Optimising selective excitation pulses to maximise saturation transfer difference NMR spectroscopy†
Nathan B. Ley,
Michelle L. Rowe,
Richard A. Williamson and
Mark J. Howard*
School of Biosciences, University of Kent, Canterbury CT2 7NJ, UK. E-mail: m.j.howard@kent.ac.uk; Tel: +44 (0)1227 3274730
First published on 8th January 2014
Abstract
A simple method is presented that optimizes the STD NMR Gaussian pulse to deliver significant increases in STD amplification factors with minimal perturbation of the ligand. This approach is practically demonstrated using the wheat-germ agglutinin/N-acetyl-D-glucosamine protein–ligand system.
Saturation transfer difference (STD) NMR is a popular method for identifying small ligand molecules that interact with a particular protein of interest1–6 and is used extensively to identify chemical fragments that bind to target biomolecules in drug discovery.7–14 However, more recently STD NMR has been used to investigate the binding mode of samples containing a single ligand from the outset or as a secondary screen and it is important to obtain optimum results for such experiments.15–18 1H STD NMR is initiated by the saturation of protein magnetization that is created using a specific shaped excitation pulse (Fig. 1).
Although the shaped excitation pulse initially saturates a small proportion of 1H, typically upfield methyl protons, this saturation disperses across the protein quickly via spin-diffusion to saturate many protons. 1H nuclei of any ligand interacting with the protein will also experience this saturation as it is transferred from protein to ligand. Any magnetization transferred to the ligand before it dissociates from the protein can be measured in a 1H NMR ligand spectrum as the difference between two NMR datasets; one where the protein is saturated (on or I) and the other when it is not (off or Io). The difference spectrum ISTD is simply defined as (I−Io). To compensate for indirect saturation effects, ‘on’ saturation is achieved by placing the pulse within the protein proton spectral envelope (ca. 0 ppm) and ‘off’ saturation is achieved by placing the same pulse distant from this envelope (ca. 30 ppm). It is worth noting that for STD NMR involving cells or virus-like particles, the ‘off’ saturation position has to be much further up- or downfield (ca. ±300 ppm) to prevent accidental excitation during the ‘off’ condition brought about by the very large molecular weight of these systems.19 Indirect effects are further minimized by interleaving ‘on’ and ‘off’ experiments and the data split into their respective spectra after acquisition. The saturation of the protein is typically achieved by repeatedly pulsing a shaped excitation pulse that is typically 20 to 50 milliseconds in length over a period of 1 to 10 seconds. The pulse is shaped in nature (e.g. Gaussian20 or E-burp21) to further limit its excitation profile and prevent accidental excitation of ligand. Gaussian or E-burp pulses are preferred to hard pulses for STD NMR because of the virtual absence of side-lobes and low excitation levels at large offsets from the pulse.3,20,21 It is crucial that saturation pulses are applied to provide efficient saturation of the protein without accidental excitation of ligand protons that can distort results.
We communicate here that Gaussian shaped excitation pulses can be shorter than 50 ms and rationally placed to minimise direct ligand excitation. This approach provides maximal saturation of the protein to deliver optimal STD amplification factors from any difference spectra obtained. This optimisation is demonstrated using the Gaussian pulse, as this is currently the most commonly used shaped-pulse for STD in both academia and industry. However, the approach describe herein could equally be applied to E-burp or other pulse schemes. Optimising any STD shaped-pulse will dramatically improve the sensitivity of STD NMR data and we can demonstrate +10-fold increases in amplification factor for the Gaussian pulse scheme. The demonstration datasets presented were obtained using a known STD NMR system of wheat-germ agglutinin (WGA) protein from Triticum vulgaris, with an N-acetyl-D-glucosamine (GlcNAc) ligand.4 All components were purchased from Sigma-Aldrich. Samples were prepared using 20 μM WGA, 1 mM GlcNAc in deuterium oxide corrected to pH 7.4 in a buffer of 10 mM sodium phosphate and 10 mM sodium chloride. 1 mM raffinose was used in addition to GlcNAc and WGA as a negative STD control for Fig. 2 but was omitted from all other experiments. All NMR experiments were run at 283 K using a Bruker AV3 600 MHz NMR spectrometer equipped with a QCI-F cryoprobe. Datasets were processed and analysed using Bruker Topspin 3.0 and 1H spectra were referenced to 4,4-dimethyl-4-silapentane-1-sulphonic acid (DSS). Shaped pulses were generated and optimized using Bruker Shape Tool. STD NMR datasets were obtained over 512 scans (256 scans ‘on’ and 256 scans ‘off’ saturation) with 2.5, 5, 10, 25 or 50 ms Gaussian shaped pulses and variable ‘on’ saturation positions but with ‘off’ saturation set to −30 ppm in all experiments. The GlcNAc methyl proton absolute intensity was obtained using MestReNova (Mnova) and used to calculate the STD amplification factor (STDamp) from STD difference spectra [ISTD = (I − Io)] and STD control spectra (Io) as previously described using the equation:3
| STDamp = (ISTD/I0) × ligand excess. |
Fig. 2 shows the STD NMR difference spectra for WGA/GlcNAc where the methyl protons of GlcNAc (at 1.8 ppm relative to DSS) provide a simple but robust system to monitor the effect of changing the Gaussian pulse length and ‘on’ saturation position for a constant saturation period of 2 seconds.
