Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The versatile enzyme Araf51 allowed efficient synthesis of rare pathogen-related β-D-galactofuranosyl-pyranoside disaccharides
Ilona
Chlubnová
abc,
Blanka
Králová
c,
Hana
Dvořáková
d,
Petr
Hošek
c,
Vojtěch
Spiwok
c,
Dominik
Filipp
e,
Caroline
Nugier-Chauvin
*ab,
Richard
Daniellou†
ab and
Vincent
Ferrières
*ab
aEcole Nationale Supérieure de Chimie de Rennes, CNRS, UMR 6226, 11 Allée de Beaulieu, CS 50837, 35708 Rennes Cedex 7, France. E-mail: vincent.ferrieres@ensc-rennes.fr; Tel: +33 223238058
bUniversité européenne de Bretagne, France
cDepartment of Biochemistry and Microbiology, Institute of Chemical Technology of Prague, Techniká 3, 166 28 Prague 6, Czech Republic
dLaboratory of NMR spectroscopy, Institute of Chemical Technology of Prague, Techniká 3, 166 28 Prague 6, Czech Republic
eLaboratory of Immunobiology, Institute of Molecular Genetics AS CR, Videnska 1083, 142 20 Prague 4, Czech Republic
First published on 21st February 2014
Abstract
The preparation of galactofuranosyl-containing disaccharidic parts of natural glycoconjugates was performed according to a chemo-enzymatic synthesis. Our goals were firstly to develop an alternative approach to standard chemical strategies by limiting the number of reaction and purification steps, and secondly to evaluate the scope of the Araf51 biocatalyst to transfer a galactofuranosyl moiety to a set of pyranosidic acceptors differing from each other by the series, the anomeric configuration as well as the conformation. The study of binding mode of the resulting disaccharides was also performed by molecular modeling and showed significant differences between (1→2)- and (1→6)-linked disaccharides.
Introduction
The complex heterogeneity of carbohydrates in living systems is a direct result of several carbohydrate characteristics: the ability of different types and numbers of sugar residues to form glycosidic bonds with one another, the type of anomeric linkage, the position and the absence or presence of branching, the more or less flexible conformations of the resulting oligo-, polysaccharides or glycoconjugates,1,2 and the size of the monosaccharidic ring.3,4 Indeed, sugars exhibit significant differences depending on whether they are present as pyranosides or as furanosides. It is now well established that the importance of the furanose ring in biology can no longer be understated. A key characteristic of furanose ring systems is their higher flexibility compared to that of their pyranosidic counterparts,5 and this profoundly influences their role in biological processes.6 While furanosyl-containing oligosaccharides are crucial constituents of surface glycoconjugates in cell walls of bacteria,7–9 fungi10 and parasitic11,12 microorganisms, including some clinically significant pathogens, with an exception of 2-deoxy-D-ribose and D-ribose, furanosides are completely absent from mammals. This makes them interesting targets for development of new therapeutics.A major limitation preventing the use of oligosaccharides as therapeutics is the difficulty in producing sufficient amounts of these molecules in a desired purity. The isolation of these compounds from biological sources tends to be low yielding and presents the risk of contamination from infectious agents.13 Although progress in the chemical synthesis of oligosaccharides has been made14–20 and synthesis of docosanaarabinofuranoside realized by Lowary's research team21 is impressive evidence, this approach still remains a challenge. The chemical synthesis requires stereo- and regioselective control of glycosidic bond formation, thus multiple protection and deprotection schemes are needed to achieve the required selectivity. Low yields of the desired products are also a result of the difficulty in purifying the deprotected compound along with product loss during each step of a multistep synthesis.13,22 Nature's solution to the assembly of glycosidic bonds are enzymes belonging to the families of glycosyl transferases and of glycosidases. These enzymes have therefore enormous potential for the synthesis of biologically relevant carbohydrate structures.23 These biocatalysts offer a significant advantage over their chemical counterparts in their ability to form a specific glycosidic linkage in the presence of other reactive functional groups.22
