Exploring structural requirements of leads for improving activity and selectivity against CDK5/p25 in Alzheimer's disease: an in silico approach†
Abstract
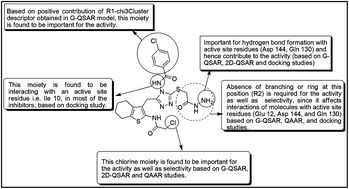
A congeneric series of 224 cyclin-dependant kinase 5/p25 (CDK5/p25) inhibitors was exploited to understand the structural requirements for improving activity against CDK5/p25 and selectivity over CDK2. The CDK5/p25 enzyme complex plays a significant role in the formation of neurofibrillary tangles in Alzheimer's disease. In the present study, 2D-quantitative structure–activity relationship (2D-QSAR), group or fragment based QSAR (G-QSAR), and quantitative activity–activity relationship (QAAR) models were developed and validated with satisfactory performance as evidenced from statistical metrics, indicating the reliability and robustness of the models. The 2D-QSAR and G-QSAR models explore the structural requirements for improving activity, while the QAAR model facilitates the better understanding of features required for selectivity of the inhibitors. The docking study further provides information regarding the key active site residues and structural features important for proper binding in the active site of the CDK5/p25 complex.
- This article is part of the themed collection: Towards understanding and treating Alzheimer’s disease

 Please wait while we load your content...
Please wait while we load your content...