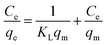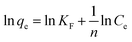Two-dimensional ultrathin nanosheets of Ni–In-layered double hydroxides prepared in water: enhanced performance for DNA adsorption†
Yongjuan Wang,
Yuming Zhou*,
Tao Zhang,
Man He and
Xiaohai Bu
School of Chemistry and Chemical Engineering, Southeast University, Jiangsu Optoelectronic Functional Materials and Engineering Laboratory, Nanjing 211189, P.R. China. E-mail: ymzhou@seu.edu.cn; Fax: +86-025-52090617; Tel: +86-025-52090617
First published on 27th May 2014
Abstract
This work describes a facile approach for the delaminating of Ni–In-layered double hydroxides (LDHs) by the intercalation of acetate anions in the LDHs gallery using co-precipitation method. The acetate-intercalated Ni–In LDHs exhibited swelling behavior in water and delaminated into semitransparent suspensions on the action of water. The transmission electron microscopy (TEM) and atomic force microscopic (AFM) observations revealed the formation of unilamellar LDHs nanosheets. This simple method did not need reflux at high temperatures, overcoming the deficiency of using organic solvents, resulting in a steady colloidal suspension of 2D nanosheets. The unique properties of 2D ultrathin nanosheets with full access to the positively charged activity sites make them attractive in terms of the improvement of DNA adsorption. The effect of dosage on the adsorption of DNA and the adsorption isotherm were investigated in detail. The equilibrium data were found to be well described by both the Langmuir and Freundlich models. Based on the mechanism of adsorption, the adsorption sites of exfoliated nanosheets can be easily occupied by highly negatively charged small anions through electrostatic interactions. The adsorbed DNA can be released via the addition of a hydrogen phosphate solution.
Introduction
Currently, nanosheets with ultimate two-dimensional structures, as a new class of nanoscale materials, have attracted significant attention because novel physical and chemical properties, e.g. surface effects, have indeed been gradually revealed in 2D systems.1 Many inorganically layered compounds with 2D surfaces, such as metal chalcogenides,2 metal phosphates and phosphonates3 and layered metal oxides,4,5 give rise to renewed research. Delaminating these materials could realize more excellent properties. Layered double hydroxides, also known as hydrotalcite-like clays, are unusual members of the layered materials family and have drawn increasing research attention in the past few decades.6,7 The chemical composition of LDHs could be expressed by the general formula [MII1−xMIIIx(OH)2][An−x/n]·xH2O, in which MII and MIII represent the divalent and trivalent metal cations, respectively, and interlayer anions An− balance the charge.8Intensive research has been focused on LDHs due to their broad applications as catalysts,9 fire retardants,10 drug-delivery hosts,11,12 and electrochemical materials.13–16 In particular, the application of LDHs as adsorbents for biomolecular adsorption17,18 has attracted much attention due to their positively charged surfaces and significant structures. Because DNA has become a central molecule in bionanotechnology, materials that can absorb DNA are useful for developing biosensors and gene delivery, and for DNA extraction and separation. Chan et al.19 have immobilized LDHs on the polycarbonate substrate as the media to extract the specific DNA molecules through a fluidic system, and the results show that the immobilized LDHs extract the specific DNA molecules from the biomolecular solution with high efficiency. Kim et al.20 investigated the selective DNA adsorption on LDH nanoparticles, and they found that the LDHs have advantages in the selective adsorption of DNA. As DNA is highly negatively charged, the adsorption of DNA on positively charged LDHs surfaces is based on electrostatic interactions. However, all the LDHs used in these studies were unexfoliated, which did not sufficiently express their positively charged active sites. Consequently, the delamination of the bulk LDHs was required to expose the positively charged sites of the host layer completely.
So far, extensive efforts have been made to exfoliate LDHs into 2D ultrathin nanosheets with positive charges, which would improve the accessibility to the inner surfaces of host layers and extend the physical and chemical properties of anisotropic nanosheets. The delamination of LDHs in formamide was observed to be instant and spontaneous and did not need any heat or refluxing.21 It is regarded as a very simple and effective method for the delamination of LDHs containing different anions. However, this method was also accompanied by some disadvantages. For example, formamide was difficult to remove due to its high boiling point (210 °C), and exfoliated LDHs prepared in formamide would restack when in contact with water.22 These problems restricted the applications of exfoliated LDHs nanosheets.