In this context, only few groups have been interested in the chemo-enzymatic synthesis of furanosyl-containing conjugates.3,24–27 Furanosyl transferases are however rare and require both nucleotide-diphospho (NDP)-sugars as donors and suitable acceptors.28–30 Another approach, recently proposed by Thorson, is based on the glycosyl transferase reversibility, that uses simple glycosides as precursors of NDP-sugars for further transfer onto more complex natural products.31,32 To the best of our knowledge, this strategy was not applied for the synthesis of furanosides. Recently, our team developed an enzyme-based protocol for the preparation of di- and oligofuranosides using the thermophilic Araf51 as biocatalyst.33 Considering the structural similarity between L-arabino- and D-galactofuranosyl (D-Galf) entities, and the wide presence of D-Galf residues in natural glycoconjugates and polysaccharides, we now propose to expand the chemo-enzymatic methodology to transglycosylation reactions in order to transfer a galactofuranosyl entity to a variety of pyranosidic acceptors (Fig. 1). These substrates were chosen so as to perform the synthesis of furanosyl-pyranoside sequences relevant to pathogenic microorganisms3 as well as to estimate the specificity of this enzyme towards carbohydrate acceptors. As we expected that such pyranosides are resistant to hydrolysis by the furanosidase Araf51, the presence of the pNP group in the anomeric position was envisaged to enable simple UV detection of reaction products. Moreover, the D-gluco- (D-Glcp), D-galacto- (D-Galp) and D-mannopyranosidic (D-Manp) epimers, as well as their anomeric configurations, offer stereochemical variations that may influence the fate of the coupling process. Finally, we made a focus on the L-rhamnopyranoside (L-Rhap) 8 for structural and biological reasons but also because it displays a 1C4 conformation instead a 4C1 one observed for other acceptors 2–7.
Results
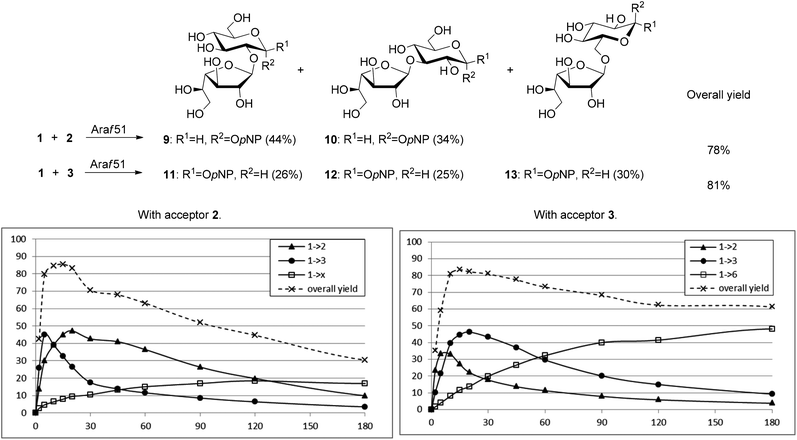
In the first place, unavailability of the −1 subsite of Araf51 to pyranosidic acceptors was confirmed spectrophotometrically (405 nm) by incubation of individual pNP pyranosides 2–8 with the enzyme for 2 h at 60 °C in a phosphate buffer at pH 7.4. No release of p-nitrophenolate was recorded, indicating that these compounds cannot act as donors, thus confirming our first hypothesis. Subsequently, to screen enzyme readiness to catalyze transglycosylation, individual analytical-scale reactions were performed starting from donor 1 and each of pNP glycopyranoside, and in the presence of Araf51. Monitoring by thin layer chromatography showed the formation of disaccharides when the donor 1 was incubated with an equal molar quantity or with a 5-fold molar excess of pyranosidic acceptor, after only short incubation periods (5–20 min). These compounds resulted from both self-condensation of 1 and desired transglycosylation reactions.As a model of biocatalyzed coupling, the progress of the reaction of 1 with 2 (pNP α-D-Glcp) was monitored by HPLC (Fig. 2). Between 5 and 20 minutes, digalactofuranosides constituted the main reaction products.33 At the same time, these products of self-condensation were hydrolyzed more rapidly than furanosyl-pyranoside disaccharides, which resisted to hydrolysis for more than 3 h. This could be explained by the structures of the resulting disaccharides which significantly differ from those of natural substrates of the enzyme. In the present model reaction, traces of trisaccharides were observed from the early stage of the reaction (5–10 min) and were confirmed by mass spectrum analysis, but precise structures could not be elucidated. Importantly, when the ratio donor/acceptor was increased to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, no products of self-condensation were detected. Moreover, synthesis of