Fig. 3 demonstrates the effect on STD amplification factor when altering the on-resonance position with respect to the upfield methyl GlcNAc resonance and shortening the length of the Gaussian pulse. Our analysis concentrates on data using 2.5, 5 and 10 ms Gaussian pulses in preference to longer 20 and 50 ms pulses that have been used in many studies (e.g. see ref. 3, 15, 22–27). Inspection of Fig. 3 suggests that 2.5 ms Gaussian pulse placed at −1.8 ppm (1080 Hz) upfield from the ligand resonance provides the optimum STD result with an amplification factor 7 times greater than a 5 ms pulse and 19 times greater than a 10 ms pulse. This is not surprising and it has been noted previously that STD difference spectra display a high-dependence on the power level of the shaped pulse.21 The 2.5 ms Gaussian pulse surpasses 5 ms and 10 ms pulses over the entire on-resonance range in Fig. 3 and the measured amplification factor when using this pulse increases dramatically when it is applied with an offset below 2.8 ppm (1680 Hz) upfield from the GlcNAc methyl resonance.
 | ||
| Fig. 3 GlcNAc methyl 1H STD amplification factor in the presence of WGA for 2.5, 5 and 10 ms Gaussian pulses over a range of ‘on’ saturation points. The ‘on’ resonance position is shown as a ppm offset (600 MHz 1H) from the ligand resonance; e.g. an offset of −1.8 ppm is at 0 ppm. NMR spectra associated with these data are shown in ESI† Fig. S1–3. | ||
The increase observed by the 2.5 ms Gaussian pulse could be attributed to either accidental excitation of the free-ligand or excitation of the bound ligand resonance. The latter case is extremely unlikely because the GlcNAc/WGA system does not demonstrate large ligand shifts upon binding. The interpretation of accidental excitation with small on-resonances offsets for a 2.5 ms Gaussian pulse can be tested in two ways. First, by obtaining practical NMR Gaussian excitation profiles (Fig. 4) from a 2 seconds comb of 2.5 ms and 5.0 ms pulses. Second, by measuring a ‘virtual’ STD amplification factor for GlcNAc methyl resonances in the absence of the protein WGA for a range of on-resonances offsets. The second process would detect excitation of the ligand by a Gaussian pulse for a particular offset (Fig. 5). This accidental excitation would manifest as a non-zero STDamp value because STD ‘I’ and ‘Io’ experiments should yield the same result in the absence of protein. As ISTD = (I − Io), the difference spectrum should be blank and yield an STDamp value of zero. If a difference spectrum with signals is obtained, this is because ISTD > 0, therefore I ≠ Io and as no protein is present to facilitate saturation transfer, the I ≠ Io scenario has to be due to excitation by the on-resonance pulse. Ultimately, Fig. 5 demonstrates both correct (ISTD = 0) and incorrect (ISTD ≠ 0) results for this negative control experiment. The combination of Fig. 4 and 5 will allow us to identify offset values at which the excitation profile creates accidental excitation. This also provides the identification of a maximum allowable percentage excitation that would not create a false-positive spectrum.