Therefore, the exfoliation of LDHs in water has been desired. The pioneering work was conducted by Hibino and Kobayashi.23 They reported that LDHs containing lactate swelled and exfoliated in water. Then, Kumar et al.24 simulated the molecule dynamics of LDHs containing the monocarboxylic acids formate, acetate and propanoate as the charge-balancing interlayer anions. The results support the concept that carboxylate sites interact relatively weakly with LDH layers and that large expansions leading to delamination in water can be energetically favorable. Iyi et al.25 synthesized these monocarboxylic acid-intercalated LDHs by the anion exchange method. They found that the acetate- and propanoate-intercalated LDHs did exhibit swelling behavior in water, whereas energy-intensive multiple procedures were required by the anion exchange method. Recently, carboxylate intercalated LDHs have been prepared by carboxylamide hydrolysis, but the obtained LDHs undergo only partial exfoliation with a thickness of several nanometers.26,27 Thus, in this work, we report a facile method to prepare the acetate-containing Ni–In LDHs without any carbonate impurity. The obtained LDHs delaminated well in water. The characteristics of the exfoliated 2D nanosheets were investigated in detail to verify the ultrathin property of the nanosheets. The colloidal suspension of the 2D ultrathin nanosheets was used as an adsorbent for DNA separation. The effect of dosage on the adsorption of DNA and the adsorption isotherm were investigated in detail. DNA desorption was then realized via the addition of a salt solution.
Experimental
Materials
Fish sperm DNA was acquired from Sigma Chemical Co. Nickel hydroxide and indium hydroxide were obtained from Aladdin Reagent, Shanghai, China. Acetic acid, sodium hydroxide, sodium chloric and sodium hydrogen phosphate were purchased from Sinopharm Chemical Reagent Co., Ltd., Shanghai, China. All the aforementioned reagents were of analytical reagent grade and were used without further purification. Deionized and decarbonated water was used in all solutions.Synthesis and delamination of acetate-intercalated Ni–In LDHs
Typically, 0.03 mol of nickel hydroxide and 0.01 mol of indium hydroxide were dissolved in 20 mL of 10 mM acetate acid solution. The mixtures were stirred at 80 °C until a clear and transparent green solution was obtained. NaOH solution (4 M) was then additionally introduced until the pH of the reaction mixture reached 10. The resulting slurry was sealed in an autoclave and heated at 80 °C for 24 h. The solid was centrifuged and washed thoroughly with deionized water to pH = 7. The sample previously synthesized was denoted as Ni–In–CH3COO− LDHs.The wet gel was suspended in deionized water, and the suspension was laid aside for about 24 h; a translucent colloidal solution was then produced. The content of LDH nanosheets in the colloidal solution is estimated to be 4 mg mL−1. The exfoliated LDHs were named as Ni–In ELDHs nanosheets. The entire procedure was carried out under N2 atmosphere to avoid contamination by atmospheric CO2. The deionized water for the preparation of all aqueous solutions was decarbonated by boiling prior to use.
In comparison, Ni–In–CO32− LDHs were prepared by the following procedure: 0.03 mol of nickel nitrate and 0.01 mol of indium nitrate were dissolved in 50 mL deionized water. Further, a mixture solution of NaOH (2 M) and Na2CO3 (0.2 M) were additionally introduced dropwise until the pH of the reaction mixture reached 10. The resulting slurry was stirred at 80 °C for 24 h. The solid was centrifuged, washed thoroughly with deionized water to pH = 7 and dried at 60 °C for 12 h.
Adsorption of DNA with colloidal Ni–In ELDHs nanosheets
The sorption experiments were performed by batch adsorption experiments and were conducted by mixing 10 mL colloidal suspension containing 1–10 mg Ni–In ELDH nanosheets with 10 mL water solution containing 200–2000 μg DNA. After 12 h of standing at 25 °C, the mixtures were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. DNA in the supernatant was determined by UV-vis spectrophotometry at 260 nm. The amount of DNA adsorbed by Ni–In ELDHs nanosheets was calculated by the difference between the amount of DNA added and that remaining in the supernatant.
