GTCT (mm) |
37.1 | 5.41 | 5.02 | ||
| 24 | M12_AT(Btr) | PyBtr | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GGGACG (comp) GGGACG (comp) |
62.7 | 0.0203 | 6.77 | 6.14 |
| 25 | PyBtr | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) GGGACG (mm) GGGACG (mm) |
45.6 | 1.52 | 0.92 | ||
| 26 | PyBtr | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) GGGACG (mm) GGGACG (mm) |
35.3 | 0.65 | 0.70 | ||
| 27 | PyBtr | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) GGGACG (mm) GGGACG (mm) |
42.1 | 9.47 | 5.99 | ||
| 28 | M12_AT(Btl) | PyBtl | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) GGGACG (comp) GGGACG (comp) |
65.8 | 0.0112 | 17.3 | 14.0 |
| 29 | PyBtl | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) GGGACG (mm) GGGACG (mm) |
48.0 | 1.92 | 1.32 | ||
| 30 | PyBtl | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) GGGACG (mm) GGGACG (mm) |
37.1 | 3.94 | 1.67 | ||
| 31 | PyBtl | TCAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) GGGACG (mm) GGGACG (mm) |
46.9 | 15.0 | 7.07 |
| Entry | PNA | DNA (3′ → 5′) | Tm (°C) | ΦF (ss) | ΦF(ds)/ΦF(ss) | F(ds)/F(ss) |
|---|---|---|---|---|---|---|
| 1 | M11_AAAA | CGTAT![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) TATG (comp) TATG (comp) |
>80 | 0.0760 | 3.89 | 3.17 |
| 2 | CGTAT![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) TATG (mm) TATG (mm) |
35.2 | 1.97 | 1.48 | ||
| 3 | CGTAT![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) TATG (mm) TATG (mm) |
<25 | 0.76 | 0.56 | ||
| 4 | CGTAT![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) TATG (mm) TATG (mm) |
29.4 | 0.85 | 0.58 | ||
| 5 | M11_ATTA | CGTAT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TATG (comp) TATG (comp) |
75.7 | 0.0648 | 3.57 | 3.89 |
| 6 | CGTAT![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) TATG (mm) TATG (mm) |
35.2 | 0.55 | 0.37 | ||
| 7 | CGTAT![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) TATG (mm) TATG (mm) |
28.2 | 0.77 | 0.56 | ||
| 8 | CGTAT![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) TATG (mm) TATG (mm) |
27.4 | 0.92 | 0.73 | ||
| 9 | M11_ACCA | CGTAT![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TATG (comp) TATG (comp) |
63.4 | 0.0709 | 4.23 | 3.74 |
| 10 | CGTAT![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/i_char_0043_0332.gif) TATG (mm) TATG (mm) |
31.7 | 0.34 | 0.18 | ||
| 11 | CGTAT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TATG (mm) TATG (mm) |
34.7 | 1.02 | 0.83 | ||
| 12 | CGTAT![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) TATG (mm) TATG (mm) |
27.3 | 0.64 | 0.68 | ||
| 13 | M11_AGGA | CGTAT![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) ![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) TATG (comp) TATG (comp) |
68.9 | 0.0693 | 3.06 | 3.28 |
| 14 | CGTAT![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/i_char_0047_0332.gif) TATG (mm) TATG (mm) |
33.2 | 0.80 | 0.63 | ||
| 15 | CGTAT![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) TATG (mm) TATG (mm) |
25.5 | 0.82 | 0.67 | ||
| 16 | CGTAT![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) TATG (mm) TATG (mm) |
28.4 | 0.83 | 0.67 | ||
| 17 | M11_TAAT | CGTAA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) AATG (comp) AATG (comp) |
70.7 | 0.0742 | 3.24 | 7.08 |
| 18 | CGTAA![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) AATG (mm) AATG (mm) |
<25 | 1.16 | 1.58 | ||
| 19 | M11_TTTT | CGTAA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) AATG (comp) AATG (comp) |
75.3 | 0.0194 | 16.4 | 19.4 |
| 20 | CGTAA![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) ![[T with combining low line]](https://www.rsc.org/images/entities/i_char_0054_0332.gif) AATG (mm) AATG (mm) |
27.1 | 2.26 | 2.48 |
| Entry | PNA | DNA (3′ → 5′) | Tm (°C) | ΦF (ss) | ΦF(ds)/ΦF(ss) | F(ds)/F(ss) |
|---|---|---|---|---|---|---|
| 1 | T9 | AAAAAAAAA (comp) | >80 | 0.0030 | 30.8 | 46.1 |
| 2 | T9 | AAAATAAAA (mm) | 52.6 | 15.6 | 14.2 | |
| 3 | T9 | TAAAAAAAA (mm) | >80 | 1.00 | 3.56 | |
| 4 | M10 | CATCTAGTGA (comp) | 64.3 | 0.0470 | 0.21 | 0.23 |
| 5 | M10 | CATCCAGTGA (mm) | 42.2 | 0.26 | 0.34 | |
| 6 | M11 | GATTTAAGTCT (comp) | >80 | 0.0262 | 0.36 | 0.46 |
| 7 | M11 | GATTTCAGTCT (mm) | 78.5 | 0.12 | 0.22 | |
| 8 | M11 | AATTTAAGTCT (mm) | 77.3 | 2.09 | 2.49 | |
| 9 | M11 | TGGGATTTAAGTCT (comp with hanging 3′ end) | >80 | 0.59 | 0.62 | |
| 10 | M12 | TCAATAGGGACG (comp) | 73.5 | 0.0956 | 0.21 | 0.33 |
| 11 | M12 | TCAATCGGGACG (mm) | 50.2 | 0.22 | 0.24 | |
| 12 | M12 | CCAATAGGGACG (mm) | 70.6 | 1.58 | 2.48 | |
| 13 | M12 | GCAATAGGGACG (mm) | 72.1 | 1.62 | 2.29 | |
| 14 | M12 | ACAATAGGGACG (mm) | 71.6 | 2.03 | 1.98 | |
| 15 | M12 | TTCAATAGGGACG(comp with hanging T at 3′ end) | n.d. | 0.10 | 0.13 | |
| 16 | M12 | CTCAATAGGGACG (comp with hanging C at 3′ end) | n.d. | 0.35 | 0.14 | |