furano-pyrano-disaccharides was strongly favored over hydrolysis of 1. After careful chromatographic purification, two main disaccharides 9 and 10 were isolated and their structures were elucidated by NMR spectroscopy. The major product 9 was obtained in the yield of 44%. It exhibited an intense three-bond coupling in the 13C–1H (HMBC) spectrum between H-1′ (5.09 ppm) and C-2 (79.2 ppm) as well as C-1′ (109.4 ppm) and H-2 (3.69 ppm) and was identified as pNP β-D-galactofuranosyl-(1→2)-α-D-glucopyranoside 9 (Fig. 2). The second regioisomer, isolated in 34% yield, presented a correlation between H-1′ (5.24 ppm) and C-3 (79.4 ppm) as well as between C-1′ (108.3 ppm) and H-3 (3.96 ppm). The compound was identified as (1→3)-linked disaccharide 10. The time-course analysis monitored by HPLC revealed that the (1→3)-disaccharide 10 was kinetically synthesized, followed by the formation of the (1→2)-isomer 9. This monitoring also showed that these compounds disappeared with time in favor of a third regioisomer. The yield of the latter reached its maximum after 4 hours of reaction and it was still present after 24 hours. The precise nature of the corresponding glycosidic linkage could however not be clearly elucidated. As a result, the overall yield of transglycosylation reached a maximum, slightly greater than 80%, between 5 and 20 minutes. In the present model reaction, traces of trisaccharides were observed from the early stage of the reaction (5–10 minutes) and confirmed by mass spectrum analysis, but precise structures could not be elucidated.
10, no products of self-condensation were detected. Moreover, synthesis of furano-pyrano-disaccharides was strongly favored over hydrolysis of 1. After careful chromatographic purification, two main disaccharides 9 and 10 were isolated and their structures were elucidated by NMR spectroscopy. The major product 9 was obtained in the yield of 44%. It exhibited an intense three-bond coupling in the 13C–1H (HMBC) spectrum between H-1′ (5.09 ppm) and C-2 (79.2 ppm) as well as C-1′ (109.4 ppm) and H-2 (3.69 ppm) and was identified as pNP β-D-galactofuranosyl-(1→2)-α-D-glucopyranoside 9 (Fig. 2). The second regioisomer, isolated in 34% yield, presented a correlation between H-1′ (5.24 ppm) and C-3 (79.4 ppm) as well as between C-1′ (108.3 ppm) and H-3 (3.96 ppm). The compound was identified as (1→3)-linked disaccharide 10. The time-course analysis monitored by HPLC revealed that the (1→3)-disaccharide 10 was kinetically synthesized, followed by the formation of the (1→2)-isomer 9. This monitoring also showed that these compounds disappeared with time in favor of a third regioisomer. The yield of the latter reached its maximum after 4 hours of reaction and it was still present after 24 hours. The precise nature of the corresponding glycosidic linkage could however not be clearly elucidated. As a result, the overall yield of transglycosylation reached a maximum, slightly greater than 80%, between 5 and 20 minutes. In the present model reaction, traces of trisaccharides were observed from the early stage of the reaction (5–10 minutes) and confirmed by mass spectrum analysis, but precise structures could not be elucidated.
Under similar conditions, using the β-glucopyranoside 3 as an acceptor (Fig. 2), and after 20 minutes of reaction, three disaccharides were chromatographically separated: pNP β-D-Galf-(1→2)-β-D-Glcp11, pNP β-D-Galf-(1→3)-β-D-Glcp12, and pNP β-D-Galf-(1→6)-β-D-Glcp13 were isolated in 26%, 25% and 30% yields, respectively. The time-course analysis revealed that the (1→2)-linked regioisomer 11 was the major kinetic product and is present together with the (1→3)-linked regioisomer 12 from the first minutes of the reaction. From about 15 minutes, the (1→6)-linked regioisomer 13 was formed and became the prevalent one after 1 hour. It was still detected in 15% yield after 24 hours. It is interesting to note that, when 12 was incubated with the biocatalyst and an excess of 3, it was transformed into 13. This emphasizes the striking stability of this (1→6)-bond in the presence of the furanosyl hydrolase Araf51.
