 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Aryl ethynyl anthraquinones: a useful platform for targeting telomeric G-quadruplex structures†
Claudia
Percivalle
a,
Claudia
Sissi
b,
Maria Laura
Greco
b,
Caterina
Musetti
b,
Angelica
Mariani
a,
Anna
Artese
c,
Giosuè
Costa
c,
Maria Lucia
Perrore
a,
Stefano
Alcaro
c and
Mauro
Freccero
*a
aDipartimento di Chimica, Università di Pavia, V.le Taramelli 10, 27100, Pavia, Italy. E-mail: claudia.percivalle01@universitadipavia.it; angy88@tiscali.it; marialucia.perrone01@universitadipavia.it; mauro.freccero@unipv.it
bDepartment of Pharmaceutical and Pharmacological Sciences, v. Marzolo 5, 35131 Padova, Italy. E-mail: claudia.sissi@unipd.it; marialaura.greco@studenti.unipd.it; caterina.musetti@gmail.com
cDipartimento di Scienze della Salute, Universitá di Catanzaro, Campus “Salvatore Venuta”, Viale Europa, 88100 Catanzaro, Italy. E-mail: alcaro@unicz.it; artese@unicz.it; gcosta@unicz.it
First published on 4th April 2014
Abstract
Aryl ethynyl anthraquinones have been synthesized by Sonogashira cross-coupling and evaluated as telomeric G-quadruplex ligands, by the FRET melting assay, circular dichroism, the DNA synthesis arrest assay and molecular docking. Both the binding properties and G-quadruplex vs. duplex selectivity are controlled by the structures of the aryl ethynyl moieties.
Introduction
Small molecule-mediated DNA targeting represents one of the most effective approaches for the development of chemotherapeutics. The ability of DNA to fold into highly stable secondary structures could be exploited for the design of anticancer agents interacting with nucleic acids in a sequence or structural selective fashion.1,2 One such target is represented by G-quadruplex (G4) DNA and RNA motifs.3,4 G4 is a four-stranded nucleic acid structure that can be formed in guanine-rich nucleic acid sequences via Hoogsten hydrogen bond formation and cation coordination.5 G4 stabilization has been proposed to interfere with important biological processes for cellular homeostasis, such as DNA damage response activation,6–8 oncogene expression3,9–11 and genomic stability.12 Putative quadruplex sequences (PQS) are highly spread in the genome13 and transcriptome,14 including gene promoter regions or gene bodies and telomeres, providing these structures with the potential to act as regulatory elements of different processes. A general lack of evidence of the formation and the existence of G4 in vivo and its real biological functions made the G4 relevance as therapeutic targets controversial.15 However, the existence of G4 structures in cells has recently been demonstrated by means of immuno-fluorescence staining with an engineered structure-specific antibody16 and by Chromatin Immuno-Precipitation (ChIP-Seq).7 The potential therapeutic opportunities offered by the targeting of these structures prompted the design of a large number of ligands that specifically interact with the terminal tetrads, G4 loops and grooves.17 In the last two decades, several selective G4 ligands have been reported and in most of the cases they share a large planar aromatic surface that provide the ligands with a π-stacking surface for binding with the external tetrads of the G4.17,18 Cationic side chains, at physiological pH, further increase the ligand binding properties, providing an additional electrostatic interaction with the phosphate backbone.17 Dissecting the function of a specific G4 family over the others could be achieved by developing small molecule ligands that can discriminate not only over duplex DNA but also over different G4 architectures.19 The development of these compounds would provide unprecedented tools to analyze cells for the functions of G4s present in a specific genomic region.Anthraquinone derivatives (AQs) represent an interesting scaffold to develop selective and multifunctional G4 ligands, with many potential applications, because of their well characterised DNA-binding properties,20,21 fairly low redox potential and their ability to act as photosensitizers by one-electron oxidation.22 Structurally, AQs are strictly related to the anthracycline antibiotics like doxorubicin and daunomycin.23–26
It has been shown that doxorubicin and daunomycin can interact with telomeric DNA via G4 stabilization, mediated by the anthraquinone scaffold and demonstrated by the crystal structure of a complex between the telomeric G4 DNA and daunomycin.27 With the aim to optimize G4 recognition the synthesis of several 1,4-1,5-1,8-2,6- and 2,7-difunctionalized amidoanthracene-9,10-diones has been performed and the resulting compounds have been tested as G4 ligands.28–31 The five different regio-isomers showed different abilities to recognize G4 telomeric structures according to the nature and the position of the substituent side chains. Consistently, conjugation of the anthraquinone core with amino sugars32 or amino acids33,34 was applied to modulate their G4 binding properties. One such example is a neomycin–anthraquinone conjugate that exhibits a nanomolar affinity for telomeric G4 DNA, which is 1000-fold higher when compared to its constituent units.32 This higher affinity is ascribed to the dual binding mode of the conjugate which can interact with the grooves (neomycin) and with the guanines of the G4 (anthraquinone) via π-stacking interactions.
For AQ-amino acid conjugates, the combination of a basic amino acid (Lys) with a more hydrophobic residue (Phe) has provided a better G4 selectivity versus the duplex DNA.34
Unlike the large majority of G4 ligands, AQs exhibit interesting redox properties, as they easily generate radical anions and di-anions by bio-compatible reduction.35 We demonstrated that formation of stable radical anions can be exploited to generate alkylating agents such as Quinone Methides (QMs, generated from o- or p-benzyl substituted phenols).36,37 We anticipate that AQs could be similarly exploited for in situ generation of QMs at G4 sites, thus enabling G4 covalent targeting.38 Alkylation has been proposed as an alternative approach to physically lock the DNA G4 in its folded conformation, enabling the investigation of the biological implications associated with G4s stabilization.38–42
Effective electronic conjugation between the AQ core and the aryl moiety (embedding the QM precursor) will ensure generation of the alkylating QM under reductive conditions. Moreover, we postulated that increasing the aromatic surface and the structural constraints by introducing aryl side chains would have been beneficial for both the AQs G4 binding properties and for G4 vs. duplex selectivity. Therefore, we explored conjugation of suitable QM precursors to the AQ scaffold introducing ethynyl spacers by means of the Sonogashira cross-coupling. Our synthetic effort resulted in a small library of aryl ethynyl anthraquinones (AQs, 1–6, Scheme 1). It involved the symmetric functionalization of the anthraquinone core at 2 and 7 positions with chemically diverse aryl moieties, such as negatively charged phenolates arising from 1 and 2, positively charged secondary amines (3,4) and zwitterionic Mannich bases (5,6).
Then, all the ligands here synthesized have been tested for their G4 binding properties in comparison with double stranded DNA. As G4 model sequences, we selected the human telomeric DNA. Telomeres consist of a hexameric nucleotide repeat unit d(TTAGGG) and several four repeat sequences are currently extensively used as mimics, since they allow exploration of different G4 conformations.
Due to the differential binding properties observed on the selected DNA substrates, the chemical versatility of the Sonogashira cross-coupling involved in the preparation of these compounds, and their redox properties,35 we believe that our aryl ethynyl anthraquinones represent a promising platform for the development of a new generation of multifunctional G4 interacting ligands.
Results and discussion
Chemistry
Final products 1–6 were synthesized starting from the commercially available 1,8-dihydroxyanthraquinone 7. The bromination reaction was performed at r.t. using NBS in dichloromethane and NH(iPr)2. The high reactivity of the hydroxyanthraquinone under these conditions resulted in a poorly selective bromination of 7, affording the anthraquinone 8 (Scheme 2) as the major product of the mixture (30% yield).The structure of the most abundant stereoisomer 8 has been tentatively assigned according to literature data, which suggest that direct bromination of the unprotected 1,8-dihydroxyanthraquinone such as aloe-emodin and chrysophanol analogues takes place at the desired 2- and 7-positions (see Scheme 2 for numbering), in the presence of a catalytic amount of a secondary amine.43–45 The presence of the two OH groups is not compatible with the Sonogashira cross-coupling reaction, therefore these groups were protected as methyl ethers. This step was conducted directly on the bromination crude, which was extremely challenging to purify under standard chromatographic conditions. This crude was suspended in acetone and heated to reflux overnight in the presence of dimethyl sulfate and K2CO3. The chromatographic purification of the resulting dimethyl ether 9 was much more efficient (70%) and straightforward than 8. Unfortunately, the unambiguous assignment of the corrected bromination regioselectivity could not be achieved by NMR through 1H, 13C-HMBC (Heteronuclear Multiple Bond Correlation) experiments as the chemical shifts of the two carbonyls were too close to each other (180.9 vs. 181.8 ppm). Therefore, such a task was carried out on the further synthesized anthraquinone 11. A first Sonogashira cross-coupling was conducted with compound 9 in the presence of a large excess (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) of trimethylsilylacetylene (TMSA) in anhydrous THF, TEA and Pd(PPh3)2Cl2 and CuI (20 mol% each). These reaction conditions provided 10 in reasonably good yield (60%). The yield of this key step was significantly affected by the sequence of the reactants addition. Adding TMSA immediately after TEA was found to provide the best reaction yield (60%). Deprotection of TMS groups was achieved quantitatively using K2CO3 in MeOH–DCM at 0 °C, affording the bis-terminal alkyne 11, which was used without further purification. This synthetic strategy provided a facile route to the synthesis of the building block 11 in only 4 steps and fairly good yields. The functionalization of the anthraquinone core at 2- and 7-positions was finally and unambiguously assigned using HMBC interactions of the terminal alkyne hydrogens (3.56 ppm) with the quaternary carbons on the methoxy substituents (C-1 and C-8, 161.7 ppm). In addition, both H-4 and H-5 exhibit the HMBC interaction with the most de-shielded carbonyl C-10 (ESI†).