| 17 | M12 | GTCAATAGGGACG (comp with hanging G at 3′ end) | n.d. | 0.17 | 0.37 | |
| 18 | M12 | ATCAATAGGGACG (comp with hanging A at 3′ end) | 79.4 | 0.50 | 0.46 | |
| 19 | M12 | _CAATAGGGACG (comp with 3′ terminal deletion) | 67.3 | 1.76 | 2.10 | |
| 20 | M12 | _AATAGGGACG (comp with 3′ terminal deletion) | 56.6 | 1.90 | 2.12 |
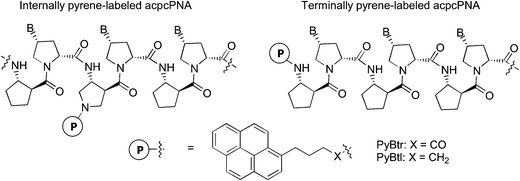
All single stranded internally pyrene-labeled acpcPNAs are weakly fluorescent as shown by the rather small quantum yields (ranging from 0.003 to 0.140) (Table 2). The weak fluorescence of pyrene-labeled acpcPNAs is likely a consequence of quenching by neighboring nucleobases.15,16 Close contact between the pyrene and the nucleobases is facilitated by the tendency of hydrophobic PNA to fold into a compact conformation in aqueous environments.6 To obtain further insights on the effect of neighboring bases on fluorescence, some simple internally pyrene-labeled acpcPNA dimers were synthesized. These model acpcPNAs contain a PyBtr label flanked by two DNA bases (A/A, T/T, C/C, G/G and abasic acpcPNA). Quantum yield values of the acpcPNA dimers suggest that the efficiency of pyrene quenching by nucleobases decreases in the order T > C ≫ G > A (Fig. 1). This quenching order corresponds well with the previously reported redox potentials of nucleobases with pyrene.16 Remarkably, base A exhibited almost no quenching effect when compared to the abasic acpcPNA.
Upon hybridization with complementary DNA targets, the fluorescence of all internally pyrene-labeled acpcPNAs significantly increased (Table 2). The values of F(ds)/F(ss) ranged from 2.9 (M11_TC), to 73 fold (T9_TT). Slightly different figures were obtained for ΦF(ds)/ΦF(ss) (2.7 for M11_TC and >100 fold for T9_TT), due to a small change in the absorption spectra in the pyrene region upon hybridization. It was reasoned that in the double-stranded state, the pyrene label may no longer favorably interact with the nucleobases, resulting in less quenching. It should be noted that the fluorescence quantum yields of various acpcPNA–complementary DNA hybrids [ΦF(ds)] were similar and almost independent of neighboring bases (ranging from 0.187–0.338), indicating less significant interactions between the pyrene label and the nucleobases compared to the single stranded acpcPNA, which is in good agreement with the proposed model.
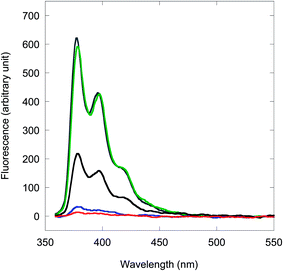
When a single mismatch base is incorporated into the DNA target, the fluorescence of the resulting mismatched acpcPNA·DNA hybrid is dependent on the position and type of the mismatch base. From our results, if the mismatch base is distant from the pyrene label, the fluorescence of the duplex proved almost as large as the complementary duplexes (see Table 2 entries 9–11, 18–23). On the other hand, if the position of the mismatch base is in the vicinity of the pyrene label, the fluorescence of the duplex was generally small (with few exceptions, vide infra). In some cases, the fluorescence values were similar to the single-stranded acpcPNA (e.g. Table 2 entries 16, 25, 29 and 30) while in others, the fluorescence showed a significant decrease (e.g. Table 2 entries 6–8, 13, 15 and 26). Examples of fluorescence spectra of PyBtr-labeled acpcPNA M11_TC and its hybrids with various DNAs are shown in Fig. 2. The marked difference in the fluorescence properties suggest that the internally pyrene-labeled acpcPNA can discriminate between complementary and single-mismatched DNA targets not by the ability/inability to form hybrids, but rather by probing the local environment in the vicinity of the pyrene label. The decrease in pyrene fluorescence in the mismatched hybrids suggests that the pyrene chromophore undergoes stacking interactions with the nucleobases, possibly through intercalation or groove binding. UV absorption spectra of the mismatched duplexes (Fig. 2, inset) support this hypothesis as evidenced by the presence of red-shifted and hypochromic pyrene absorption bands relative to single stranded acpcPNA. In contrast, blue-shifted pyrene absorption bands were observed with the complementary duplex, or when the position of the mismatch was further away from the pyrene label, thus preventing the stacking interaction. The driving force for this process is the ability of the hydrophobic pyrene label to intercalate within the base stacks in DNA duplexes.17 Intercalation of pyrene in DNA–DNA duplexes is accompanied by fluorescence quenching, therefore internally pyrene-labeled DNAs are not generally useful as DNA hybridization probes,18 an exception being when the pyrene label is linked to the DNA via a rigid linker.4c,e For acpcPNA·DNA duplexes, UV and fluorescence spectra indicate that the intercalation may be accommodated only when there is a mismatch base pair nearby. These results highlight the distinct properties of acpcPNA in comparison to DNA.