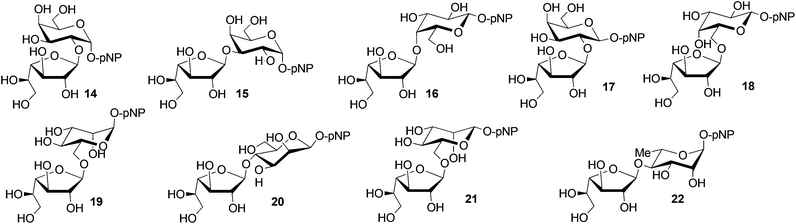
To go further with this methodology, we applied the Araf-51-assisted synthesis of furanosyl-containing disaccharides to three other families of acceptors. The use of galactosidic acceptors 4 and 5 did not fundamentally change the observations previously exposed with the gluco series. Three disaccharides with (1→2)- (14), (1→3)- (15), and (1→4)-linkage (16) were obtained from the α-anomer 4 and isolated in 73% overall yield (Table 1, entry 1). Substrate 5, characterized by a β-configuration, afforded only the (1→2)- (17) and (1→6)-disaccharides (18) in a near equimolecular ratio and in a 52% overall yield (entry 2).
On the other hand, the reaction with the α-mannopyranoside 6 (entry 3) was relatively rapid compared to other pyranosidic acceptors and the degree of hydrolysis of formed disaccharides was slightly higher. The reaction in a preparative scale was thus carried out for only 10 minutes. pNP β-D-Galf-(1→6)-α-D-Manp19 was obtained in 15% yield. 2-D NMR analyses confirmed that three other regioisomers were obtained as a mixture in an overall yield of 43%. The time-course analysis monitored by TLC revealed that these regioisomers are readily formed from the first minutes of reaction with the maximum yields reached between 5 and 10 minutes. The (1→6)-linked regioisomer 19 was formed from about 5 minutes of reaction.
The transglycosylation with the β-anomer 7 was apparently slower than that of the α-anomer and the conversion of pNP β-D-Galf still did not reach a maximum after two hours of reaction. However, the degree of hydrolysis was quite low, similarly to the other pNP pyranosides. After 25 minutes of the preparative-scale reaction, two regioisomers were isolated (entry 4). pNP β-D-Galf-(1→4)-β-D-Manp20 was obtained in a very high yield of 49%. The second regioisomer 21, identified as the (1→6)-disaccharide, was isolated in 16% yield. The time-course analysis confirmed kinetic preferences for the (1→4)-linked disaccharide 20. In about 40 minutes of reaction, both regioisomers were present equivalently. At the end of 2 hours incubation, the favoured (1→6)-linked regioisomer 21 prevailed. Structures were unambiguously established according to NMR data.
Finally, we also studied the L-rhamnopyranoside 8 since it presents significantly different reactivity and because it is widely found in Nature. While three disaccharides were observed after 20 minutes of the biocatalytic process, exclusively one regioisomer 22, characterized by a (1→4)-connection, was indeed isolated and in a fairly good 38% yield (entry 5). Evidence of the precise structure was established on the basis of NMR data. More specifically, the C-4 signal in the starting substrate 8 (δC-4 = 71.4 ppm) was shifted to 77.8 ppm within the disaccharide 22, thus demonstrating the (1→4)-coupling, and this result was corroborated by 2D NMR experiments.
Discussion
All results obtained for the transfer of a galactofuranosyl residue to pyranosidic acceptors mediated by the arabinofuranosidase Araf51 underline the ability of this enzyme to accept within its +1 subsite all the seven tested pyranosidic acceptors. Importantly, the anomeric configuration of the latter modulated the behavior of the furanosyl transfer. Usually, the (1→2)-disaccharides were the most common in the gluco and galacto series, and readily formed and stable. The (1→3) linkage was formed subsequently and was also frequently presented in these series. Interestingly, none of these α-anomers displayed the (1→6)-connection although for the β-anomers, it represents the major thermodynamically formed linkage. In Nature, exo-acting α-L-arabinofuranosidases release the arabinosyl decorations at C-2 and C-3 position of arabinogalactans and arabinoxylans from a range of plant structural polysaccharides, where both the galactose and xylose moieties are presented in a β-D-pyranose form. Thus, considering the structural similarity of xylose and glucose, the transglycosylation preferences correspond to the hydrolytic activity on natural substrates.Starting from β-D-mannopyranoside and α-L-rhamnopyranoside, the kinetically preferred linkage was the (1→4). It results from this observation that the adopted conformations within the active site of Manp and Rhap derivatives vary to some extent compared to the glucosidic and galactosidic monosaccharides. Thus the axial C-2 hydroxyl group in mannose, in contrast to glucose, seems to impact the conformation within the active site more markedly than the orientation of C-4 OH group distinguishing glucose from galactose. Overall, in all the three series where both anomers were tested (D-Glcp, D-Galp, D-Manp), the reactions involving the α-anomer proceeded markedly faster than those of their β-anomeric counterparts. The time-course analyses also revealed the crucial effect of the reaction time on the ratio of regioisomers formed in individual reactions. Thus, in most of cases, it is possible to isolate either kinetic or thermodynamic disaccharide with a very low quantity of the other regioisomer with respect to the time-course of the reaction.