000 rpm for 15 min. DNA in the supernatant was determined by UV-vis spectrophotometry at 260 nm. The amount of DNA adsorbed by Ni–In ELDHs nanosheets was calculated by the difference between the amount of DNA added and that remaining in the supernatant.
In comparison, 10–100 mg of Ni–In–CO32− LDHs were dispersed in 10 mL deionized water and then mixed with 10 mL water solution containing 1000 μg DNA. The procedures followed were the same as those previously described for Ni–In ELDHs nanosheets.
Desorption of DNA from Ni–In ELDHs nanosheets
10 mL of colloidal Ni–In ELDHs nanosheets (0.5 mg mL−1) was mixed with 10 mL of water solution containing 1000 μg DNA. The mixture was laid aside for 12 h at 25 °C and centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The amount of DNA adsorbed was determined as previously mentioned. The DNA–LDHs complexes formed after equilibrium adsorption were dispersed in 100 mL of 0.1 M NaCl solution (dissolved in 10 mM Tris buffer) and 100 mL of 0.1 M hydrogen phosphate at pH = 7, sequentially. After 3 h of desorption, the mixtures were centrifuged at 15
000 rpm for 15 min. The amount of DNA adsorbed was determined as previously mentioned. The DNA–LDHs complexes formed after equilibrium adsorption were dispersed in 100 mL of 0.1 M NaCl solution (dissolved in 10 mM Tris buffer) and 100 mL of 0.1 M hydrogen phosphate at pH = 7, sequentially. After 3 h of desorption, the mixtures were centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. The amount of DNA desorbed was determined by UV-vis spectrophotometry at 260 nm in the supernatant. The percentage of DNA desorbed was calculated according to the following expression: the percentage of DNA desorption (%) = the total amount of DNA desorbed/the amount of DNA adsorbed.
000 rpm for 15 min. The amount of DNA desorbed was determined by UV-vis spectrophotometry at 260 nm in the supernatant. The percentage of DNA desorbed was calculated according to the following expression: the percentage of DNA desorption (%) = the total amount of DNA desorbed/the amount of DNA adsorbed.
Characterization
X-ray diffraction (XRD) data were collected by a Bruker D8 instrument (Cu Kα radiation, λ = 0.15405 nm, 40 kV per 30 mA). Thermogravimetry (TG) and differential scanning calorimetry (DSC) curves were carried out with a Rigaku ThermoPlus TG8120 at 10 °C min−1 and under nitrogen flow from 50 °C to 800 °C. Fourier transform infrared (FT-IR) spectra of all the samples were recorded using a Bruker Alpha-P FT-IR spectrometer (ATR mode, Diamond crystal, 400–4000 cm−1, 4 cm−1 resolution). The morphologies and dimensions of the particles were examined with a Hitachi H-600 transmission electron microscopy. Tapping-mode atomic-force microscopic images were acquired under ambient conditions using a DimensionIcon system (Bruker AXS) with NanoScope software to examine the surface topography of nanosheets deposited onto a mica substrate.Results and discussion
Synthesis of acetate-intercalated Ni–In LDHs
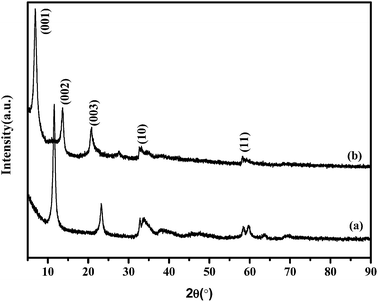
Acetate-intercalated Ni–In LDHs were synthesized by the co-precipitation method followed by hydrothermal treatment. The products were very fine green gels and could be well dispersed in water. The morphology of the obtained Ni–In–CH3COO− LDHs was shown in Fig. 1. As can be seen, the material exhibited a homogeneous distribution of well-defined monodispersed plate-like particles. Unlike the acetate-intercalated LDHs prepared using hydrolysis of metal–salt precursors in a polyol medium, which leads to highly compact particle aggregation,28 individual hexagonal particles can be distinguished clearly using the co-precipitation method with water as the medium. Fig. 