Since the fluorescence enhancement ratio [F(ds)/F(ss)] appears to be determined mainly by the original fluorescence of the single stranded acpcPNA [F(ss)], attempts were made to understand the effects of the base sequence on the fluorescence quantum yield of the single-stranded acpcPNA. A series of 11 mer acpcPNAs were synthesized, with a systematic variation of the base sequence around the pyrene label, and their optical properties studied (Table 3). Unexpectedly, the fluorescence quantum yields of the single-stranded acpcPNAs M11_AXXA (X = A, T, C, G) are quite similar (ΦF = 0.065–0.076) regardless of the nature of the flanking bases. The data suggests that the immediate neighboring bases may not be the only factor responsible for the quenching of the pyrene label in the single stranded acpcPNA.15b In another series of experiments, the next-nearest neighboring bases were changed from A to T. It was observed that the single stranded M11_TAAT gave a similar fluorescence quantum yield to the M11_AXXA sequences (ΦF = 0.074), while the M11_TTTT gave much weaker fluorescence (ΦF = 0.019), suggesting possible involvement of the next-nearest neighboring T bases in pyrene label quenching. In all cases, hybridization with complementary DNA resulted in fluorescence increases ranging from 3.2–19.4 fold (or 3.0–16.4 fold in terms of ΦF increase, see Table 3, entries 1, 5, 9, 13, 17 and 19). With few exceptions (e.g. Table 3 entries 2, 18 and 20) hybridization with non-complementary DNA (double mismatched) resulted in mostly decreased fluorescence relative to single stranded acpcPNA. In one case (Table 3, entry 10) the decrease was more than fivefold [F(ds)/F(ss) = 0.18)]. It should be noted that in most of these mismatched cases, the hybrids were still formed albeit with low stability according to UV melting analysis.
Although it is not yet possible to completely understand the factors that determine the brightness of the single stranded internally pyrene-labeled acpcPNA from the available data, our results clearly demonstrate that: (1) the fluorescence of internally pyrene-labeled acpcPNA is low in the single stranded state due to quenching by neighboring nucleobases, especially T and C; (2) the fluorescence of internally pyrene-labeled acpcPNA consistently increases upon hybridization with complementary DNA targets, regardless of the nature of flanking bases and (3) the fluorescence of the mismatched hybrids may be unchanged, decreased or increased, depending on the position and type of mismatch involved. Hybridization-induced fluorescence changes in pyrene-labeled DNA and analogues are generally highly dependent on the nature of the flanking nucleobases. In many cases, the fluorescence increases were observed only in limited sequence contexts.4e,19 The internally pyrene-labeled acpcPNAs described here are rather well-behaved in regards to the fluorescence increase, which was consistently observed in all sequences tested.
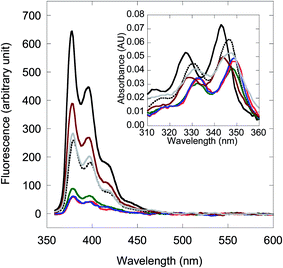
Perhaps the most undesirable aspect of the presently described internally pyrene-labeled acpcPNA probes are the variable responses to mismatched DNA hybrids. Although the internally pyrene-labeled acpcPNA generally yielded unchanged or decreased fluorescence upon hybridization with single-mismatched DNA targets having the mismatch position close to the pyrene label, exceptions to this general trend were observed in a few cases (mostly with T as a partner of the mismatch pair, see Table 2 entries 14, 17, 27 and 31). Furthermore, the fluorescence of the mismatched duplex is large when the mismatch base is distant from the pyrene label. This can be problematic when applying to DNA targets having unknown mismatch positions. If one observes unchanged, or lower fluorescence after hybridization, it is clear that the DNA target cannot be complementary with the probe (i.e., contains more than one mismatch, is fully non-complementary, or contains a single mismatch base close to the pyrene label). However, a fluorescence increase is not conclusive as to whether the DNA target is fully complementary, contains a mismatched base that is located further away from the pyrene label, or contains a special mismatched base close to the pyrene label. While this information can be useful for probing the DNA sequence in question, it may not be desirable in applications such as SNP typing. To address this issue, the enzymatic digestion by S1 nuclease previously successfully applied for increasing the specificity of related probes13b,20 was applicable in this study. The underlying principle is that complementary acpcPNA·DNA hybrids are more resistant to S1 nuclease degradation than mismatched hybrids. As a result, even though the fluorescence of the mismatched acpcPNA·DNA hybrids was initially high [F(ds)/F(ss) ranging from 2–6 depending to the type and position of the mismatch], the signal returned to the same as that seen in single stranded acpcPNA [F(ds)/F(ss) around 1] after 10 min digestion with S1 nuclease (Fig. 3 and S22†). The F(ds)/F(ss) value for the complementary duplex also decreased, but to a much smaller extent (<30% after 10 minutes of digestion). Hence, the S1 nuclease digestion can be used to unambiguously differentiate false positives due to the presence of mismatches away from the location of the pyrene label, thus improving the specificity of the pyrene-labeled acpcPNA probes.