In order to elucidate binding modes of acceptors 2–7 in transglycosylation reactions, complexes of Araf51 with all putative transglycosylation products were subjected to molecular dynamics simulations (24 × 10 ns). Each putative product was docked into the active site by rigid fit of its Galf moiety onto the Araf moiety in the experimental structure. The pyranosyl and pNP-moieties were adjusted manually before the simulation. The results of simulations for disaccharides 11, 14, 17 and 18 are shown in Fig. 3. These selected products showed stable binding in the active site illustrated by low root-mean-square deviation (RMSD). The results also demonstrated that binding modes of (1→2) products are very similar. The pNP-moiety is oriented perpendicularly to the access of the active site. On the other hand, the pNP-moiety in 18 [β-D-Galf-(1→6)-β-D-Galp], as an example of (1→6) product, is placed along the axis of the active site. The fact that the pNP-moiety may orient in different angles explains relatively broad acceptor specificity. Most other putative products have shown high RMSD which indicate either inability of the product to bind into the active site or incorrect docking. Unfortunately, attempts to quantitatively predict acceptor preferences from molecular simulations or free-energy methods were not successful (not shown).
Among the disaccharides prepared throughout this part, several display a configuration reported from uncommon cell wall glycoconjugates of some pathogenic species. Galactofuranosyl residue connected to D-Glcp moiety through the (1→3)-linkage has been identified e.g. in Streptococcus species,34 the one with the (1→6)-linkage is reported from Escherichia coli K-12 strain.35 The (1→3)-linkage to D-Galp was identified in Fibrobacter succinogenes36 or in the O-antigen repeating unit of Klebsiella pneumoniae.36 In Mycobacterium species37–40 and Plesiomonas shigelloides,41 β-D-Galf is connected to L-Rhap through the (1→4)-linkage. The sequence β-D-Galf-(1→2)-D-Manp is present in many microorganisms, among the pathogens notably Cryphonectria parasitica42 or Trichoderma.43 β-D-Galf-(1→3)-D-Manp is frequent in Aspergillus,44Trypanosoma cruzi12,45 and Leishmania,11 β-D-Galf-(1→6)-D-Manp e.g. in Aspergillus,44Leishmania11 or Paracoccidioides brasiliensis.46
Conclusions
A chemo-enzymatic synthesis of galactofuranosyl-containing disaccharides was proposed mediated by the thermophilic arabinofuranosidase Araf51. We have first demonstrated that a large excess of acceptor allowed overcoming the self-condensation side reaction. Secondly, transglycosylation products were obtained in the early stage of the biocatalyzed process and increased reaction times did not affect the target furanosyl-pyranoside disaccharides since the degree of hydrolysis of the latter remained very low. These factors together were fruitful from a synthetic point of view. Consequently, and with regard to the very good yields obtained with the assistance of the hydrolytic furanosidase Araf51, this chemo-enzymatic approach constitutes a very interesting alternative to multi-step chemical synthesis of various mimetics of biologically significant structures. The +1 subsite of Araf51 was able to recognize simple glycopyranosides as acceptors, even a Rhap derivative and its 1C4 conformation. Among targets were prepared disaccharides biosynthesized by Mycobacteria, Leishmania, Trypanosoma or Paracoccidioides microorganisms.Experimental section
General remarks
