Distribution of label free cationic polymer-coated gold nanorods in live macrophage cells reveals formation of groups of intracellular SERS signals of probe nanoparticles†
Abstract
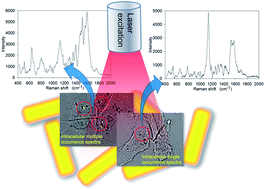
Gold nanorods coated with poly(diallydimethylammonium chloride) (PDAC-GNRs) were used to observe the distribution of surface-enhanced Raman signals in live cells and generate distinct groups of surface-enhanced Raman scattering (SERS) spectra in different regions of the cells. Spectra with unique features were clustered into sets of molecules in live murine macrophage cells (Raw 264.7). The distribution of biological substances detected by SERS signals of PDAC-GNRs is also discussed.

 Please wait while we load your content...
Please wait while we load your content...