DNA binding and optical properties of terminally pyrene-labeled acpcPNA
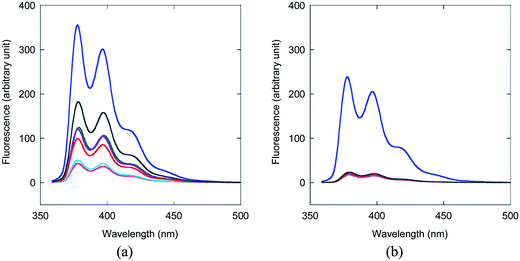
In contrast to the internally pyrene-labeled acpcPNA which exhibited low fluorescence in the single stranded state and strong fluorescence enhancement after hybridization with a complementary DNA target, the corresponding terminally pyrene-labeled acpcPNA behaved rather differently. The Tm and fluorescence data for N-terminally pyrene-labeled acpcPNA are summarized in Table 4. As shown by the Tm measurements these terminally-labeled acpcPNAs exhibited a strong affinity and sequence specificity to complementary DNA targets.In accordance with the internally-labeled acpcPNA, the fluorescence quantum yields of the single stranded terminally-labeled acpcPNA were also small (0.003 to 0.096) (Table 4). The brightness of the single stranded terminally-labeled acpcPNA correlates well with the ability of the adjacent nucleobase (i.e. the N-terminal base in the acpcPNA strand) to quench the pyrene label [ΦF(ss) M12(A) > M10(G) > M11(C) > T9(T)]. In contrast to the internally pyrene-labeled acpcPNA, hybridization of the terminally pyrene-labeled acpcPNA to their complementary DNA targets, with or without hanging 3′-ends, resulted in further quenching of the pyrene fluorescence [ΦF(ds)/ΦF(ss) in the range of 0.10–0.59] (Table 4, entries 4, 6, 9, 10, 15–18). The only exception is the case of the T9 sequence, which gave a 30-fold increase in ΦF (Table 4, entry 1). Hybridization with DNA targets carrying internal mismatched bases results in similar fluorescence changes to the complementary targets (Table 4, entries 2, 5 and 7) because both can form duplexes with reasonable stability at room temperature, and the position of the mismatch is quite distant from the pyrene label. On the other hand, if a mismatched base was present at the 3′-end of the DNA target, the fluorescence was shown to marginally increase [ΦF(ds)/ΦF(ss) in the range of 1.6–2.1] (Table 4, entries 8, 12–14). Hybridization with complementary DNA targets with 3′-terminal base deletion also resulted in small fluorescence increases [ΦF(ds)/ΦF(ss) in the range of 1.8–1.9] (Table 4, entries 19 and 20). The lower fluorescence values of complementary duplexes can be rationalized by quenching of the terminal pyrene label through stacking interactions with the N-terminal base pairs, which exists in both duplexes having exactly complementary DNA, and with DNA having 3′-hanging ends.15a Such stacking interactions would not occur in duplexes with 3′-terminal base deletion, and should be weaker in terminally mismatched duplexes. The small fluorescence increase in these cases can therefore be explained by decreased levels of interactions between the pyrene labels and the nucleobases, as a result of conformational change from single stranded (interaction possible with several bases) to double stranded states (interaction possible only with the terminal base). In the case of terminally-labeled T9, the relatively large fluorescence enhancement (30 fold) after hybridization can be explained by the improved quenching effect of having multiple T bases in the single stranded acpcPNA, relative to a single T–A base pair in the complementary duplex.