Prior to NMR analysis, fractions were exchanged in D2O (99.9% purity) at room temperature with intermediate freeze-drying, and then dissolved in 400 μL of D2O. 1H, 13C, COSY, HSQC, HMBC, TOCSY and NOESY NMR spectra were recorded at the Laboratory of NMR spectroscopy (ICT Prague, Czech Republic) on a Bruker 600 Avance spectrometer equipped with a cryoprobe at 600 MHz for 1H and 125 MHz for 13C, and on a Bruker ARX 400 at 400 MHz for 1H, 100 MHz for 13C at ENSCR (France). Chemical shifts are given in δ-units (ppm). Coupling constants J are given in Hz.The HRMS were measured at the Centre Régional de Mesures Physiques de l'Ouest (CRMPO, Université de Rennes 1, France) with a MS/MS ZabSpec TOF Macromass using m-nitrobenzylic alcohol as the matrix and accelerated caesium ions for ionization and at the Laboratory of Mass Spectrometry (ICT Prague, Czech Republic) using a Q-TOF Micro (Waters, USA), where electrospray-ionisation mass spectra (ESI-MS) were recorded on samples dissolved in MeOH injected in a volume of 2–5 μL into a flow (100 μL min−1) of MeOH. Sample cone voltage was 42 V and the source temperature was 150 °C. Measurements were performed in positive ([M + Na]+ ion detection) mode in the range of 100–1000 Da. Finally, at the Mass Spectrometry Group (Institute of Organic Chemistry and Biochemistry, Academy of Sciences of the Czech Republic) on LTQ-Orbitrap XL (THERMO), where ESI+ spectra were recorded on samples dissolved in MeOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), sample cone voltage was 40 V.
1), sample cone voltage was 40 V.
Thin layer chromatography (TLC) analyses were conducted on Kieselgel 60 F254 (Merck) plates with 0.2 mm layer thickness. Spots were visualized by UV (254 nm) and by exposure to 0.2% w/v orcinol in H2SO4 (20% v/v) in ethanol. For column chromatography, either Si 60 (40–63 μm) Silica gel or pre-packed Chromabond®Flash RS 15 SiOH columns (Macherey-Nagel) were used.
Gel permeation chromatography was performed on P-2 Bio-Gel (Bio-Rad) using FPLC system consisting of a solvent delivery system Biologic F40 DuoFlow, Biologic QuadTec UV-Vis Detector and Biologic BioFrac Fraction Collector (all Bio-Rad). Deionized filtrated (0.22 μm PVDF membrane, Millipore) water was used as a mobile phase with a flow rate of 0.15 mL min−1. Separation was monitored by UV absorbance at 280 and 405 nm by operating software Biologic DuoFlow. Collected fractions were lyophilized (FreeZone Freeze Dry System, Labconco).
Recombinant arabinofuranosidase Araf51 was produced in E. coli and purified as already described.47 The hydrolytic activity toward p-nitrophenyl pyranosides was tested by incubation of the solution of enzyme (obtained after affinity Ni-NTA chromatography and exchanged to 50 mM PBS pH 7.4 by PD10 gel chromatography) with individual glycopyranosides (5 mM) in 50 mM potassium phosphate buffer pH 7.4 at 60 °C. The release of p-nitrophenolate was continuously measured at 405 nm (Microplate Spectrophotomerer PowerWave XS/XS2, BioTek) and data evaluated with Gen5 Data Analysis Software (BioTek). pNP α-D-Galp, pNP β-D-Galp, pNP α-D-Glcp, pNP β-D-Glcp, pNP α-D-Manp, pNP β-D-Manp and pNP α-L-Rhap are commercially available.
General procedure for the transglycosylation reactions
Prior to preparative-scale reactions, the time-course analyses were performed in the analytical scale (5 μmol of individual pNP pyranosides, 0.5 μmol of pNP β-D-galactofuranoside and 36 U of Araf51 in 100 μL of 50 mM PBS pH 7.4). The reaction was monitored by TLC (7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 EtOAc–AcOH–H2O). Preparative-scale reactions were performed at 60 °C by incubation of the pNP β-D-galactofuranoside (20 mg, 66 μmol; or 10 mg, 33 μmol) with individual pNP pyranosides (200 mg, 660 μmol for pNP D-Glcp and pNP D-Galp; 100 mg, 330 μmol for pNP D-Manp and pNP L-Rhap) and 4800 U of Araf51 (for reactions with pNP D-Glcp and pNP D-Galp) or 2400 U of Araf51 (for reactions with pNP D-Manp and pNP L-Rhap) in 50 mM PBS pH 7.4 in a total volume with regard to the solubility of substrates: 5 mL (reactions with α- and β-D-Galp and β-D-Glcp) or 10 mL (α-D-Glcp and α- and β-D-Manp) or 15 mL (α-L-Rhap).