2b showed the X-ray diffraction pattern of Ni–In–CH3COO− LDHs. The patterns were typical of LDHs phases displaying a series of 00l peaks at low 2θ values, which were indicative of the inorganic layer spacing. Broad and asymmetrically shaped peaks are observed at higher angles, a result of turbostratic effects emanating from the translational disorder of the metal hydroxide sheets along the a and c axes, which destroys the line shapes of the (hk0) reflections.29,30 In comparison with the Ni–In–CO32− LDHs (Fig. 2a), the 00l reflection peaks of Ni–In–CH3COO− LDHs shifted to lower 2θ values. Correspondingly, the basal spacing of Ni–In–CH3COO− LDHs was estimated to be 1.28 nm, larger than the CO32− intercalated LDHs (0.78 nm). Considering the size of acetate anion (0.48 nm × 0.39 nm), the bilayer arrangement of acetate anions in the interlayer was proposed, which was in agreement with the acetate intercalated LDHs prepared in polyol.28 In our latest study,31 acetate intercalated Ni–In LDHs were also successfully prepared by the anion exchange method, in which the basal spacing of d003 = 0.85 nm, corresponding to the monolayer arrangement of the acetate anion in the interlayer. As a result, the distance between the host layers of Ni–In–CH3COO− LDHs was increased by the coprecipitation method resulting in the decrease of the interactions between the layers, which were more favorable for delamination. The FT-IR spectra of Ni–In–CH3COO− LDHs were shown in Fig. S1a.† The vibrations at 1570 and 1402 cm−1 corresponded to the antisymmetric and symmetric stretching modes of the COO− group, respectively. The band at 1355 cm−1 was due to the bending mode of the –CH3 group of the acetate anion. All these results indicated the successful synthesis of the acetate-intercalated Ni–In LDHs.Delaminating of acetate-intercalated Ni–In LDHs
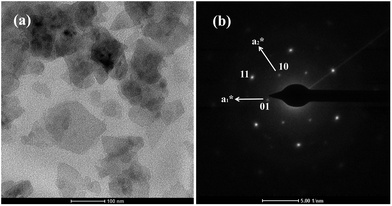
The exfoliation of Ni–In–CH3COO− LDHs proceeded in the dispersion of the wet gels of the as-prepared LDHs in water. A green colloidal suspension was obtained without shaking and heating, exhibiting a Tyndall effect on laser exposure (Fig. S2†). A clear Tyndall light scattering was observed, indicating the presence of exfoliated LDH nanosheets dispersed in water. The resulting colloidal suspension was highly stable, and no sediment was observed upon long-term standing. Fig. 3a gives the TEM image of the Ni–In ELDHs nanosheets. Large amounts of nanosheets are translucent, indicating the ultrathin thickness of nanosheets. The morphology of the Ni–In ELDHs nanosheets was irregular, indicating that the delamination process broke the macrostructure of nanosheets. However, the micro-crystal structure still remained, which can be observed in Fig. 3b. The SAED pattern of individual sheets displays hexagonally arranged spots, suggesting their single-crystal nature. The hexagonal lattice with a = 0.305 nm was compatible with the in-plane structural parameter of the Ni–In–CH3COO− LDHs precursor crystals determined from the XRD characterization, indicating that the basic architecture of the layer remained unchanged after exfoliation. Fig. 4 displays an AFM image collected by dropping the fresh emulsion of Ni–In ELDHs nanosheets on a clean mica substrate. The image covering an 800 nm × 800 nm area (Fig. 4a) shows the presence of particles with morphology and sizes coincident with the TEM images. The thicknesses of these nanosheets were studied with greater detail in several areas. According to Fig. 4b, we observe the presence of an average height profile of 1.18 nm, corresponding to the presence of a single unilamellar nanosheet. The observed deviation with respect to the crystallographic thickness of a single brucite layer (0.48 nm) has been already observed in the AFM studies of exfoliated acetate-intercalated Mg–Al LDHs.25 It is generally attributed to the adsorption of water molecules and residual acetate anions present in the medium.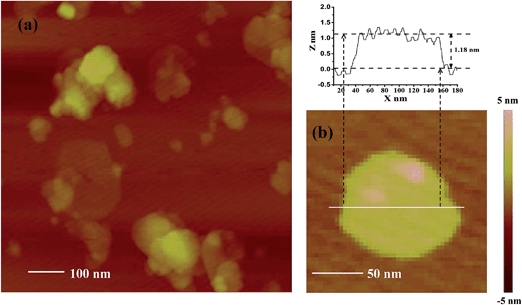 | ||