Structural basis of the different fluorescence behavior of internally and terminally pyrene-labeled acpcPNA as revealed by MD simulations
To obtain further insight into the mechanism of fluorescence change on hybridization of internally- and terminally-labeled acpcPNA with DNA, molecular dynamics (MD) simulations were performed. The results from the MD simulations of the complementary hybrid of the internally PyBtr-labeled acpcPNA M11_TC showed that bases pT7, pC8 (acpcPNA), and dA5, dG4 (DNA)21 formed normal Watson–Crick base pairs and tightly stacked with neighboring base pairs. Even though the MD simulation was initiated with the pyrene motif stacked between the base pairs, the PyBtr label was forced out into the bulk solvent in the final structure although a part of the pyrene structure was located in the minor groove albeit with no stacking interactions with any nearby base pairs (Fig. 4a). This would indicate reduced interaction between the pyrene and the nucleobases in this structure, relative to the single stranded acpcPNA. On the other hand, in the mismatched duplex case, the pC8 (acpcPNA) and the mismatched dA4 (DNA) bases could not form a stable base pair. As a result, the base dA4 is mislocated in the major groove, and the pyrene remains stacked with neighboring base pairs during the 10 ns simulation time as in the starting structure (Fig. 4b) leading to a larger interaction between the pyrene and the neighboring bases in this case. For the terminally PyBtr-labeled acpcPNA·DNA system M11, the pyrene shows stacking with the pC1 (acpcPNA)-dG11 (DNA) pair in the complementary DNA hybrid (Fig. 5a). However, for the terminal mismatched DNA hybrid, the pyrene was found to stack only with base dA11 on the DNA strand whereas pC1 on the acpcPNA strand twisted towards the major groove (Fig. 5b). This should result in a less favourable interaction between the pyrene and nucleobases in this instance, compared to the case of the complementary hybrid.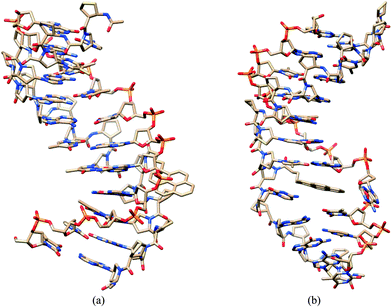 | ||
Fig. 4 MD structures of internally pyrene-labeled acpcPNA M11_TC·DNA hybrids. (a) Complementary DNA and (b) single mismatched DNA averaged over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures. 000 structures. | ||
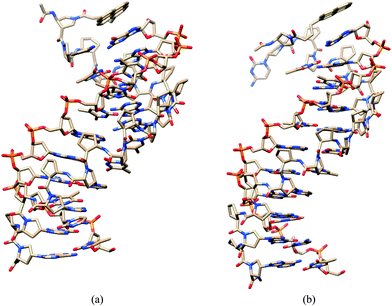 | ||
Fig. 5 MD structures of terminally pyrene-labeled acpcPNA M11·DNA hybrids. (a) Complementary DNA and (b) single mismatched DNA averaged over 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 structures. 000 structures. | ||
Binding free energy and interaction of pyrene
The thermodynamic stabilities of all four acpcPNA·DNA duplexes were estimated for the MD structures in terms of the binding free energy, ΔGbinding, using molecular mechanics combined with generalized Born and surface area (MM-GBSA) methods.14b,22 In order to evaluate the interaction on pyrene affected by nucleotides, pairwise energies were decomposed for each pyrene–nucleotide pair. Note that the nucleotide refers to not only the monomeric unit of DNA, but also that of acpcPNA. The summation of all pairwise energies, excluding pyrene–pyrene pairwise energy, reflects the total interaction on pyrene (ΔEpyrene). Both ΔGbinding and ΔEpyrene were calculated using the AMBER 12 package.23As shown in Table 5, for both internally and terminally pyrene-labeled acpcPNA·DNA duplexes, the ΔGbinding values of complementary duplexes are more negative than those of mismatched duplexes. This indicates higher stabilities for complementary systems, which is in agreement with the experimentally obtained Tm values. For the interaction of pyrene with nucleobases where the pyrene label is internal, the ΔEpyrene of the mismatched duplex is significantly more negative than that of the complementary DNA, which corresponds to the stacking configuration of pyrene as discussed above. This larger interaction between pyrene and nucleobases is consistent with its lower fluorescence quantum yield. In contrast, for the terminal pyrene case a stronger interaction between the pyrene and the nucleobase was observed in the complementary compared to the mismatched duplexes, thus explaining the lower fluorescence quantum yield in this system.
| Position of pyrene | PNA | DNA (3′ → 5′) | ΔGbinding | ΔEpyrene |
|---|---|---|---|---|
| Internal | M11_TC | GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) TCT (comp) TCT (comp) |
−75.8 | −6.2 |
GATTTA![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) ![[A with combining low line]](https://www.rsc.org/images/entities/i_char_0041_0332.gif) TCT (mm) TCT (mm) |
−64.9 | −15.4 | ||
| Terminal | M11 | GATTTAAGTCT (comp) | −85.6 | −8.1 |
| AATTTAAGTCT (mm) | −71.7 | −5.2 |
Experimental
General
All reagent grade chemicals and solvents were purchased from standard suppliers and were used as received without further purification. Methanol used for HPLC experiments was HPLC grade and was filtered through a membrane filter (13 mm ϕ, 0.45 μm) before use. Anhydrous N,N-dimethylformamide (H2O ≤ 0.01%) for solid phase peptide synthesis was obtained from RCI LabScan (Thailand). MilliQ water was obtained from a Millipore (USA) ultrapure water system fitted with a Millipak® 40 filter unit (0.22 μm). Oligonucleotides were purchased from BioDesign or Pacific Science (Thailand) and were used as received.Synthesis of internally- and terminally pyrene-labeled acpcPNA
Synthesis of internally pyrene-labeled acpcPNA was carried out by acylation or reductive alkylation of APC-modified acpcPNA on the solid support as described previously.13 For terminally pyrene-labeled acpcPNA, no APC-modification was necessary and the acpcPNA was acylated at the N-termini with 1-pyrenebutyric acid. All syntheses were performed at 0.5 μmol scale. Cleavage and reverse HPLC purification were performed as previously reported.13Melting temperature measurement
Tm curves were measured at 260 nm using a CARY 100 Bio UV-Vis spectrophotometer (Varian) equipped with a thermal melt system. The sample for Tm experiment was prepared by mixing the calculated amounts of stock oligonucleotide and PNA solutions with sodium phosphate buffer (pH 7.0) in a 10 mm quartz cell with a Teflon stopper, and the final volume was adjusted to 1000 μL with degassed, deionized water. The sample was equilibrated at the starting temperature for 10 min. The A260 was recorded from 20–90 °C (block temperature) with a temperature ramp of 1 °C min−1. The temperature recorded was corrected by a calibration graph created using the built-in temperature probe. The melting temperature was determined from the maximum of the first derivative after smoothing using KaliedaGraph 4.1 (Synergy Software). Tm values obtained from independent experiments were accurate to within ±0.5 °C.UV-Vis absorption spectroscopy and fluorescence experiments
Absorption spectra were performed on a CARY 100 Bio UV-Vis spectrophotometer (Varian). The sample for both measurements was prepared by mixing the calculated amounts of stock oligonucleotide, PNA and sodium phosphate buffer (pH 7.0) solutions together in a 10 mm quartz cell with a Teflon stopper. The final volumes were adjusted to 1000 μL with degassed, deionized water.Fluorescence experiments were performed on a Cary Eclipse Fluorescence Spectrophotometer (Varian/Agilent Technologies). The sample for fluorescence experiment was prepared as described for the UV-Vis experiments. Both the excitation and emission slits were set to 5 nm and the photomultiplier tube (PMT) voltage was set to either medium or high.