2 EtOAc–AcOH–H2O). Preparative-scale reactions were performed at 60 °C by incubation of the pNP β-D-galactofuranoside (20 mg, 66 μmol; or 10 mg, 33 μmol) with individual pNP pyranosides (200 mg, 660 μmol for pNP D-Glcp and pNP D-Galp; 100 mg, 330 μmol for pNP D-Manp and pNP L-Rhap) and 4800 U of Araf51 (for reactions with pNP D-Glcp and pNP D-Galp) or 2400 U of Araf51 (for reactions with pNP D-Manp and pNP L-Rhap) in 50 mM PBS pH 7.4 in a total volume with regard to the solubility of substrates: 5 mL (reactions with α- and β-D-Galp and β-D-Glcp) or 10 mL (α-D-Glcp and α- and β-D-Manp) or 15 mL (α-L-Rhap).
Based on the results from analytical-scale reactions, individual reactions proceeded for 20 min with the exception of α-D-Manp (15 min) and β-D-Manp (25 min) to obtain the maximum ratio of transglycosylation/hydrolysis. The reactions were stopped by enzyme denaturation at 100 °C for 10 min. The reaction products were repeatedly separated by silica gel flash chromatography using EtOAc–AcOH–H2O in ratios from 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 40
1 to 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Separations were monitored by TLC, fractions corresponding to individual regioisomers were collected, evaporated and lyophilized several times with intermediate dissolving in H2O, finally in D2O and subjected to structural analyses.
1. Separations were monitored by TLC, fractions corresponding to individual regioisomers were collected, evaporated and lyophilized several times with intermediate dissolving in H2O, finally in D2O and subjected to structural analyses.
Molecular modeling
All simulations were performed in Gromacs 4.5.5.48 Each system included one molecule of Araf51 (from the experimental structure, PDB I.D. 2C8N),49 one molecule of putative transglycosylation product, ∼18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 water molecules and 15 sodium counter-ions. Protein was modelled using Amber99SB force field,50 transglycosylation products were modelled using Glycam0651 (carbohydrates) and General Amber Force Field52 (pNP-moiety). Charges were calculated by RESP method on the basis of HF/6-31G*//HF/6-31G* wavefunction calculated for individual fragments in Gaussian 03.53 Molecules of putative products were placed to the active site by 3D-alignment of the Galf moiety with the Araf moiety in the experimental structure. This was followed by manual adjustment of glycosidic bond torsions to avoid steric clashes. Finally, the system was minimized (3000 steps of L-BFGS and 500 steps of steepest descent) and simulated by 1 ns of equilibration and 10 ns of production simulation. Non-hydrogen atoms of the protein and Galf were restrained during the equilibration run by a harmonic restraint potential.
300 water molecules and 15 sodium counter-ions. Protein was modelled using Amber99SB force field,50 transglycosylation products were modelled using Glycam0651 (carbohydrates) and General Amber Force Field52 (pNP-moiety). Charges were calculated by RESP method on the basis of HF/6-31G*//HF/6-31G* wavefunction calculated for individual fragments in Gaussian 03.53 Molecules of putative products were placed to the active site by 3D-alignment of the Galf moiety with the Araf moiety in the experimental structure. This was followed by manual adjustment of glycosidic bond torsions to avoid steric clashes. Finally, the system was minimized (3000 steps of L-BFGS and 500 steps of steepest descent) and simulated by 1 ns of equilibration and 10 ns of production simulation. Non-hydrogen atoms of the protein and Galf were restrained during the equilibration run by a harmonic restraint potential.
Acknowledgements
I.C. thanks the Czech Ministry of Education for a grant (MSM: 6046137305). Financial support by the Ministère Français des Affaires Etrangères, the Programme Hubert Curien (Barrande), the Agence Nationale de la Recherche (JCJC06_140075), and the GDR-RDR2 “Aller vers une chimie éco-compatible” are gratefully acknowledged, as well as Prof. G. Davies for supplying the plasmid of Araf51. Molecular modeling was supported by access to high performance computing facilities (MetaCentrum – LM2010005 and CERIT-SC – CZ. 1.05/3.2.00/08.0144), by COST (CM1102 – MultiGlycoNano) and the Ministry of Education, Youth and Sports of the Czech Republic (LD13024 and 7AMB13FR014). The authors are finally grateful to the GlycoOuest network, supported by the Région Bretagne and the Région Pays de la Loire.Notes and references
- R. Mody, S. H. A. Joshi and W. Chaney, J. Pharmacol. Toxicol. Methods, 1995, 33, 1–10 CrossRef CAS.