| Fig. 4 (a) Tapping-mode AFM image of Ni–In ELDHs nanosheets deposited on mica substrate, and (b) height profile concordant with that expected for a single layer. | ||
Adsorption of DNA with colloidal Ni–In ELDHs nanosheets
The Ni–In ELDHs nanosheets prepared in this work were positively charged (Fig. S3†) with the inner surfaces of the LDHs host layers exposed outside completely and dispersed well in water, which were favorable for the adsorption for the negatively charged DNA biomolecules. The DNA adsorption behavior of the Ni–In ELDHs nanosheets was studied, and the unexfoliated Ni–In LDHs were also used as an adsorbent in comparison. The effect of adsorbent dosage on the amount of DNA adsorbed was studied at an adsorption temperature of 25 °C. As can be seen in Fig. 5a, the concentration of DNA decreased with an increased amount of Ni–In ELDHs nanosheets, reaching a minimum concentration of 7.6 μg mL−1 at an adsorbent dosage of 5 mg. By continuing to increase the adsorbent dosage, the concentration of DNA remained constant. However, it was difficult to reduce the DNA concentration below 7 μg mL−1, which may be ascribed to the adsorption equilibrium between free DNA and Ni–In ELDHs nanosheets. It was also noticed that adsorption capacity decreased with increasing adsorbent dosage. The DNA adsorption capacity decreased from 256.0 to 84.6 μg mg−1 with the Ni–In ELDHs nanosheets dosage increased from 1 to 10 mg. | ||
| Fig. 5 Effect of the dosages of (a) Ni–In ELDHs nanosheets, (b) Ni–In–CO32− LDHs on DNA adsorption (conditions: adsorption time, 12 h; initial DNA amount, 1000 μg; adsorption temperature, 25 °C). | ||
It is obvious that by increasing the adsorbent dosage, the amount adsorbed per unit mass decreases. It is readily understood that the number of available adsorption sites increases by increasing the adsorption dosage. The decrease in adsorption capacity with an increase in the adsorbent dosage is then mainly due to the unsaturation of adsorption sites through the adsorption reaction.32,33 Considering the efficiency of DNA separation, 5 mg Ni–In ELDHs nanosheets were fixed as optimum dosage. The adsorption behavior of Ni–In–CO32− LDHs was studied with the same initial amount of DNA as shown in Fig. 5b. Whereas the amount of Ni–In–CO32− LDHs increased from 5 to 60 mg, the concentration of DNA decreased, reaching a minimum number of 25.6 μg mL−1, which was obviously higher than that adsorbed by the Ni–In ELDHs nanosheets. The minimum amount of Ni–In–CO32− LDHs used for adsorption equilibrium was 30 mg, five times higher than the Ni–In ELDHs nanosheets used. Correspondingly, the DNA adsorption capacity of the unexfoliated LDHs with a maximum of 41.7 μg mg−1 was much lower than Ni–In ELDHs nanosheets. It was difficult for the Ni–In–CO32− LDHs to disperse homogeneously in water to form a steady colloidal suspension, resulting in an incomplete interaction between the LDHs particles and DNA. Moreover, the inaccessibility to the positively charged inner surfaces of the host layers of the unexfoliated LDHs restricted the adsorption of the negatively charged DNA. Whereas the Ni–In ELDHs nanosheets dispersed well in water and supplied full access to the inner surfaces of the positively charged host layers, higher DNA adsorption capacity was obtained. This evidence showed that the adsorption activity of Ni–In ELDHs was obviously enhanced by the formation of 2D nanosheets.