Fluorescence quantum yield (ΦF) measurement
The fluorescence quantum yield (ΦF) of the single stranded-PNA and PNA·DNA duplex were calculated using quinine sulfate (ΦF = 0.54)24 as the standard. A stock solution of quinine sulfate (1.0 mM) was prepared by dissolution in 0.1 M H2SO4. For calculation of quantum yield, five concentrations of quinine sulfate standard were prepared, all of which had <0.1 A350. Absorption spectra of the quinine sulfate standards and samples were measured on a CARY 100 Bio UV-Vis spectrophotometer (Varian) and fluorescence spectra were measured on a Cary Eclipse Fluorescence Spectrophotometer (Varian/Agilent Technologies). The integrated fluorescence intensities (excited at 345 nm) and the absorbance values (at 345 nm) of the quinine sulfate standard and the samples were plotted and the slopes were determined to give mstandard and msample, respectively.The quantum yield of sample was then calculated according to eqn (1):
| Φsample = Φstandard(msample/mstandard)(ηsample2/ηstandard2) | (1) |
S1 nuclease digestion experiment
The sample for digestion by S1 nuclease was prepared by mixing the calculated amounts of stock oligonucleotide, acpcPNA and other solutions together to obtain the final concentrations at 1.0 μM acpcPNA, 1.0 μM DNA in 30 mM sodium acetate buffer (pH 4.6) containing 1 mM zinc acetate and 5% glycerol (total volume = 500 μL) in a 10 mm quartz cell. The enzyme stock (S1 nuclease from Aspergillus oryzae obtained from Sigma, 1000 unit per mL) was added to give a final concentration of 5 unit per mL. The progress of the enzymatic digestion was monitored by fluorescence spectrophotometry (kinetic mode). The fluorescence spectra were recorded every 5 min until no further change was observed.Molecular dynamics simulation
Molecular dynamics (MD) simulations of internally and terminally pyrene-labeled acpcPNA·DNA duplexes were performed for 4 systems, Table 2 entries 5 and 8, and Table 4 entries 6 and 8. Since the force field parameters of pyrene and acpcPNA are not available, they were generated as follows. Each monomeric unit of acpcPNA as well as pyrene was optimized with B3LYP/6-31G* level using the Gaussian 09 program.25 The atomic point charge was calculated using HF/6-31G* level, and this was applied in the RESP method26 using the Antechamber suite in AmberTools 12 program.23 The starting structure of the duplex was generated according to previous work.14b,27 For terminally pyrene-labeled acpcPNA·DNA systems, the pyrene was initially placed and stacked with the pC1-dG11 pair (for complementary duplex, Table 4 entry 6), or with the pC1-dA11 pair (for single mismatched duplex, Table 4 entry 8). For the complementary duplex in the internal pyrene systems (Table 2 entry 5), the pyrene was initially placed between pT7-dA5 and pA9-dT3 base pairs so that it was stacked between these two base pairs. The pC8 and pG4 bases were twisted into the major groove. The geometries of the pyrene, pC8 and dG4 bases were then optimized using molecular mechanics in order to relax any steric geometry. The starting structures for the single mismatched system (Table 2 entry 8) was built similarly.Each generated duplex was immersed into the rectangular TIP3P water box28 extended by 10 Å in each direction from the duplex. The DNA strand was neutralized by adding Na+ counter-ions. The unrestrained MD simulations were performed using a well-established protocol and standard conditions (T = 300 K, P = 1 atm, 1 fs integration step, generated pyrene and acpcPNA parameters complemented with parm10 force field, periodic boundary conditions, cutoff of 9 Å for nonbonded interactions, and particle-mesh Ewald algorithm29 for treatment of electrostatic interactions). The MD trajectories were performed for 10 ns for each duplex using the AMBER 12 package. The coordinates of the system were saved every 1 ps for further analysis.
