Yan Fu, Xiaoli Duan, Xiongfei Chen, Jinli Zhang and Wei Li
RSC Adv., 2014,4, 1329-1333
DOI:
10.1039/C3RA43251C,
Paper
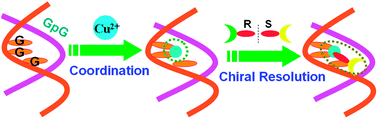
The DNA-based selector for discriminating chiral ofloxacin with high enantioselectivity and affinity is constructed through Cu(II)-coordination with G-rich duplex containing successive guanines. Using this chiral selector, R- and S-ofloxacin can be directly enriched from the racemate, with the enantiomeric excess of 85% (R) and 78% (S) individually by three operational stages.