1) of trimethylsilylacetylene (TMSA) in anhydrous THF, TEA and Pd(PPh3)2Cl2 and CuI (20 mol% each). These reaction conditions provided 10 in reasonably good yield (60%). The yield of this key step was significantly affected by the sequence of the reactants addition. Adding TMSA immediately after TEA was found to provide the best reaction yield (60%). Deprotection of TMS groups was achieved quantitatively using K2CO3 in MeOH–DCM at 0 °C, affording the bis-terminal alkyne 11, which was used without further purification. This synthetic strategy provided a facile route to the synthesis of the building block 11 in only 4 steps and fairly good yields. The functionalization of the anthraquinone core at 2- and 7-positions was finally and unambiguously assigned using HMBC interactions of the terminal alkyne hydrogens (3.56 ppm) with the quaternary carbons on the methoxy substituents (C-1 and C-8, 161.7 ppm). In addition, both H-4 and H-5 exhibit the HMBC interaction with the most de-shielded carbonyl C-10 (ESI†).
With this building block in hand we sought to investigate convenient synthetic strategies for the preparation of the final products: 1–6. The aryliodides (14–19, Table 1) required for the Sonogashira reaction were synthesized starting from the p- and m-iodophenol (ESI, Scheme S1†). For the synthesis of compounds 1 and 2 a protection/deprotection strategy of the phenol was required, as expected, while for the Mannich base the cross-coupling could be carried out using the free phenol derivatives (18, 19). Such an unexpected difference may be related to the formation of an intramolecular H-bond within the Mannich bases 18 and 19, which could introduce a sort of “self-protective” effect on the phenol OH acidity, recovering the typical reactivity of an “OH-free” aryl iodide.46 The yields of the Sonogashira coupling with Mannich bases (18, 19) are still lower compared to the reaction with iodoacetylphenol (14, 15), but the opportunity to avoid the protection and deprotection steps justified somehow the choice of our synthetic strategy.
The Sonogashira coupling conditions between the bis-terminal alkyne 11 and the aryl iodides have been optimized with respect to the solvent, base and catalysts (THF anhydrous, TEA, Pd(PPh3)2Cl2, CuI) for each single aryliodide substrate (see the Experimental section for the procedure and yields). The final products 3–6 were purified as bis-hydrochloride salts by reverse phase HPLC followed by trifluoroacetate/chloride exchange. Compounds 12 and 13 required an additional deprotection step, which was performed in aqueous methanol in the presence of K2CO3 at r.t. HPLC purification afforded the final products 1 and 2 (ESI, Scheme S2†).
The new ligands 1–6 were characterized by absorption spectroscopy and their molar extinction coefficients were calculated in 10 mM Tris, 50 mM KCl at pH 7.5 (Experimental section).
The cationic 3–6 showed a linear correlation between absorption and concentration of up to 50 μM thus confirming good solubility and the lack of extensive aggregation. The only exception was provided by derivatives 1 and 2, which evidenced a relevant deviation starting from the 15 μM ligand concentration followed by precipitation at higher levels. Therefore, all the subsequent analyses for these two derivatives were performed at concentrations lower than 15 μM.
Fluorescence melting assay
To assess the potential of the AQs 1–6 to stabilize peculiar G4 topologies we screened them by fluorescence melting using DNA telomeric sequences properly labelled at the 5′-end with a quencher (dabcyl) and at the 3′-end with a fluorophore (fluorescein).47 An increase of the oligonucleotide melting temperature upon addition of the tested compound relies on the ability of the ligands to stabilize the DNA G4 folded structure. Since the human telomeric G4 is characterized by a large conformational flexibility, the analysis was performed under different conditions and with different sequences known to promote distinct folding: HTS (d[AG3(T2AG3)3T]) which in the presence of K+ folds mainly in a population of prevalently hybrid conformations, whereas in Na+ it assumes a defined antiparallel folding,48 and Tel24 [d(T2AG3)4] which adopts a hybrid-1 folding in K+ containing solutions.49 The same analysis was additionally performed using a double stranded random sequence (dsDNA) to check for duplex vs. quadruplex selectivity.To summarize our results we report the variation of the oligonucleotide melting temperature as a function of ligand concentration (Fig. 1).
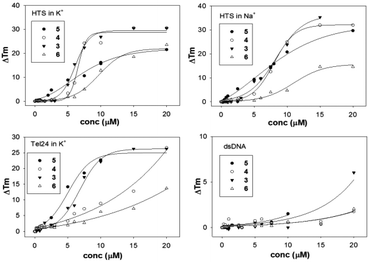 | ||
| Fig. 1 Variation of the melting temperature of HTS, Tel24 and double stranded DNA (dsDNA) by increasing concentrations of the tested ligands (3–6) in K+ or Na+ containing buffer. | ||
Among the tested ligands only 1 and 2 did not induce any modification of the melting profile of the tested DNA sequences. This sustained the fundamental requirement of protonable groups in the side chain to grant effective nucleic acid recognition. Although the ΔTm values remain quite low at 1 μM ligand concentration, all the other compounds (AQs 3–6) stabilized the G4 forms. In particular, a sigmoidal correlation emerged between the observed ΔTm and ligand concentrations, which suggested the presence of cooperative binding events. Among the active derivatives, at low micromolar ligand concentration, 5 and 6 turned out to be the most and the least effective, respectively. Conversely, at concentrations higher than 5 μM 3 and 4 behaved as better ligands for Tel22 in K+. If we compare the behavior of each anthraquinone derivative for the different tested G4 targets, we did not observe prominent selectivity for any of them. The only exception was 4, which was less active on Tel24. Thus, the presence of oxygen in the meta position on the aromatic ring of the side chains seems to negatively perturb the DNA recognition process. Interestingly, the regio-isomers 5 and 6 showed very different binding profiles and this can suggest a peculiar binding interaction for the para isomer 5 with the G4 structures. Finally, all tested compounds almost did not affect the thermal stability of the double stranded DNA (Fig. 1), indicating their use as potential G4 selective binders.
Circular dichroism
CD titrations were performed to investigate the ability of the novel compounds to induce structural modifications to the tested G4. Thus, the study was performed using the same oligonucleotide sequences used for thermal stabilization experiments. Moreover, we extended our analysis to wtTel26: d[(T2AG3)4T2] which, in the presence of K+, folds into a hybrid-2 type of arrangement.50The recorded dichroic spectra of the oligonucleotides in the presence of potassium are all characterized by two positive bands, one centered at 290 nm and the other at 265–268 nm which reflect the principal 3 + 1 hybrid arrangement assumed. Conversely, in the presence of sodium, the dichroic spectrum of Tel22 shows a negative band at 260 nm and a positive band at 290 nm, typical of the antiparallel conformation signature identified by NMR spectroscopy.51
Variations of the intensity of the dichroic features of all tested G4 folded DNA sequences were detected upon addition of the ligands (Fig. 2).
This confirmed the occurrence of a DNA–ligand interaction, which does not affect the G4 topology to a large extent. Interestingly, the most relevant CD variations occurred generally with 5. In this instance, induced dichroic bands (ICD) in the ligand absorption range were also observed (Fig. 2). Such contribution should derive from the insertion of the ligand chromophore into the chiral environment provided by the nucleic acid. Since this effect is a function of the mutual orientation of the AQ chromophore and DNA, we can assume that its presence/lack among the tested derivatives is linked to a significant repositioning of the ligand in the complex as a consequence of the side chain nature and position. Thus, distinct binding modes for 5vs.3, 4 and 6 can be further inferred.