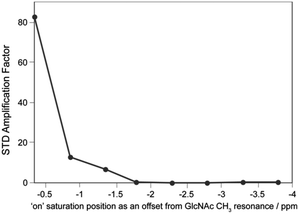 | ||
| Fig. 5 1H STD NMR amplification factor using a 2.5 ms Gaussian pulse over a range of ‘on’ saturation points for 1 mM GlcNAc control with no WGA protein present in the sample. ESI† Fig. S5 shows the GlcNAc methyl STD difference. | ||
Although spectrometer software enables users to evaluate shaped pulses for excitation and width, it was considered prudent to obtain practically obtained excitation profiles for 2 seconds trains of 2.5 ms and 5 ms Gaussian pulses. These profiles were produced using identical pulse trains that created the STD NMR data shown in Fig. 1–3 and 5. The standard approach to providing saturation for STD is to loop the pulse continually without any inter-pulse delay over the saturation period. Therefore, a 2 seconds saturation period would then use 400 5 ms pulses or 800 2.5 ms pulses. Measuring the residual 1H resonance (HDO) in 2H2O provides a single resonance with a narrow half peak height below 0.003 ppm (2 Hz). This minimises potential measurement discrepancies caused by B1 inhomogeneity that can distort the efficiency of composite pulses. Profiles were created by sweeping the carrier frequency in a pulse-acquire experiment containing the shaped pulse of interest and measuring the signal intensity from the residual 1H resonance.
Fig. 4 demonstrates the expected result of shortening the Gaussian pulse length to create a wider excitation profile. In addition, Fig. 4 illustrates the profile obtained from a 2 seconds train of pulses is significantly different from a single Gaussian pulse as shown in ESI† Fig. S4. The pulse train profiles confirm a 2 ppm offset is sufficient for a 5 ms Gaussian pulse but a 2.5 ms Gaussian pulse should be placed at least 2.5 ppm from the nearest resonance to prevent accidental excitation. Referring to Fig. 3 suggests STD performed with a 5 ms Gaussian pulse rather than a 10 ms pulse provides ca. 3-fold increase in STDamp for a 2.0 ppm offset. Equally, Fig. 3 also informs that STD experiments using a 2.5 ms Gaussian pulse in preference to 10 ms provides ca. 15-fold increase in STDamp when using a 2.5 ppm offset. These data confirm the merit of using shorter Gaussian pulses that are optimally positioned with respect to the closest ligand resonance.
The effect of significant sidebands that occur away from the pulse centre was investigated using a control experiment where no WGA protein was added to the sample (Fig. 5). Any significant sideband was defined for where the intensity was greater 1% of the maximum; this dictates the farthest significant side band for the 2.5 ms Gaussian pulse at ± 2.0 ppm. Fig. 5 displays the STDamp results of a control experiment without protein where STD signal must be due to excitation of the ligand by the 2.5 ms Gaussian shaped pulse in the on-resonance position. The STDamp values fall to zero when the 2.5 ms pulse is placed at offsets greater than or equal to 1.8 ppm and Fig. 4 confirms the 1.8 ppm position as providing a ‘valley’ in the excitation profile in between two excitation bands at offsets of 2.0 ppm and 1.4 ppm. Interestingly, despite 2.5 ms Gaussian excitation sideband at 1.4 ppm delivering over 85% of the maximum excitation intensity, the equivalent point provided a modest STDamp of less than 10 in Fig. 5. In contrast, the sideband at 0.6 ppm that defines the 100% profile intensity point is responsible for a much higher control STDamp. This suggests that although Gaussian side bands are present in pulse trains, their influence on saturation and creating accidental excitation in STD could be limited. Furthermore, the excitation sideband at 2.0 ppm appears not to provide any control amplification value at the same location in Fig. 5 and suggests this small sideband does not significantly excite the ligand resonance. The 1.8 ppm and 2.0 ppm offsets in Fig. 4 correlate to 1.5% and 6.4% of the maximum excitation for a 2.5 ms Gaussian pulse train. As both of these offsets provide zero STDamp in the control experiment (Fig. 5), they must be below the lower excitation limit where false-positive STD data could occur in a 256-scan STD NMR experiment.