Conclusion
In conclusion, we successfully synthesized internally and terminally pyrene-labeled pyrrolidinyl peptide nucleic acids and evaluated their fluorescence properties. In the single stranded state, both internally and terminally pyrene-labeled acpcPNA show weak fluorescence due to quenching by neighboring nucleobases. Thymine was found to be the most effective quencher, and the quenching effects probably extend beyond the nearest neighbors. The internally pyrene-labeled acpcPNA showed moderate to strong fluorescence enhancement (2.9–73 fold) upon hybridization with the complementary DNA target, on comparison with single stranded acpcPNA. The fluorescence is generally low in duplexes carrying a mismatched base pair close to the pyrene label in the case of internally pyrene-labeled acpcPNA. On the other hand, terminally pyrene-labeled acpcPNA generally gave lower fluorescence upon hybridization with complementary DNA and slightly increased fluorescence upon hybridization with DNA carrying terminal mismatches or deletions. The different fluorescence properties of internally, and terminally pyrene-labeled acpcPNA were rationalized by the different arrangements of the pyrene label, which was supported by optical spectroscopy and MD simulations. These results emphasize the active role of the label and provide a basis to assist in the design of more effective singly-labeled beacons in the future.Acknowledgements
Financial support of this work from the Thailand Research Fund (RTA5280002), the Ratchadaphiseksomphot Endowment Fund of Chulalongkorn University (RES560530126-AM), the Thai government stimulus package 2 (TKK2555, SP2) under the Project for Establishment of Comprehensive Center for Innovative Food, Health Products and Agriculture, the Development and Promotion of Science and Technology Talents Project (to CB), and Computational Science Research Group, Faculty of Science, Khon Kaen University (to KS) is gratefully acknowledged. Support from the National e-Science Infrastructure Consortium (URL: http://www.e-science.in.th), Bangkok, is acknowledged for computing resources.References and Notes
- (a) S. Tyagi and F. R. Kramer, Nat. Biotechnol., 1996, 14, 303 CrossRef CAS PubMed; (b) S. Tyagi, D. P. Bratu and F. R. Kramer, Nat. Biotechnol., 1998, 16, 49 CrossRef CAS PubMed.
- Reviews: (a) S. A. E. Marras, Mol. Biotechnol., 2008, 38, 247 CrossRef CAS PubMed; (b) J. Guo, J. Ju and N. J. Turro, Anal. Bioanal. Chem., 2012, 402, 3115 CrossRef CAS PubMed.
- (a) R. T. Ranasinghe and T. Brown, Chem. Commun., 2005, 5487 RSC; (b) R. Nutiu and Y. Li, Nucleic Acids Res., 2002, 30, e94 CrossRef PubMed; (c) J. Cheng, Y. Zhang and Q. Li, Nucleic Acids Res., 2004, 32, e61 CrossRef PubMed; (d) N. Dobson, D. G. McDowell, D. J. French, L. J. Brown, J. M. Mellor and T. Brown, Chem. Commun., 2003, 1234 RSC; (e) T. N. Grossmann, L. Röglin and O. Seitz, Angew. Chem., Int. Ed., 2007, 46, 5223 CrossRef CAS PubMed; (f) C. Holzhauser and H.-A. Wagenknecht, Angew. Chem., Int. Ed., 2011, 50, 7268 CrossRef CAS PubMed; (g) K. Stöhr, B. Häfner, O. Nolte, J. Wolfrum, M. Sauer and D.-P. Herten, Anal. Chem., 2005, 77, 7195 CrossRef PubMed.
- (a) N. Venkatesan, Y. J. Seo and B. H. Kim, Chem. Soc. Rev., 2008, 37, 648 RSC; (b) I. Nazarenko, B. Lowe, M. Darfler, P. Ikonomi, D. Schuster and A. Rashtchian, Nucleic Acids Res., 2002, 30, e37 CrossRef PubMed; (c) A. Okamoto, K. Kanatani and I. Saito, J. Am. Chem. Soc., 2004, 126, 4820 CrossRef CAS PubMed; (d) G. T. Hwang, Y. J. Seo and B. H. Kim, J. Am. Chem. Soc., 2004, 126, 6528 CrossRef CAS PubMed; (e) M. E. Østergaard, D. C. Guenther, P. Kumar, B. Baral, L. Deobald, A. J. Paszczynski, P. K. Sharma and P. J. Hrdlicka, Chem. Commun., 2010, 46, 4929 RSC; (f) M. E. Østergaard and P. J. Hrdlicka, Chem. Soc. Rev., 2011, 40, 5771 RSC.
- P. E. Nielsen, Chem. Biodiversity, 2010, 7, 786 CAS.
- (a) H. Kuhn, V. V. Demidov, J. M. Coull, M. J. Fiandaca, B. D. Gildea and M. D. Frank-Kamenetskii, J. Am. Chem. Soc., 2002, 124, 1097 CrossRef CAS PubMed; (b) E. Socher, L. Bethge, A. Knoll, N. Jungnick, A. Herrmann and O. Seitz, Angew. Chem., Int. Ed., 2008, 47, 9555 CrossRef CAS PubMed; (c) E. Socher, A. Knoll and O. Seitz, Org. Biomol. Chem., 2012, 10, 7363 RSC.
- E. A. Englund and D. H. Appella, Org. Lett., 2005, 7, 3465 CrossRef CAS PubMed.
- (a) N. Svanvik, G. Westman, D. Wang and M. Kubista, Anal. Biochem., 2000, 281, 26 CrossRef CAS PubMed; (b) N. Svanvik, J. Nygren, G. Westman and M. Kubista, J. Am. Chem. Soc., 2001, 123, 803 CrossRef CAS PubMed.
- O. Köhler, D. V. Jarikote and O. Seitz, ChemBioChem, 2005, 6, 69 CrossRef PubMed.