When the DNA substrate was arranged into a double helix, the most prominent effect was reduction of the 275 and 245 nm DNA dichroic bands. This should exclude the occurrence of an efficient intercalation binding mode for these ligands, since this process usually causes an increment of these optical contributions. This result is in agreement with the above reported lack of thermal stabilization induced by the tested ligands in this nucleic acid conformation.
Enzymatic assays
The above described results were collected to evaluate the recognition of a G4 structure by the tested ligands. Additionally, we analyzed whether they can promote G4 folding generating species sufficiently stable to interfere with enzymes devoted to the processing of the nucleic acid. Thus, the DNA polymerase stop assay was performed using a template containing a four-repeat human telomeric sequence (HT4-temp). In a typical experiment, if the compounds under investigation promote G4 formation by the template, the formation of truncated products due to the collision of polymerase with the folded G-rich tract (see the cartoon on the right side of gel reported in Fig. 3) appears.52The results summarized in Fig. 3 showed that increasing concentrations of the tested ligands lead to a slight decrease of the intensity of the band relative to the fully processed oligonucleotide, which is more evident for 5. At the same time, the tested compounds blocked, by some other means, the primer extension by DNA polymerase starting from 5 to 10 μM, the same concentration range in which they stabilized the G-quadruplex as determined by the melting assay. The observed stop occurs at a well-defined site corresponding to the template G-rich stretch. Interestingly, 3 and 6 tend to arrest the enzyme at position-1 with reference to the G-rich tract at 5 μM concentration. This behavior is not shared by 5, thus further sustaining the different binding mode of this derivative.
Conversely, the same reaction performed on a DNA sequence not G-rich (HT4sc-temp) failed to evidence any interference in the enzymatic activity by tested ligands up to 40 μM. This result correlates with the increased ability of the tested compound to recognize G4 over other nucleic acid arrangements.
Evaluation of the best fitting ligand by docking
The conformational polymorphism of the DNA human telomeric repeat sequence Tel22 prompted us to generate poses of our anthraquinone derivatives using Induced Fit Docking (IFD)53 simulations following our recent experience,54 in order to take into account the target flexibility and to optimize the network of DNA–ligand interactions as compared to rigid docking. In particular, as already reported in our recent modeling work,55 we included in our study the Protein Data Bank [The Research Collaboratory for Structural Bioinformatics (RCSB) Protein Data Bank (PDB); http://www.rcsb.org/pdb] entries with the codes 1KF1,56143D,512HY9,572JPZ,582JSL59 and 2JSM59 taking into account all available X-ray and NMR telomeric structures. The binding energy (IFD score) related to the docking generated ensembles indicated a different theoretical ligand affinity toward the six used G4 folds (Table S1†).Recent studies indicated the hybrid-type intramolecular G4 structures as the major conformations formed in human telomeric sequences in K+ containing solution, with a dynamic equilibrium between hybrid-1 and hybrid-2 folds.57,58,60–63 However in K+ solution the parallel structure is also found. Remarkably, we obtained better docking results with these G4 folds. These data are in agreement with the human telomeric stabilization reported in Fig. 1 which highlighted a favourable contribution of K+ in G4 stabilization by tested ligands. Since this cation is much more abundant than Na+ in cellular environments, such a finding highlights the preference toward the physiologically relevant G4 conformations.
Among the analyzed compounds, 5 showed the best average affinity (consensus score, Table 2) with respect to the others, in particular against 1KF1, 2JSM and 2JSL models (Table S1†).
| AQ | Consensus score (kcal mol−1) |
|---|---|
| 1 | −6.42 |
| 2 | −6.79 |
| 3 | −8.84 |
| 4 | −9.01 |
| 5 | −9.29 |
| 6 | −9.00 |
Interestingly, solution studies evidenced a striking difference between the two regio-isomers 5 and 6. This experimental observation is in agreement with our theoretical results, since 5 showed an improved affinity compared to 6 in almost all the considered folds. Such a finding is particularly evident in the recognition of these ligands towards the G4 2JSM hybrid-1 model, as indicated in Fig. 4. Specifically 5 was better embedded in the DNA structure, since it is accommodated in a kind of internal pocket and is involved in a wide stacking interaction network. By contrast 6 is able to recognize only the bottom site of the 2JSM model, probably due to the different position of the phenolic hydroxyl moiety.
 | ||
| Fig. 4 Best pose of (A) 5 and (B) 6 against 2JSM hybrid-1 model of the DNA human telomeric repeat sequence d[AG3(T2AG3)3]. 5 and 6 are indicated as green carbon stick representation, while the DNA is shown as transparent surface. Nonpolar hydrogen atoms are omitted for sake of clarity. | ||
Moreover IFD simulations revealed the ability of 5 to establish one pivotal hydrogen bond between its hydroxyl group with the phosphate oxygen of guanine at position 9 and another one between the hydrogen atom of its amino moiety with the phosphate oxygen of guanine at position 2, thus allowing the ligand to better anchor to the G4 structure.
The best poses of the studied anthraquinone derivatives in complex with all the G4 considered folds are reported in the ESI (Fig. S2–S35†).
Conclusions
In conclusion, we reported the synthesis of several aryl ethynyl anthraquinones (1–6) via the optimized Sonogashira cross-couplings. This synthetic protocol is flexible and can be exploited to introduce further chemical diversity into the ethynyl-AQ scaffold. The ethynyl-AQ derivatives demonstrated to interact and stabilize G4 structures of the telomeric DNA sequence. Their binding properties and quadruplex vs. duplex selectivity have been characterized by FRET melting, CD, stop assay as well as IF docking experiments. Compound 5 was the most effective ligand according to all of the assays performed. Varying the relative position of the substituents on the phenolic aromatic ring from para to ortho (5 and 6, respectively) provided compounds with quite different G4 binding and stabilization properties. Our investigation highlights that structural positioning of Mannich bases is crucial for efficient G4 binding. Although our best candidate (5) is not the most efficient G4 binder among the AQ derivatives tested so far,28–31 it lacks significant intercalation into the double helix. This finding highlights the potential to exploit 5 and its analogues as precursors of alkylating QMs, targeting G4s. Although further structural refinement is required to increase the affinity of this scaffold towards telomeric G4-DNA, these preliminary results are encouraging. We are currently working on the development of a larger aryl (QM precursor) ethynyl anthraquinone library as G4 bi-modal ligands, acting on a selective reversible recognition and subsequent alkylation upon reductive activation.Experimental section
Synthesis of 2,7-dibromo-1,8-dihydroxyanthracene-9,10-dione (8)
10 g of 1,8-dihydroxyanthraquinone 7 (0.042 mol) were dissolved in 300 ml of dichloromethane and 7.35 ml of diisopropylamine NH(iPr)2 were added under stirring. A solution of N-bromosuccinimide (18.5 g, 0.104 mol in 800 ml of DCM) was added dropwise over 30 min and the mixture was stirred for 4–6 h at room temperature. After that time the reaction was quenched in slightly acidic water (500 ml, 1% HCl) and the organic layer was separated. The aqueous solution was then washed with DCM (2 × 250 ml); the organic phases were recombined and dried over Na2SO4. The solvent was removed under vacuum to afford an orange solid. The crude product can be used directly for the next protection step. To determine the reaction yield and characterize the product, 8 was isolated by column chromatography in cyclohexane–toluene 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 affording a yellow-orange solid. Yield 30%. Mp >300 °C. 1H-NMR (300 MHz, DMSO-d6): δ 7.60 (d, 2H, J = 7.9 Hz), 8.17 (d, 2H, J = 7.9 Hz), 12.41 (br s, 2H).
1 affording a yellow-orange solid. Yield 30%. Mp >300 °C. 1H-NMR (300 MHz, DMSO-d6): δ 7.60 (d, 2H, J = 7.9 Hz), 8.17 (d, 2H, J = 7.9 Hz), 12.41 (br s, 2H).
Anal. found: C, 42.2; H, 1.6. Calc. for C14H6Br2O4: C, 42.2; H, 1.5%.