Equilibrium adsorption of DNA on Ni–In ELDHs nanosheets
The adsorption isotherms of DNA on Ni–In ELDHs nanosheets were shown in Fig. 6. The DNA adsorption capacity increased with increasing initial concentrations. In order to further investigate the adsorption process of DNA, the equilibrium experimental data were further analyzed by two commonly used isotherm models: the Langmuir and Freundlich equations.34 The forms of these isotherm models are presented mathematically as follows:where qe is the amount of DNA adsorbed per unit mass of Ni–In ELDH nanosheets (μg mg−1), qm is the maximum amount of DNA adsorbed (μg mg−1), KL is a constant related to the adsorption energy (mL μg−1) and Ce stands for the concentration of DNA in the equilibrium solution (μg mL−1). KF and n are the Freundlich isotherm constants, which indicate the extent of the adsorption and the degree of nonlinearity between the solution concentration and the adsorption.
 | ||
| Fig. 6 Adsorption isotherms of DNA adsorption onto Ni–In ELDH nanosheets (conditions: adsorption time, 12 h; initial DNA amount, 200–2000 μg; adsorption temperature, 25 °C). | ||
As shown in Table 1, the experimental data were fitted both to the Langmuir and Freundlich models, which can generally be used to describe the adsorption processes of proteins35,36 with the constraint that the Langmuir isotherm as the simplest model cannot describe all phenomena occurring during protein adsorption.37 For the Langmuir isotherm, the adsorbate is monolayer adsorbed, and the surface is homogeneous with all surface sites energetically equivalent. Moreover, there are no interactions between adjacent adsorption sites or adsorbed molecules. The Freundlich isotherm is usually applied to non-ideal sorption on heterogeneous surfaces, as well as multilayer sorption. As can be seen, the correlation coefficients R2 of the Langmuir and Freundlich equations were 0.9685 and 0.9763, respectively. Within the experimental errors, it is not possible to distinguish which model describes the data more accurately. The relatively high correlation coefficient value of the Freundlich isotherm suggested the existence of different adsorption energies on the surface. It may be that DNA molecules first adsorb on the surfaces in the monolayer, then the interactions between the DNA molecules occur, followed by the adsorption onto previously adsorbed DNA layer resulted in the partial formation of multilayer adsorption. Thus, both the Langmuir and Freundlich models described the equilibrium data well.
| KL (mL μg−1) | qm (μg mg−1) | R2 | |
|---|---|---|---|
| Langmuir | 0.0514 | 625.0 | 0.9685 |
| KF (μg mg−1 (mL μg−1)1/n) | n | R2 | |
|---|---|---|---|
| Freundlich | 43.3367 | 1.4928 | 0.9763 |
Desorption of DNA from Ni–In ELDHs nanosheets
Desorption was conducted after the adsorption of DNA on Ni–In ELDHs nanosheets. The results of desorption were presented in Table 2. It had been observed that more than 75% DNA was released in a hydrogen phosphate solution, whereas the low desorption efficiency of 2.7% was observed in the sodium chloride solution. The DNA desorption capacity in the presence of HPO42− was higher than that in the presence of Cl−. The adsorbed DNA was easily released, mainly due to the electrostatic interactions between Ni–In ELDHs nanosheets, and DNA was much weaker than the interactions between the Ni–In ELDHs nanosheets and competitive anions. Hence, the anions were easily absorbed by the surplus positive charges of Ni–In ELDHs nanosheets. The adsorption sites of the nanosheets were easily occupied by those small anions, resulting in the release of DNA. The electrostatic interactions played an important role in DNA adsorption and desorption, illustrating that the mechanism of adsorption was governed by physisorption, which was confirmed by FT-IR analysis (Fig. S1c†). There was no new band in FT-IR spectra of the DNA–adsorbed complexes, indicating that no chemical reaction between DNA and Ni–In ELDHs nanosheets occurred. Hence, the mechanism of DNA adsorption was mainly governed by electrostatic attraction.| Initial DNA concentration (μg mL−1) | Adsorbed DNA (μg) | Competitive anion | Desorbed DNA (μg) | Desorption percentage (%) |
|---|---|---|---|---|
| 50 | 846.7 | Cl− | 23.2 | 2.7 |
| HPO42− | 639.9 | 75.6 |
Finally, in order to determine the restacking ability of the exfoliated nanosheets in the presence of small anions, the precipitate that appeared after DNA desorption by HPO42− was collected and analyzed. On the basis of the XRD data (Fig. S4†), the restacked material exhibits the typical profile for LDHs materials with intense peaks at low theta values, whereas they become weaker and less defined at higher angular values. The basal spacing is estimated to be about 1.07 nm, which is consistent with the hydrogen phosphate intercalated LDHs as reported elsewhere.38,39 The results indicated that the presence of intense electrostatic interactions between the positively charged layers and the HPO42− mediates the re-assembling of the original layered structure after the desorption progress.