- (a) T. Vilaivan, C. Suparpprom, P. Harnyuttanakorn and G. Lowe, Tetrahedron Lett., 2001, 42, 5533 CrossRef CAS; (b) P. Ngamwiriyawong and T. Vilaivan, Nucleosides, Nucleotides Nucleic Acids, 2011, 30, 97 CrossRef CAS PubMed; (c) W. Mansawat, C. Vilaivan, A. Ba’lazs, D. J. Aitken and T. Vilaivan, Org. Lett., 2012, 14, 1440 CrossRef CAS PubMed.
- (a) T. Vilaivan and C. Srisuwannaket, Org. Lett., 2006, 8, 1897 CrossRef CAS PubMed; (b) C. Vilaivan, C. Srisuwannaket, C. Ananthanawat, C. Suparpprom, J. Kawakami, Y. Yamaguchi, Y. Tanaka and T. Vilaivan, Artificial DNA: PNA & XNA, 2011, 2, 50 Search PubMed.
- (a) B. Boontha, J. Nakkuntod, N. Hirankarn, P. Chaumpluk and T. Vilaivan, Anal. Chem., 2008, 80, 8178 CrossRef CAS PubMed; (b) C. Ananthanawat, T. Vilaivan, V. P. Hoven and X. D. Su, Biosens. Bioelectron., 2010, 25, 1064 CrossRef CAS PubMed; (c) P. S. Laopa, T. Vilaivan and V. P. Hoven, Analyst, 2013, 138, 269 RSC; (d) S. Jampasa, W. Wonsawat, N. Rodthongkum, W. Siangproh, P. Yanatatsaneejit, T. Vilaivan and O. Chailapakul, Biosens. Bioelectron., 2014, 54, 428 CrossRef CAS PubMed.
- (a) N. Reenabthue, C. Boonlua, C. Vilaivan, T. Vilaivan and C. Suparpprom, Bioorg. Med. Chem. Lett., 2011, 21, 6465 CrossRef CAS PubMed; (b) B. Ditmangklo, C. Boonlua, C. Suparpprom and T. Vilaivan, Bioconjugate Chem., 2013, 24, 614 CrossRef CAS PubMed; (c) N. Maneelun and T. Vilaivan, Tetrahedron, 2013, 69, 10805 CrossRef CAS PubMed.
- (a) C. Boonlua, C. Vilaivan, H.-A. Wagenknecht and T. Vilaivan, Chem. – Asian J., 2011, 6, 3251 CrossRef CAS PubMed; (b) W. Mansawat, C. Boonlua, K. Siriwong and T. Vilaivan, Tetrahedron, 2012, 68, 3988 CrossRef CAS PubMed.
- (a) J. S. Mann, Y. Shibata and T. Meehan, Bioconjugate Chem., 1992, 3, 554 CrossRef CAS; (b) J. N. Wilson, Y. Cho, S. Tan, A. Cuppoletti and E. T. Kool, ChemBioChem, 2008, 9, 279 CrossRef CAS PubMed.
- (a) M. Manoharan, K. L. Tivel, M. Zhao, K. Nafisi and T. L. Netzel, J. Phys. Chem., 1995, 99, 17461 CrossRef CAS; (b) C. A. M. Seidel, A. Schulz and M. H. M. Sauer, J. Phys. Chem., 1996, 100, 5541 CrossRef CAS.
- A. Wolfe, G. H. Shimer and T. Meehan, Biochemistry, 1987, 26, 6392 CrossRef CAS.
- (a) K. Yamana, R. Iwase, S. Furutani, H. Tsuchida, H. Zako, T. Yamaoka and A. Murakami, Nucleic Acids Res., 1999, 27, 2387 CrossRef CAS PubMed; (b) M. Nakamura, Y. Fukunaga, K. Sasa, Y. Ohtoshi, K. Kanaori, H. Hayashi, H. Nakano and K. Yamana, Nucleic Acids Res., 2005, 33, 5887 CrossRef CAS PubMed.
- J. H. Ryu, Y. J. Seo, G. T. Hwang, J. Y. Lee and B. H. Kim, Tetrahedron, 2007, 63, 3538 CrossRef CAS PubMed.
- (a) S. Ye, Y. Miyajima, T. Ohnishi, Y. Yamamoto and M. Komiyama, Anal. Biochem., 2007, 363, 300 CrossRef CAS PubMed; (b) M. Komiyama, S. Ye, X. Liang, Y. Yamamoto, T. Tomita, J.-M. Zhou and H. Aburatani, J. Am. Chem. Soc., 2003, 125, 3758 CrossRef CAS PubMed.
- The numbering starts from N-terminal base of acpcPNA and 5′-base of DNA.
- K. Siriwong, P. Chuichay, S. Saen-oon, C. Suparpprom, T. Vilaivan and S. Hannongbua, Biochem. Biophys. Res. Commun., 2008, 372, 765 CrossRef CAS PubMed.
- AMBER 12, University of California, San Francisco, 2012 Search PubMed.
- W. H. Melhuish, J. Phys. Chem., 1961, 65, 229 CrossRef CAS.
- Gaussian 09, Revision A.1, Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- C. I. Bayly, P. Cieplak, W. D. Cornell and P. A. Kollman, J. Phys. Chem., 1993, 97, 10269 CrossRef CAS.
- N. Poomsuk and K. Siriwong, Chem. Phys. Lett., 2013, 588, 237 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Am. Chem. Soc., 1981, 103, 335 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, J. Chem. Phys., 1993, 98, 10089 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ra47997h |
| This journal is © The Royal Society of Chemistry 2014 |