Synthesis of 2,7-dibromo-1,8-dimethoxyanthracene-9,10-dione (9)
18.0 g of the crude product 8 were suspended in acetone (900 ml) and K2CO3 (19.1 g, 0.128 mol) and Me2SO4 (44 ml, 0.461 mol) was added. The mixture was heated to reflux overnight while stirring under argon (18–20 h). During the reaction the mixture became dark while at the end of it a yellow solid crashed out. After 20 h the suspension was cooled down and the solvent was removed under reduced pressure. The crude product was dissolved in DCM (200 ml) and an ammonia aqueous solution (5%) was added. The biphasic mixture was stirred at room temperature for 1 h. After this period the organic phase was separated and washed twice (2 × 200 ml) with an acidic aqueous solution (1% HCl), while the aqueous phase was washed with DCM to recover all the product traces (2 × 200 ml). The organic phases were then recombined, dried over Na2SO4 and the solvent removed under vacuum to afford a brown solid. The crude product was purified by flash chromatography (MPLC) with a cyclohexane–ethyl acetate gradient (TLC eluent cyclohexane–ethyl acetate 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) affording a yellow solid. Yield 70%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 4.07 (s, 6H), 7.90–7.99 (m, 4H). 13C-NMR (CDCl3): δ 62.3, 123.7, 127.5, 128.8, 133.6, 138.1, 156.8, 180.9, 181.8. Anal. found: C, 45.0; H, 2.4. Calc. for C16H10Br2O4: C, 45.1; H, 2.4%.
3) affording a yellow solid. Yield 70%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 4.07 (s, 6H), 7.90–7.99 (m, 4H). 13C-NMR (CDCl3): δ 62.3, 123.7, 127.5, 128.8, 133.6, 138.1, 156.8, 180.9, 181.8. Anal. found: C, 45.0; H, 2.4. Calc. for C16H10Br2O4: C, 45.1; H, 2.4%.
Synthesis of 1,8-dimethoxy-2,7-bis((trimethylsilyl)ethynyl)anthracene-9,10-dione (10)
3.0 g of 9 (7.30 mmol) were dissolved in anhydrous THF (400 ml) and then in order Pd(PPh3)2Cl2 (20 mol%, 1.02 g, 1.46 mmol) and CuI (20 mol%, 0.278 g, 1.46 mmol) were added under stirring while bubbling the solution with an argon flow. 10.1 ml of TEA (10 equiv., 73 mmol) were added followed immediately by 10.4 ml of TMSA (10 equiv., 73 mmol). The stirred solution was heated to reflux for 16 h under an argon atmosphere. After this period the dark solution was cooled and poured in 200 ml of water. The mixture was then extracted with DCM (3 × 250 ml); the organic phase was collected and dried over Na2SO4. The solvent was removed under reduced pressure affording the crude product as a dark solid. The crude was purified by flash chromatography with a hexane–ethyl acetate gradient (TLC eluent hexane–ethyl acetate, 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 10 as a yellow product. Yield 60%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 0.31 (s, 18H), 4.13 (s, 6H), 7.74 (d, 2H, J = 8.0 Hz), 7.94 (d, 2H, J = 8.0 Hz).
1) to give 10 as a yellow product. Yield 60%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 0.31 (s, 18H), 4.13 (s, 6H), 7.74 (d, 2H, J = 8.0 Hz), 7.94 (d, 2H, J = 8.0 Hz).
13C-NMR (CDCl3): δ −0.4, 62.0, 99.7, 104.5, 122.0, 125.5, 128.4, 133.8, 137.7, 161.4, 181.6, 182.1. Anal. found: C, 67.8; H, 6.2. Calc. for C26H28O4Si2: C, 67.8; H, 6.1%.
Synthesis of 2,7-diethynyl-1,8-dimethoxyanthracene-9,10-dione (11)
1.13 g of 10 (2.46 mmol) was dissolved in a MeOH–DCM 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture (167
1 mixture (167![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 ml) and the solution obtained was cooled at 0 °C. 0.60 g of K2CO3 was added and the mixture was stirred at 0 °C for 1 h under Ar. The solution was then allowed to reach room temperature and quenched in 100 ml of water. The aqueous solution was extracted with DCM (3 × 150 ml) and the organic phase was collected and dried over Na2SO4. The solvent was then removed under vacuum to afford quantitatively 11 as a yellow solid. Yield ≥98%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 3.56 (s, 2H), 4.13 (s, 6H), 7.79 (d, 2H, J = 8.0 Hz), 7.97 (d, 2H, J = 8.0 Hz). 13C-NMR (CDCl3): δ 62.5, 78.6, 85.8, 122.2, 124.7, 128.3, 133.2, 138.2, 161.7, 181.4, 182.0. Anal. found: C, 75.8; H, 3.8. Calc. for C20H12O4: C, 75.9; H, 3.8%.
33 ml) and the solution obtained was cooled at 0 °C. 0.60 g of K2CO3 was added and the mixture was stirred at 0 °C for 1 h under Ar. The solution was then allowed to reach room temperature and quenched in 100 ml of water. The aqueous solution was extracted with DCM (3 × 150 ml) and the organic phase was collected and dried over Na2SO4. The solvent was then removed under vacuum to afford quantitatively 11 as a yellow solid. Yield ≥98%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 3.56 (s, 2H), 4.13 (s, 6H), 7.79 (d, 2H, J = 8.0 Hz), 7.97 (d, 2H, J = 8.0 Hz). 13C-NMR (CDCl3): δ 62.5, 78.6, 85.8, 122.2, 124.7, 128.3, 133.2, 138.2, 161.7, 181.4, 182.0. Anal. found: C, 75.8; H, 3.8. Calc. for C20H12O4: C, 75.9; H, 3.8%.
General procedure for the synthesis of 12–13
The corresponding aryl iodide 14 or 15 (2.0 mmol, 0.53 g) was dissolved in anhydrous THF (60 ml) and Pd(PPh3)2Cl2 (5 mol%, 70.4 mg, 0.10 mmol) and CuI (10 mol%, 38.2 mg, 0.2 mmol) were added under stirring while bubbling the solution with an argon flow. TEA (0.56 ml, 2 equiv., 4.0 mmol) was then added immediately followed by a solution of 11 (0.4 equiv., 254 mg, 0.80 mmol) in a degassed anhydrous THF solution (15 ml).The mixture was heated to reflux for 5 h under Ar, and then cooled down and poured into water (100 ml). The aqueous solution was extracted with DCM (3 × 100 ml) and the organic phases were collected and dried over Na2SO4. The solvent was removed under reduced pressure to afford an orange solid. The crude products were purified by flash chromatography (eluent: cyclohexane–acetate) to give 12 (17%) and 13 (20%).
2,7-Bis((p-acetoxylphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione (12)
Orange powder. Yield 17%. Mp >300 °C. 1H-NMR (300 MHz, CDCl3): δ 2.34 (s, 6H), 4.20 (s, 6H), 7.16 (d, 4H, J = 8.6 Hz), 7.62 (d, 4H, J = 8.6 Hz), 7.83 (d, 2H, J = 8.0 Hz), 8.02 (d, 2H, J = 8.0 Hz). 13C-NMR (CDCl3): δ 21.0, 62.3, 84.8, 97.3, 120.1, 121.8, 122.3, 125.7, 128.4, 132.9, 133.7, 137.3, 151.1, 160.9, 168.9, 181.7, 182.0. Anal. found: C, 73.9; H, 4.1. Calc. for C36H24O8: C, 74.0; H, 4.1%.2,7-Bis((m-acetoxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione (13)
Yellow powder. Yield 20%. Mp >300° C. 1H-NMR (300 MHz, CDCl3): δ 2.35 (s, 6H), 4.20 (s, 6H), 7.15–7.18 (m, 2H), 7.34–7.35 (m, 2H), 7.40–7.50 (m, 4H), 7.83 (d, 2H, J = 8.0 Hz), 8.03 (d, 2H, J = 8.0 Hz). 13C-NMR (CDCl3): δ 20.9, 62.4, 85.3, 97.0, 122.3, 122.6, 123.7, 124.7, 125.6, 128.4, 129.2, 129.5, 133.9, 137.4, 150.5, 169.1, 171.3, 181.7, 182.0. Anal. found: C, 74.1; H, 4.1. Calc. for C36H24O8: C, 74.0; H, 4.1%.General procedure for the synthesis of 1–2
Compounds 12 or 13 (0.59 g, 1 mmol) were dissolved in a mixture of MeOH–H2O 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (80
1 (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 ml). K2CO3 was added (1.68 g, 12.1 mmol) and the mixture was stirred at room temperature under Ar for 5 h (12) or 20 h (13). After the indicated time, the solution was poured in 50 ml of water and methanol was eliminated by evaporation. The aqueous solution was then acidified with HCl 10% and extracted with CHCl3 (3 × 250 ml). The organic phase was collected and dried over Na2SO4. The solvent was removed under reduced pressure to afford the crude products.