The ‘on’ saturation shaped pulse is usually positioned around 0 ppm to excite protons of upshifted methyl groups within the protein. The efficiency of protons excitation will clearly influence saturation of the protein and transfer to the ligand. Optimal positioning of the shaped excitation pulse with respect to protein methyl protons will boost efficiency. Fig. 3 demonstrates this effect through the observed increase in STD amplification factor as the shaped-pulse offset is reduced. Therefore, the protein target used in STD NMR experiments has an influence on the optimization of the shape pulse and when methyl protons are significantly upshifted they can be excited for saturation by a shaped pulse with a larger offset. This effect is further accentuated for larger proteins with greater numbers of methyl groups and additional dipolar line broadening to extend the protein excitation envelope. Therefore, Fig. 3 is specific to the WGA/GlcNAc system and STD optimisation should be considered for any new target-protein ligand system, particularly if using quantitative STD NMR.16,26 STD optimisation curves, as in Fig. 3, provide cursory identification of direct on-resonance ligand excitation as a significant increase in observed STD amplification factor. However, a control STD curve, such as that shown in Fig. 5, provides the ultimate assessment and confirms the offsets where excitation is avoided when protein is absent. In our WGA/GlcNAc system, Fig. 5 also confirms that an offset of −1.8 ppm from the ligand reference is safe for 256 scan STD experiments. It is important to reiterate the scan dependence on this information and it is crucial that scan number is identical when creating curves equivalent to Fig. 3 and 5.
Conclusions
Our 14.1 T based study concluded that for 256-scan ‘on’ saturation STD experiments, a 2.5 ms Gaussian pulse can be placed as near as −1.8 ppm upfield from the closest ligand 1H resonance to provide a 19-fold improvement in STDamp compared to a 10 ms Gaussian pulse train. However, 5 ms Gaussian pulse still delivers a 3-fold improvement over the 10 ms pulse should the user want to exercise caution. The overall message is that Gaussian pulses within a 2 seconds pulse train can be shortened to provide significant enhancements in STD NMR. This approach would be beneficial when optimising STD NMR for single ligand/target systems or when applying quantitative analysis.1H STD NMR can be optimized by using short Gaussian shaped pulses that are rationally placed at relatively short offset distances from the closest ligand resonance. Our example measured ligand STDamp values over a range of offsets for the ligand in the presence and absence of protein to identify the optimum offset condition. The increased efficiency in saturating the protein was due to a wider-targeted Gaussian pulse that excites a larger population of upshifted methyl groups in the protein. This does suggest that the widespread use of 20 and 50 ms Gaussian pulses in STD NMR to be disadvantageous and the application of shorter pulses can be evaluated easily using the methods described. The shaped pulse length is an experimental parameter that can be easily modified within modern spectrometers and the power level adjustment required is easily obtained using spectrometer manufacturer software tools within the acquisition software. This approach need not be limited to Gaussian pulses and our method could be utilized with any STD shaped pulse configuration (e.g. E-BURP 90° pulses).
Acknowledgements
We would like to thank the BBSRC for the allocation of a CASE studentship (BB/F016719/1) to NBL and MJH and RAW would also like to thank the Wellcome Trust for equipment award 091163/Z/10/Z.Notes and references
- M. Mayer and B. Meyer, J. Am. Chem. Soc., 2001, 123, 6108–6117 CrossRef CAS PubMed.
- M. Mayer and B. Meyer, Angew. Chem., Int. Ed., 1999, 38, 1784–1788 CrossRef CAS.
- B. Meyer and T. Peters, Angew. Chem., Int. Ed. Engl., 2003, 42, 864–890 CrossRef CAS PubMed.
- M. Mayer and B. Meyer, Angew. Chem., Int. Ed., 1999, 38, 1784–1788 CrossRef CAS.
- J. L. Wagstaff, S. L. Taylor and M. J. Howard, Mol. Biosyst., 2013, 9, 571–577 RSC.
- T. Biet and T. Peters, Angew. Chem., Int. Ed., 2001, 40, 4189–4192 CrossRef CAS.
- B. J. Stockman and C. Dalvit, Prog. Nucl. Magn. Reson. Spectrosc., 2002, 41, 187–231 CrossRef CAS.