Conclusion
In conclusion, we exfoliated the Ni–In LDHs in water by the intercalation of acetate anions in the LDHs gallery by the co-precipitation method. The acetate-intercalated Ni–In LDHs exhibited swelling behavior in water and delaminated into semitransparent suspensions. All the results revealed the formation of unilamellar 2D nanosheets. This simple method did not need reflux at high temperatures, overcoming the deficiency of using organic solvents, resulting in a steady colloidal suspension of 2D nanosheets. The delaminated 2D nanosheets with positive charges showed an enhanced ability for DNA adsorption, compared with the unexfoliated bulk lamellar LDHs. The effect of the dosage on the adsorption of DNA and the adsorption isotherm were investigated. The appropriate nanosheet dosage was 5 mg for the initial DNA amount of 1000 μg. Both the Langmuir and Freundlich models described the equilibrium data well. The adsorbed DNA can be released by the addition of the salt solution. This work not only provides a simple approach to delaminate the Ni–In LDHs but also gives an excellent adsorbent for DNA adsorption.Acknowledgements
The authors are grateful to the National Nature Science Foundation of China (51077013), the Innovation Research Foundation of College Graduates of Jiangsu Province (CXLX12-0106), and the Fund Project for Transformation of Scientific and Technological Achievements of Jiangsu Province of China (BA2011086) for financial support.Notes and references
- R. Ma and T. Sasaki, Adv. Mater., 2010, 22, 5082–5104 CrossRef CAS PubMed.
- L. F. Nazar and A. J. Jacobson, J. Chem. Soc., Chem. Commun., 1986, 570–571 RSC.
- N. Yamamoto, T. Okuhara and T. Nakato, J. Mater. Chem., 2001, 11, 1858–1863 RSC.
- T. Sasaki, M. Watanabe, H. Hashizume, H. Yamada and H. Nakazawa, J. Am. Chem. Soc., 1996, 118, 8329–8335 CrossRef CAS.
- R. E. Schaak and T. E. Mallouk, Chem. Mater., 2002, 14, 1455–1471 CrossRef CAS.
- L. Chen, K. Sun, P. Li, X. Fan, J. Sun and S. Ai, Nanoscale, 2013, 5, 10982–10988 RSC.
- D. Pan, H. Zhang, T. Fan, J. Chen and X. Duan, Chem. Commun., 2011, 47, 908–910 RSC.
- R. Allmann, Acta Crystallogr., Sect. B: Struct. Sci., 1968, 24, 972–977 CrossRef CAS.
- W. Kagunya, Z. Hassan and W. Jones, Inorg. Chem., 1996, 35, 5970–5974 CrossRef CAS.
- C. Nyambo, P. Songtipya, E. Manias, M. M. Jimenez-Gasco and C. A. Wilkie, J. Mater. Chem., 2008, 18, 4827–4838 RSC.
- J.-H. Choy, J.-S. Jung, J.-M. Oh, M. Park, J. Jeong, Y.-K. Kang and O.-J. Han, Biomaterials, 2004, 25, 3059–3064 CrossRef CAS PubMed.
- P. Gunawan and R. Xu, Chem. Mater., 2009, 21, 781–783 CrossRef CAS.
- D. Buss, J. Bauer, W. Diembeck and O. Glemser, J. Chem. Soc., Chem. Commun., 1985, 81–82 RSC.
- C. Faure, C. Delmas and P. Willmann, J. Power Sources, 1991, 36, 497–506 CrossRef CAS.
- K. T. Ehlsissen, A. Delahaye-Vidal, P. Genin, M. Figlarz and P. Willmann, J. Mater. Chem., 1993, 3, 883–888 RSC.