20 ml). K2CO3 was added (1.68 g, 12.1 mmol) and the mixture was stirred at room temperature under Ar for 5 h (12) or 20 h (13). After the indicated time, the solution was poured in 50 ml of water and methanol was eliminated by evaporation. The aqueous solution was then acidified with HCl 10% and extracted with CHCl3 (3 × 250 ml). The organic phase was collected and dried over Na2SO4. The solvent was removed under reduced pressure to afford the crude products.
Both crude products were purified by reverse phase HPLC (gradient H2O + 0.1% TFA, CH3CN) to afford the final products 1 (15%) and 2 (10%).
2,7-Bis((p-hydroxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione (1)
Orange needles. Yield 15%. Mp >300 °C. A319(H2O) = 37![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1. 1H-NMR (300 MHz, DMSO-d6): δ 4.07 (s, 6H), 6.86 (d, 4H, J = 8.7 Hz), 7.47 (d, 4H, J = 8.7 Hz), 7.90 (br s, 4H), 10.12 (br s, 2H). 13C-NMR (DMSO-d6): δ 61.8, 83.2, 98.8, 11.7, 116.0, 122.0, 125.3, 128.3, 133.2, 133.4, 137.0, 158.9, 159.7, 181.3, 181.5. Anal. found: C, 76.8; H, 3.9. Calc. for C32H20O6: C, 76.8; H, 4.0%.
300 M−1 cm−1. 1H-NMR (300 MHz, DMSO-d6): δ 4.07 (s, 6H), 6.86 (d, 4H, J = 8.7 Hz), 7.47 (d, 4H, J = 8.7 Hz), 7.90 (br s, 4H), 10.12 (br s, 2H). 13C-NMR (DMSO-d6): δ 61.8, 83.2, 98.8, 11.7, 116.0, 122.0, 125.3, 128.3, 133.2, 133.4, 137.0, 158.9, 159.7, 181.3, 181.5. Anal. found: C, 76.8; H, 3.9. Calc. for C32H20O6: C, 76.8; H, 4.0%.
2,7-Bis((m-hydroxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione (2)
Yellow needles. Yield 10%. Mp >300 °C. A315(H2O) = 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 700 M−1 cm−1. 1H-NMR (300 MHz, DMSO-d6): δ 4.18 (s, 6H), 7.00 (d, 2H, J = 8.0 Hz), 7.10 (br s, 2H), 7.17 (d, 2H, J = 8.0 Hz), 7.39 (t, 2H, J = 8.0 Hz), 8.03 (d, 2H, J = 8.0 Hz), 8.08 (d, 2H, J = 8.0 Hz), 9.90 (s, 2H). 13C-NMR (DMSO-d6): δ 62.0, 84.2, 97.7, 111.3, 117.1, 117.8, 122.1, 122.4, 124.6, 128.3, 130.1, 133.7, 137.4, 157.5, 160.1, 181.3, 181.4. Anal. found: C, 76.7; H, 4.0. Calc. for C32H20O6: C, 76.8; H, 4.0%.
700 M−1 cm−1. 1H-NMR (300 MHz, DMSO-d6): δ 4.18 (s, 6H), 7.00 (d, 2H, J = 8.0 Hz), 7.10 (br s, 2H), 7.17 (d, 2H, J = 8.0 Hz), 7.39 (t, 2H, J = 8.0 Hz), 8.03 (d, 2H, J = 8.0 Hz), 8.08 (d, 2H, J = 8.0 Hz), 9.90 (s, 2H). 13C-NMR (DMSO-d6): δ 62.0, 84.2, 97.7, 111.3, 117.1, 117.8, 122.1, 122.4, 124.6, 128.3, 130.1, 133.7, 137.4, 157.5, 160.1, 181.3, 181.4. Anal. found: C, 76.7; H, 4.0. Calc. for C32H20O6: C, 76.8; H, 4.0%.
General procedure for the synthesis of 3–6
0.28 mmol of the corresponding aryliodide (14, 15, 16, 17) was dissolved in 10 ml of anhydrous THF. Pd(PPh3)2Cl2 (10 mol%, 20.0 mg, 0.28 mmol) and CuI (10 mol%, 5.4 mg, 0.28 mmol) were added under stirring while purging the solution with an argon flow. The mixture was heated at 50 °C and TEA (2 equiv., 78 μl, 0.57 mmol) was added.The bis-ethynyl derivative 11 (0.33 equiv., 30 mg, 0.094 mmol) was dissolved in 10 ml of THF. This solution was purged with Ar and added dropwise over 20 minutes in the reaction vessel containing the iodide and the catalysts at 50 °C. After all of the alkyne was added to the solution the reaction was stopped. The mixture was cooled to r.t. and then poured in 20 ml of water. The aqueous phase was extracted with DCM containing 20% of methanol (3 × 30 ml) to increase the solubility of the products in the organic phase.
The organic phase was dried over Na2SO4 and the solvent was removed under reduce pressure to afford the crude products.
Ethynyl-AQs (3, 4, 5 and 6) were purified by reverse phase HPLC (gradient H2O + 0.1% TFA, CH3CN). TFA salts were exchanged with HCl to afford bis-hydrochloride as final salt products. Due to the low solubility of the products the crude solid was subjected to a particular preparation method before the injection in preparative HPLC. In more detail, the crude was suspended in MeOH–H2O (slightly acidic) 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, sonicated and heated at 60 °C for 10 min. The suspension was then filtered and injected directly in HPLC, while the solid was subjected to another treatment before being wasted.
1, sonicated and heated at 60 °C for 10 min. The suspension was then filtered and injected directly in HPLC, while the solid was subjected to another treatment before being wasted.
2,7-Bis(4-(2-(dimethylamino)ethoxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione·2HCl (3)
Yellow oil. Yield 7%. A310 (H2O) = 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 3.03 (s, 12H), 3.66 (t, 4H, J = 4.6), 4.18 (s, 6H), 4.44 (t, 4H, J = 4.6), 7.13 (d, 4H, J = 8.9 Hz), 7.62 (d, 4H, J = 8.9 Hz), 7.89 (d, 2H, J = 8.0 Hz), 8.02 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 44.2, 58.0, 63.1, 63.5, 85.1, 99.3, 116.5, 117.5, 123.8, 127.6, 130.1, 134.9, 135.4, 138.8, 160.2, 162.1, 183.4, 183.8. Anal. found: C, 67.2; H, 5.7; N, 3.9. Calc. for C40H40Cl2N2O6: C, 67.1; H, 5.6; Cl, 9.9; N, 3.9%.
300 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 3.03 (s, 12H), 3.66 (t, 4H, J = 4.6), 4.18 (s, 6H), 4.44 (t, 4H, J = 4.6), 7.13 (d, 4H, J = 8.9 Hz), 7.62 (d, 4H, J = 8.9 Hz), 7.89 (d, 2H, J = 8.0 Hz), 8.02 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 44.2, 58.0, 63.1, 63.5, 85.1, 99.3, 116.5, 117.5, 123.8, 127.6, 130.1, 134.9, 135.4, 138.8, 160.2, 162.1, 183.4, 183.8. Anal. found: C, 67.2; H, 5.7; N, 3.9. Calc. for C40H40Cl2N2O6: C, 67.1; H, 5.6; Cl, 9.9; N, 3.9%.
2,7-Bis(3-(2-(dimethylamino)ethoxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione·2HCl (4)
Yellow oil. Yield 20%. A300 (H2O) = 34![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 3.02 (s, 12H), 3.64 (t, 4H, J = 4.6 Hz), 4.13 (s, 6H), 4.41 (t, 4H, J = 4.6 Hz), 7.08 (d, 2H, J = 8.2 Hz), 7.21 (br s, 4H), 7.32–7.38 (m, 2H), 7.82 (d, 2H, J = 8.0 Hz), 7.92 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 44.3, 58.0, 63.2, 63.6, 86.0, 98.8, 117.6, 118.9, 123.8, 125.3, 126.8, 127.0, 129.9, 131.4, 135.6, 139.1, 159.3, 162.3, 183.2, 183.5. Anal. Found: C, 67.1; H, 5.7; N, 4.0. Calc. for C40H40Cl2N2O6: C, 67.1; H, 5.6; N, 3.9%.
500 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 3.02 (s, 12H), 3.64 (t, 4H, J = 4.6 Hz), 4.13 (s, 6H), 4.41 (t, 4H, J = 4.6 Hz), 7.08 (d, 2H, J = 8.2 Hz), 7.21 (br s, 4H), 7.32–7.38 (m, 2H), 7.82 (d, 2H, J = 8.0 Hz), 7.92 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 44.3, 58.0, 63.2, 63.6, 86.0, 98.8, 117.6, 118.9, 123.8, 125.3, 126.8, 127.0, 129.9, 131.4, 135.6, 139.1, 159.3, 162.3, 183.2, 183.5. Anal. Found: C, 67.1; H, 5.7; N, 4.0. Calc. for C40H40Cl2N2O6: C, 67.1; H, 5.6; N, 3.9%.