- H. Jhoti, A. Cleasby, M. Verdonk and G. Williams, Curr. Opin. Chem. Biol., 2007, 11, 485–493 CrossRef CAS PubMed.
- C. A. Lepre, J. M. Moore and J. W. Peng, Chem. Rev., 2004, 104, 3641–3675 CrossRef CAS PubMed.
- J. Moore, N. Abdul-Manan, J. Fejzo, M. Jacobs, C. Lepre, J. Peng and X. L. Xie, J. Synchrotron Radiat., 2004, 11, 97–100 CrossRef CAS PubMed.
- M. Pellecchia, I. Bertini, D. Cowburn, C. Dalvit, E. Giralt, W. Jahnke, T. L. James, S. W. Homans, H. Kessler, C. Luchinat, B. Meyer, H. Oschkinat, J. Peng, H. Schwalbe and G. Siegal, Nat. Rev. Drug Discovery, 2008, 7, 738–745 CrossRef CAS PubMed.
- J. W. Peng, J. Moore and N. Abdul-Manan, Prog. Nucl. Magn. Reson. Spectrosc., 2004, 44, 225–256 CrossRef CAS PubMed.
- L. O. Sillerud and R. S. Larson, Methods Mol. Biol., 2006, 316, 227–289 Search PubMed.
- D. Wishart, Curr. Pharm. Biotechnol., 2005, 6, 105–120 CAS.
- D. Dicara, C. Rapisarda, J. L. Sutcliffe, S. M. Violette, P. H. Weinreb, I. R. Hart, M. J. Howard and J. F. Marshall, J. Biol. Chem., 2007, 282, 9657–9665 CrossRef CAS PubMed.
- S. Kemper, M. K. Patel, J. C. Errey, B. G. Davis, J. A. Jones and T. D. Claridge, J. Magn. Reson., 2010, 203, 1–10 CrossRef CAS PubMed.
- J. L. Wagstaff, S. Vallath, J. F. Marshall, R. A. Williamson and M. J. Howard, Chem. Commun., 2010, 46, 7533–7535 RSC.
- D. W. Begley, S. X. Zheng and G. Varani, Chem. Biol. Drug Des., 2010, 76, 218–233 CAS.
- C. Rademacher, N. R. Krishna, M. Palcic, F. Parra and T. Peters, J. Am. Chem. Soc., 2008, 130, 3669–3675 CrossRef CAS PubMed.
- R. Freeman, Prog. Nucl. Magn. Reson. Spectrosc., 1998, 32, 59–106 CrossRef CAS.
- B. Cutting, S. V. Shelke, Z. Dragic, B. Wagner, H. Gathje, S. Kelm and B. Ernst, Magn. Reson. Chem., 2007, 45, 720–724 CrossRef CAS PubMed.
- B. Meyer, J. Klein, M. Mayer, R. Meinecke, H. Moller, A. Neffe, O. Schuster, J. Wulfken, Y. Ding, O. Knaie, J. Labbe, M. M. Palcic, O. Hindsgaul, B. Wagner and B. Ernst, Ernst Schering Res. Found. Workshop, 2004, 149–167 CAS.
- B. Meyer, T. Weimar and T. Peters, Eur. J. Biochem., 1997, 246, 705–709 CAS.
- J. Angulo, P. M. Enriquez-Navas and P. M. Nieto, Chem.-Eur. J., 2010, 16, 7803–7812 CrossRef CAS PubMed.
- J. Angulo, B. Langpap, A. Blume, T. Biet, B. Meyer, N. R. Krishna, H. Peters, M. M. Palcic and T. Peters, J. Am. Chem. Soc., 2006, 128, 13529–13538 CrossRef CAS PubMed.
- J. Angulo and P. M. Nieto, Eur. Biophys. J., 2011, 40, 1357–1369 CrossRef CAS PubMed.
- J. Angulo, C. Rademacher, T. Biet, A. J. Benie, A. Blume, H. Peters, M. Palcic, F. Parra and T. Peters, Methods Enzymol., 2006, 416, 12–30 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ra46246c |
| This journal is © The Royal Society of Chemistry 2014 |