- L. Indira, M. Dixit and P. V. Kamath, J. Power Sources, 1994, 52, 93–97 CrossRef CAS.
- K. Ralla, U. Sohling, K. Suck, F. Sander, C. Kasper, F. Ruf and T. Scheper, Colloids Surf., B, 2011, 87, 217–225 CrossRef CAS PubMed.
- L. Jin, D. He, Z. Li and M. Wei, Mater. Lett., 2012, 77, 67–70 CrossRef CAS PubMed.
- C.-H. Chan, J.-K. Chen and F.-C. Chang, Sens. Actuators, B, 2008, 133, 327–332 CrossRef CAS PubMed.
- K.-M. Kim, C.-B. Park, A.-J. Choi, J.-H. Choy and J.-M. Oh, Bull. Korean Chem. Soc., 2011, 32, 2217–2221 CrossRef CAS.
- R. Ma, Z. Liu, L. Li, N. Iyi and T. Sasaki, J. Mater. Chem., 2006, 16, 3809–3813 RSC.
- H. Kang, G. Huang, S. Ma, Y. Bai, H. Ma, Y. Li and X. Yang, J. Phys. Chem. C, 2009, 113, 9157–9163 CAS.
- T. Hibino and M. Kobayashi, J. Mater. Chem., 2005, 15, 653–656 RSC.
- P. P. Kumar, A. G. Kalinichev and R. J. Kirkpatrick, J. Phys. Chem. C, 2007, 111, 13517–13523 CAS.
- N. Iyi, Y. Ebina and T. Sasaki, Langmuir, 2008, 24, 5591–5598 CrossRef CAS PubMed.
- G. V. Manohara, P. Vishnu Kamath and W. Milius, J. Solid State Chem., 2012, 196, 356–361 CrossRef CAS PubMed.
- G. V. Manohara, D. A. Kunz, P. V. Kamath, W. Milius and J. Breu, Langmuir, 2010, 26, 15586–15591 CrossRef CAS PubMed.
- V. Prevot, C. Forano and J. P. Besse, Chem. Mater., 2005, 17, 6695–6701 CrossRef CAS.
- A. S. Prakash, P. V. Kamath and M. S. Hegde, Mater. Res. Bull., 2000, 35, 2189–2197 CrossRef CAS.
- B. E. Warren, Phys. Rev., 1941, 59, 693–698 CrossRef CAS.
- Y. Wang, Y. Zhou, T. Zhang, M. He, X. Bu and X. Yang, Appl. Surf. Sci., 2014, 288, 710–717 CrossRef CAS PubMed.
- A. Shukla, Y.-H. Zhang, P. Dubey, J. L. Margrave and S. S. Shukla, J. Hazard. Mater., 2002, 95, 137–152 CrossRef CAS.
- W. S. Wan Ngah and M. A. K. M. Hanafiah, Biochem. Eng. J., 2008, 39, 521–530 CrossRef CAS PubMed.
- M. Chabani, A. Amrane and A. Bensmaili, Chem. Eng. J., 2006, 125, 111–117 CrossRef CAS PubMed.
- I. Lozzi, L. Calamai, P. Fusi, M. Bosetto and G. Stotzky, Soil Biol. Biochem., 2001, 33, 1021–1028 CrossRef CAS.
- B. R. Young, W. G. Pitt and S. L. Cooper, J. Colloid Interface Sci., 1988, 124, 28–43 CrossRef CAS.
- J. C. Bellot and J. S. Condoret, Process Biochem., 1993, 28, 365–376 CrossRef CAS.
- A. Shimamura, M. Jones, E. Kanezaki and J. Metson, J. Mater. Sci., 2012, 47, 1142–1147 CrossRef CAS.
- A. Shimamura, M. I. Jones and J. B. Metson, J. Colloid Interface Sci., 2013, 411, 1–7 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: XRD data and unit cell parameters for the samples, FT-IR spectrum of the materials, photograph and zeta potential curve of the colloidal suspension of Ni–In ELDHs nanosheets and XRD pattern of DNA–LDHs complexes after adsorption. See DOI: 10.1039/c3ra47728b |
| This journal is © The Royal Society of Chemistry 2014 |