2,7-Bis((3-((dimethylamino)methyl)-4-idroxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione·2HCl (5)
Orange powder. Yield 15%. Mp >300 °C. A326 (H2O) = 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 2.91 (s, 12H), 4.12 (s, 6H), 4.36 (s, 4H), 7.00 (d, 2H, J = 8.4 Hz), 7.54 (d, 2H, J = 8.4 Hz), 7.62 (s, 2H), 7.74 (d, 2H, J = 8.0 Hz), 7.88 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 43.7, 58.0, 63.2, 85.2, 99.0, 115.6, 117.3, 118.9, 123.8, 127.4, 129.8, 135.2, 136.8, 137.4, 138.7, 159.3, 162.0, 183.2, 183.6. Anal. found: C, 66.5; H, 5.3; N, 4.0. Calc. for C38H36Cl2N2O6: C, 66.4; H, 5.3; N, 4.1%.
200 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 2.91 (s, 12H), 4.12 (s, 6H), 4.36 (s, 4H), 7.00 (d, 2H, J = 8.4 Hz), 7.54 (d, 2H, J = 8.4 Hz), 7.62 (s, 2H), 7.74 (d, 2H, J = 8.0 Hz), 7.88 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 43.7, 58.0, 63.2, 85.2, 99.0, 115.6, 117.3, 118.9, 123.8, 127.4, 129.8, 135.2, 136.8, 137.4, 138.7, 159.3, 162.0, 183.2, 183.6. Anal. found: C, 66.5; H, 5.3; N, 4.0. Calc. for C38H36Cl2N2O6: C, 66.4; H, 5.3; N, 4.1%.
2,7-Bis((4-((dimethylamino)methyl)-3-idroxyphenyl)ethynyl)-1,8-dimethoxyanthracene-9,10-dione·2HCl (6)
Yellow powder. Yield 10%. Mp >300 °C. A302 = 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 2.91 (s, 12H), 4.17 (s, 6H), 4.37 (s, 4H), 7.10–7.25 (m, 4H), 7.45 (d, 2H, J = 8.2 Hz), 7.91 (d, 2H, J = 8.0 Hz), 8.01 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 43.7, 58.2, 63.3, 85.8, 98.2, 119.3, 119.5, 123.8, 124.8, 126.9, 127.4, 130.03, 134.2, 135.9, 139.1, 158.2, 162.5, 183.3, 183.6. Anal. found: C, 66.5; H, 5.3; N, 4.1. Calc. for C38H36Cl2N2O6: C, 66.4; H, 5.3; N, 4.1%.
300 M−1 cm−1. 1H-NMR (300 MHz, CD3OD): δ 2.91 (s, 12H), 4.17 (s, 6H), 4.37 (s, 4H), 7.10–7.25 (m, 4H), 7.45 (d, 2H, J = 8.2 Hz), 7.91 (d, 2H, J = 8.0 Hz), 8.01 (d, 2H, J = 8.0 Hz). 13C-NMR (CD3OD): δ 43.7, 58.2, 63.3, 85.8, 98.2, 119.3, 119.5, 123.8, 124.8, 126.9, 127.4, 130.03, 134.2, 135.9, 139.1, 158.2, 162.5, 183.3, 183.6. Anal. found: C, 66.5; H, 5.3; N, 4.1. Calc. for C38H36Cl2N2O6: C, 66.4; H, 5.3; N, 4.1%.
General procedures and synthesis of the aryliodo-derivatives 14–19 have been reported in the ESI.†
Circular dichroism measurements
Circular dichroism spectra were recorded on a Jasco J-810 spectropolarimeter equipped with a Peltier temperature controller in 10 mM Tris, 50 mM KCl at pH 7.5 using a 10 mm path-length cell. DNA substrates were the four-repeat human telomeric sequences Tel22: d[AG3(T2AG3)3], Tel24: d(T2AG3)4 and wtTel26: d[(T2AG3)4T2] provided by Eurogentec. As double stranded DNA we used calf thymus DNA (ctDNA, Sigma). Before data acquisition, G4 forming solutions (4 μM strand concentration) were heated at 95 °C for 5 min and left to cool at room temperature overnight. The spectra of the nucleic acid alone and in the presence of increasing ligand concentrations (0–20 μM) were acquired. Each reported spectrum represents the average of 3 scans recorded with 1 nm step resolution. The observed CD signals were converted to the mean residue ellipticity [Θ] = deg cm−2 dmol−1 (Mol. Ellip.).Fluorescence melting studies
Melting experiments were performed using a Roche LightCycler, using an excitation source at 488 nm and recording the fluorescence emission at 520 nm. Target DNA (Eurogentec) were the human telomeric sequence HTS d[AG3(T2AG3)3T], Tel24 d[(T2AG3)4], and a 18 bp double stranded DNA (5′-GTGAGATACCGACAGAAG) labeled with Dabcyl at the 5′ end and FAM at the 3′ end. Samples contained 0.25 μM of target DNA and increasing concentrations of tested derivatives in 50 mM potassium buffer (10 mM LiOH; 50 mM KCl pH 7.5 with H3PO4). They were first heated to 95 °C at a rate of 0.1 °C s−1, maintained at 95 °C for 5 min and then annealed by cooling to 30 °C at a rate of 0.1 °C s−1. Then, samples were maintained at 30 °C for 5 min before being slowly heated to 95 °C (1 °C min−1) and annealed at a rate of 1 °C min−1. Recordings were acquired during both these melting and annealing steps to check for hysteresis. Tm values were determined from the first derivatives of the melting profiles using the Roche LightCycler software. Each curve was repeated at least three times and errors were ±0.4 °C. ΔTm values were calculated by subtracting the Tm value recorded in the presence of the ligand from the corresponding value in the absence of the ligand.Polymerase stop assay
The DNA primer d[TAATACGACTCACTATAG], the human telomeric template sequence HT4-temp d[TC2A2CTATGTATAC(T2AG3)4ACATATCGATGA3T2GCTATAGTGAGTCGTATTA] and the control template sequence HT4sc-temp d[TC2A2CTATGTATACT2G2ATGTGAGTGTG AGTGTGAG2ACATATCGATGA3T2GCTATAGTGAGTCGTATTA] were obtained from Eurogentec. The primer was initially 5′-labeled with 32P and T4 polynucleotide kinase (Thermo Scientific), by incubating the reaction mixture at 37 °C for 30 min. The kinase activity was inactivated by heating the reaction mixture at 85 °C for 5 min, followed by two extractions with one volume of phenol–CHCl3 (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50). An equimolar mixture of the labeled primer and template (20 nM) had been annealed in the polymerase required buffer and subsequently, increasing ligand concentrations have been added. After incubation (30 min at r.t.) 2.5 U of Taq polymerase (Thermo Scientific) and 100 μM dNTP mixture were added to each sample and the resulting solutions were kept for 30 min at 55 °C. Reaction products were resolved by gel electrophoresis (12% polyacrylamide gel with 7 M urea) in 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM Na2EDTA). Gels were dried and resolved bands were visualized on a PhosphorImager (Amersham).
50). An equimolar mixture of the labeled primer and template (20 nM) had been annealed in the polymerase required buffer and subsequently, increasing ligand concentrations have been added. After incubation (30 min at r.t.) 2.5 U of Taq polymerase (Thermo Scientific) and 100 μM dNTP mixture were added to each sample and the resulting solutions were kept for 30 min at 55 °C. Reaction products were resolved by gel electrophoresis (12% polyacrylamide gel with 7 M urea) in 1× TBE (89 mM Tris base, 89 mM boric acid, 2 mM Na2EDTA). Gels were dried and resolved bands were visualized on a PhosphorImager (Amersham).
Docking experiments
In order to take into account the conformational polymorphism of the DNA human telomeric repeat sequence d[AG3(T2AG3)3], we included in our study six PDB entries (codes 1KF1,56143D,512HY9,572JPZ,582JSL59 and 2JSM59) among X-ray and NMR structures, using all the conformations stored in each experimental structure. Initially, both ligands and DNA were pretreated. For ligand preparation, the 3D structures of all the studied compounds were generated with the Maestro Build Panel [Maestro, version 9.3; Schrödinger, LLC: New York, NY, 2012] and submitted to 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 iterations of energy minimization using the Polake–Ribiere Conjugated Gradient (PRCG) algorithm, OPLS200564 as a force field with the all atoms notation, and the implicit model of solvation GB/SA water.65 Co-crystallized water molecules and counter ions were removed from the DNA X-ray structure. In their sequences, all the hybrid models presented head and tail caps, each formed by a different number of additional nucleotides. In particular, the hybrid NMR structures 2HY9 and 2JPZ resulted both formed by 26-mer, while in the hybrid models 2JSL and 2JSM they were reported sequences with, respectively, 25- and 23-mer. Thus, to obtain a similar analysis with respect to the first two models, the hybrid PDB structures were modified by deleting these caps, that is, considering them as conformational templates for the canonical 22-mer d[AG3(T2AG3)3]. The 47 experimental conformations stored in the six PDB models were energy-optimized exactly under the same conditions (force field, implicit salvation model, iterations and convergence criterion) adopted for the ligands. The energy minimization was performed until the rmsd of all heavy atoms was within 0.05 Å of the original PDB model. The evaluation of the most stable conformations of the DNA structures, for each model, has been performed after the pre-treatment. Docking studies were carried out using IFD.51 An initial Glide SP docking of each ligand was carried out using a softened potential, a van der Waals radius scaling factor of 0.50 for receptor/ligand atoms, and a number of 20 poses per ligand to be energy minimized with the OPLS-AA force field.66 The poses were saved for each ligand and submitted to the subsequent Prime side chain orientation prediction of residues with a distance cutoff of 5 Å around each ligand. After the prime minimization of the nucleobases and the ligand for each pose, a Glide SP redocking of each DNA–ligand complex structure within 30 kcal mol−1 above the global minimum was performed. Finally, each output pose was estimated by the binding energy (IFD score) and visually examined.
000 iterations of energy minimization using the Polake–Ribiere Conjugated Gradient (PRCG) algorithm, OPLS200564 as a force field with the all atoms notation, and the implicit model of solvation GB/SA water.65 Co-crystallized water molecules and counter ions were removed from the DNA X-ray structure. In their sequences, all the hybrid models presented head and tail caps, each formed by a different number of additional nucleotides. In particular, the hybrid NMR structures 2HY9 and 2JPZ resulted both formed by 26-mer, while in the hybrid models 2JSL and 2JSM they were reported sequences with, respectively, 25- and 23-mer. Thus, to obtain a similar analysis with respect to the first two models, the hybrid PDB structures were modified by deleting these caps, that is, considering them as conformational templates for the canonical 22-mer d[AG3(T2AG3)3]. The 47 experimental conformations stored in the six PDB models were energy-optimized exactly under the same conditions (force field, implicit salvation model, iterations and convergence criterion) adopted for the ligands. The energy minimization was performed until the rmsd of all heavy atoms was within 0.05 Å of the original PDB model. The evaluation of the most stable conformations of the DNA structures, for each model, has been performed after the pre-treatment. Docking studies were carried out using IFD.51 An initial Glide SP docking of each ligand was carried out using a softened potential, a van der Waals radius scaling factor of 0.50 for receptor/ligand atoms, and a number of 20 poses per ligand to be energy minimized with the OPLS-AA force field.66 The poses were saved for each ligand and submitted to the subsequent Prime side chain orientation prediction of residues with a distance cutoff of 5 Å around each ligand. After the prime minimization of the nucleobases and the ligand for each pose, a Glide SP redocking of each DNA–ligand complex structure within 30 kcal mol−1 above the global minimum was performed. Finally, each output pose was estimated by the binding energy (IFD score) and visually examined.
All the 3D figures were obtained with PyMOL graphics and the modeling package, version 0.98 [Delano W. L. The PyMOL Molecular Graphics System, 2002. http://www.pymol.org].
Acknowledgements
This research was supported by Grants from the Italian Ministry of Education, University and Research (MIUR): FIRB-IDEAS (code FIRB RBID082ATK_003), PRIN 2009 (code 2009MFRKZ8), and by AIRC, the Italian Association for Cancer Research (IG 2013 N.14708).Notes and references
- J. Choi and T. Majima, Chem. Soc. Rev., 2011, 40, 5893–5909 RSC.
- J. Zhao, A. Bacolla, G. Wang and K. M. Vasquez, Cell. Mol. Life Sci., 2010, 67, 43–62 CrossRef CAS PubMed.
- S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261–275 CrossRef CAS PubMed.
- M. L. Bochman, K. Paeschke and V. A. Zakian, Nat. Rev. Genet., 2012, 13, 770–780 CrossRef CAS PubMed.
- J. T. Davis, Angew. Chem., Int. Ed., 2004, 43, 668–698 CrossRef CAS PubMed.
- R. Rodriguez, S. Muller, J. A. Yeoman, C. Trentesaux, J. F. Riou and S. Balasubramanian, J. Am. Chem. Soc., 2008, 130, 15758–15759 CrossRef CAS PubMed.
- R. Rodriguez, K. M. Miller, J. V. Forment, C. R. Bradshaw, M. Nikan, S. Britton, T. Oelschlaegel, B. Xhemalce, S. Balasubramanian and S. Jackson, Nat. Chem. Biol., 2012, 8, 301–310 CrossRef CAS PubMed.
- C. Douarre, X. Mergui, A. Sidibe, D. Gomez, P. Alberti, P. Mailliet, C. Trentesaux and J. F. Riou, Nucleic Acids Res., 2013, 41, 3588–3599 CrossRef CAS PubMed.
- T. A. Brooks and L. H. Hurley, Genes Cancer, 2010, 1, 641–649 CrossRef CAS PubMed.
- S. Kumari, A. Bugaut, J. L. Huppert and S. Balasubramanian, Nat. Chem. Biol., 2007, 3, 218–221 CrossRef CAS PubMed.
- A. Bugaut and S. Balasubramanian, Nucleic Acids Res., 2012, 40, 4727–4741 CrossRef CAS PubMed.
- K. Paeschke, M. L. Bochman, P. D. Garcia, P. Cejka, K. L. Friedman, S. C. Kowalczykowski and V. A. Zakian, Nature, 2013, 497, 458–462 CrossRef CAS PubMed; A. Piazza, A. Serero, J. B. Boulé, P. Legoix-Né, J. Lopes and A. Nicolas, PLoS Genet., 2012, 8, 1–17 Search PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2007, 35, 406–413 CrossRef CAS PubMed.
- J. L. Huppert, A. Bugaut, S. Kumari and S. Balasubramanian, Nucleic Acids Res., 2008, 36, 6260–6268 CrossRef CAS PubMed.
- K. M. Miller and R. Rodriguez, Expert Rev. Clin. Pharmacol., 2011, 4, 139–142 CrossRef CAS PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- D. Monchaud and M. P. Teulade-Fichou, Org. Biomol. Chem., 2008, 6, 627–636 CAS.
- S. Neidle, Curr. Opin. Struct. Biol., 2009, 19, 239–250 CrossRef CAS PubMed.
- M. Di Antonio, G. Biffi, A. Mariani, E. A. Raiber, R. Rodriguez and S. Balasubramanian, Angew. Chem., Int. Ed., 2012, 51, 11073–11078 CrossRef CAS PubMed.
- D. Sun, B. Thompson, B. E. Cathers, M. Salazar, S. M. Kerwin, J. O. Trent, T. C. Jenkins, S. Neidle and L. H. Hurley, J. Med. Chem., 1997, 40, 2113–2116 CrossRef CAS PubMed.
- D. Cairns, E. Michalitsi, T. C. Jenkins and S. P. Macka, Bioorg. Med. Chem., 2002, 10, 803–807 CrossRef CAS.
- T. Ndlebe and G. B. Schuster, Org. Biomol. Chem., 2006, 4, 4015–4021 CAS.
- C. Monneret, Eur. J. Med. Chem., 2001, 36, 483–493 CrossRef CAS.
- N. Li, Y. Ma, C. Yang, L. Guo and X. Yang, Biophys. Chem., 2005, 116, 199–205 CrossRef CAS PubMed.
- Z. Hajihassan and A. Rabbani-Chadegani, J. Biomed. Sci., 2009, 11, 16–31 Search PubMed.
- H. Suzuki, T. Ikeda, T. Yamagishi, S. Nakaike, S. Nakane and M. Ohsawa, Mutat. Res., 1995, 328, 151–161 CrossRef CAS.
- G. R. Clark, P. D. Pytel, C. J. Squire and S. Neidle, J. Am. Chem. Soc., 2003, 125, 4066–4067 CrossRef CAS PubMed.
- P. J. Perry, S. M. Gowan, A. P. Reszka, P. Polucci, T. C. Jenkins, L. R. Kelland and S. Neidle, J. Med. Chem., 1998, 41, 3253–3260 CrossRef CAS PubMed.
- P. J. Perry, A. P. Reszka, A. A. Wood, M. A. Read, S. M. Gowan, H. S. Dosanjh, J. O. Trent, T. C. Jenkins, L. R. Kelland and S. Neidle, J. Med. Chem., 1998, 41, 4873–4884 CrossRef CAS PubMed.
- P. J. Perry, M. A. Read, R. T. Davies, S. M. Gowan, A. P. Reszka, A. A. Wood, L. R. Kelland and S. Neidle, J. Med. Chem., 1999, 42, 2679–2684 CrossRef CAS PubMed.
- G. Zagotto, C. Sissi, L. Lucatello, C. Pivetta, S. A. Cadamuro, K. R. Fox, S. Neidle and M. Palumbo, J. Med. Chem., 2008, 51, 5566–5574 CrossRef CAS PubMed.
- N. Ranjan, E. Davis, L. Xue and D. P. Arya, Chem. Commun., 2013, 49, 5796–5798 RSC.
- G. Zagotto, C. Sissi, S. Moro, D. Dal Ben, G. N. Parkinson, K. R. Fox, S. Neidle and M. Palumbo, Bioorg. Med. Chem., 2008, 16, 354–361 CrossRef CAS PubMed.
- G. Zagotto, A. Ricci, E. Vasquez, A. Sandoli, S. Benedetti, M. Palumbo and C. Sissi, Bioconjugate Chem., 2011, 22, 2126–2135 CrossRef CAS PubMed.
- E. H. van Dijk, D. J. T. Myles, M. H. van der Veen and J. C. Hummelen, Org. Lett., 2006, 8, 2333–2336 CrossRef CAS PubMed.
- E. Modica, R. Zanaletti, M. Freccero and M. Mella, J. Org. Chem., 2001, 66, 41–52 CrossRef CAS.
- M. Di Antonio, F. Doria, M. Mella, D. Merli, A. Profumo and M. Freccero, J. Org. Chem., 2007, 72, 8354–8360 CrossRef CAS PubMed.
- C. Percivalle, F. Doria and M. Freccero, Curr. Org. Chem., 2014, 18, 19–43 CrossRef CAS.
- M. Di Antonio, F. Doria, S. N. Richter, C. Bertipaglia, M. Mella, C. Sissi, M. Palumbo and M. Freccero, J. Am. Chem. Soc., 2009, 131, 13132–13141 CrossRef CAS PubMed.
- M. Nadai, F. Doria, M. Di Antonio, G. Sattin, L. Germani, C. Percivalle, M. Palumbo, S. N. Richter and M. Freccero, Biochimie, 2011, 93, 1328–1340 CrossRef CAS PubMed.
- F. Doria, M. Nadai, M. Folini, M. Di Antonio, L. Germani, C. Percivalle, C. Sissi, N. Zaffaroni, S. Alcaro, A. Artese, S. N. Richter and M. Freccero, Org. Biomol. Chem., 2012, 10, 2798–2806 CAS.
- F. Doria, M. Nadai, M. Folini, M. Scalabrin, L. Germani, G. Sattin, M. Mella, M. Palumbo, N. Zaffaroni, D. Fabris, M. Freccero and S. N. Richter, Chem. – Eur. J., 2013, 19, 78–81 CrossRef CAS PubMed.
- G. Bringmann and D. Menche, Angew. Chem., Int. Ed., 2001, 40, 1687–1690 CrossRef CAS.
- G. Bringmann, D. Menche, J. Kraus, J. Mühlbacher, K. Peters, E.-M. Peters, R. Brun, M. Bezabih and B. M. Abegaz, J. Org. Chem., 2002, 67, 5595–5610 CrossRef CAS PubMed.
- L. F. Tietze, K. M. Gericke and I. Schuberth, Eur. J. Org. Chem., 2007, 4563–4577 CrossRef CAS.
- E. Negishi and L. Anastasia, Chem. Rev., 2003, 103, 1979–2017 CrossRef CAS PubMed.
- R. A. J. Darby, M. Sollogoub, C. McKeen, L. Brown, A. Risitano, N. Brown, C. Barton, T. Brown and K. R. Fox, Nucleic Acids Res., 2002, 30, e39 CrossRef PubMed.
- J. M. Nicoludis, S. P. Barrett, J. L. Mergny and L. A. Yatsunyk, Nucleic Acids Res., 2012, 40, 5432–5447 CrossRef CAS PubMed.
- K. N. Luu, A. T. Phan, V. V. Kuryavyi, L. Lacroix and D. J. Patel, J. Am. Chem. Soc., 2006, 128, 9963–9970 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS PubMed.
- Y. Wang and D. J. Patel, Structure, 1993, 1, 263–282 CrossRef CAS.
- H. Han, L. H. Hurley and M. Salazar, Nucleic Acids Res., 1999, 27, 537–542 CrossRef CAS PubMed.
- W. Sherman, T. Day, M. P. Jacobson, R. A. Friesner and R. Farid, J. Med. Chem., 2006, 49, 534–553 CrossRef CAS PubMed.
- C. Varela, E. J. Tavares da Silva, C. Amaral, G. Correia da Silva, T. Baptista, S. Alcaro, G. Costa, R. A. Carvalho, N. A. Teixeira and F. M. Roleira, J. Med. Chem., 2012, 55, 3992–4002 CrossRef CAS PubMed.
- S. Alcaro, C. Musetti, S. Distinto, M. Casatti, G. Zagotto, A. Artese, L. Parrotta, F. Moraca, G. Costa, F. Ortuso, E. Maccioni and C. Sissi, J. Med. Chem., 2013, 56, 843–855 CrossRef CAS PubMed.
- G. N. Parkinson, M. P. Lee and S. Neidle, Nature, 2002, 417, 876–880 CrossRef CAS PubMed.
- J. Dai, C. Punchihewa, A. Ambrus, D. Chen, R. A. Jones and D. Yang, Nucleic Acids Res., 2007, 35, 2440–2450 CrossRef CAS PubMed.
- J. Dai, M. Carver, C. Punchihewa, R. A. Jones and D. Yang, Nucleic Acids Res., 2007, 35, 4927–4940 CrossRef CAS PubMed.
- A. T. Phan, V. Kuryavyi, K. N. Luu and D. J. Patel, Nucleic Acids Res., 2007, 35, 6517–6525 CrossRef CAS PubMed.
- P. Alberti, P. Schmitt, C. H. Nguyen, C. Rivalle, M. Hoarau, D. S. Grierson and J. L. Mergny, Bioorg. Med. Chem. Lett., 2002, 12, 1071–1074 CrossRef CAS.
- J. L. Mergny, L. Lacroix, M. P. Teulade-Fichou, C. Hounsou, L. Guittat, M. Hoarau, P. B. Arimondo, J. P. Vigneron, J. M. Lehn, J. F. Riou, T. Garestier and C. Helene, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 3062–3067 CrossRef CAS PubMed.
- L. Guittat, A. De Cian, F. Rosu, V. Gabelica, E. De Pauw, E. Delfourne and J. L. Mergny, Biochim. Biophys. Acta, 2005, 1724, 375–384 CrossRef CAS PubMed.
- A. Ambrus, D. Chen, J. Dai, T. Bialis, R. A. Jones and D. Yang, Nucleic Acids Res., 2006, 34, 2723–2735 CrossRef CAS PubMed.
- J. L. Banks, H. S. Beard, Y. Cao, A. E. Cho, W. Damm, R. Farid, A. K. Felts, T. A. Halgren, D. T. Mainz, J. R. Maple, R. Murphy, D. M. Philipp, M. P. Repasky, L. Y. Zhang, B. J. Berne, R. A. Friesner, E. Gallicchio and R. M. Levy, J. Comput. Chem., 2005, 26, 1752–1780 CrossRef CAS PubMed.
- W. C. Still, A. Tempczyk, R. C. Hawley and T. Hendrickson, J. Am. Chem. Soc., 1990, 112, 6127–6129 CrossRef CAS.
- W. L. Jorgensen, D. S. Maxwell and J. Tirado-Rives, J. Am. Chem. Soc., 1996, 11225–11236 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, procedures and materials, NMR spectra, CDs, computational data, IFD scores and best poses. See DOI: 10.1039/c4ob00220b |
| This journal is © The Royal Society of Chemistry 2014 |