- E. Gorelik, U. Galili and A. Raz, Cancer Metastasis Rev., 2001, 20, 245–277 CrossRef CAS.
- P. Peltier, R. Euzen, R. Daniellou, C. Nugier-Chauvin and V. Ferrières, Carbohydr. Res., 2008, 343, 1897–1923 CrossRef CAS PubMed.
- M. R. Richards and T. L. Lowary, ChemBioChem, 2009, 10, 1920–1938 CrossRef CAS PubMed.
- H. A. Taha, M. R. Richards and T. L. Lowary, Chem. Rev., 2013, 113, 1851–1876 CrossRef CAS PubMed.
- J. B. Houseknecht, T. L. Lowary and C. M. Hadad, J. Phys. Chem. A, 2002, 107, 372–378 CrossRef.
- P. Nassau, S. Martin, R. Brown, A. Weston, D. Monsey, M. McNeil and K. Duncan, J. Bacteriol., 1996, 178, 1047–1052 CAS.
- R. Köplin, J. R. Brisson and C. Whitfield, J. Biol. Chem., 1997, 272, 4121–4128 CrossRef PubMed.
- F. Pan, M. Jackson, Y. Ma and M. McNeil, J. Bacteriol., 2001, 183, 3991–3998 CrossRef CAS PubMed.
- S. M. Beverley, K. L. Owens, M. Showalter, C. L. Griffith, T. L. Doering, V. C. Jones and M. R. McNeil, Eukaryot. Cell, 2005, 4, 1147–1154 CrossRef CAS PubMed.
- M. J. McConville, S. W. Homans, J. E. Thomas-Oates, A. Dell and A. Bacic, J. Biol. Chem., 1990, 265, 7385–7394 CAS.
- R. M. de Lederkremer and W. Colli, Glycobiology, 1995, 5, 547–552 CrossRef CAS PubMed.
- F. A. Shaikh and S. G. Withers, Biochem. Cell Biol., 2008, 86, 169–177 CrossRef CAS PubMed.
- M. L. Sinnott, Chem. Rev., 1990, 90, 1171–1202 CrossRef CAS.
- K. N. Jayaprakash, J. Lu and B. Fraser-Reid, Angew. Chem., Int. Ed., 2005, 117, 6044–6048 CrossRef PubMed.
- B. Fraser-Reid, J. Lu, K. N. Jayaprakash and J. C. López, Tetrahedron: Asymmetry, 2006, 17, 2449–2463 CrossRef CAS PubMed.
- A. Hölemann, B. L. Stocker and P. H. Seeberger, J. Org. Chem., 2006, 71, 8071–8088 CrossRef PubMed.
- Y. Ito and S. Hanashima, in Experimental Glycoscience, ed. N. Taniguchi, A. Suzuki, Y. Ito, H. Narimatsu, T. Kawasaki and S. Hase, Springer, Japan, 2008, pp. 210–216 Search PubMed.
- P. Bojarova and V. Kren, Trends Biotechnol., 2009, 27, 199–209 CrossRef CAS PubMed.
- R. W. Gantt, P. Peltier-Pain and J. S. Thorson, Nat. Prod. Rep., 2011, 28, 1811–1853 RSC.
- M. Joe, Y. Bai, R. C. Nacario and T. L. Lowary, J. Am. Chem. Soc., 2007, 129, 9885–9901 CrossRef CAS PubMed.
- M. McGeachy, Y. Chen, C. Tato, A. Laurence, B. Joyce-Shaikh, W. Blumenschein, T. McClanahan, J. O'Shea and D. Cua, Nat. Immunol., 2009, 10, 314–324 CrossRef CAS PubMed.
- C. A. G. M. Weijers, M. C. R. Franssen and G. M. Visser, Biotechnol. Adv., 2008, 26, 436–456 CrossRef CAS PubMed.
- I. Chlubnova, L. Legentil, R. Dureau, A. Pennec, M. Almendros, R. Daniellou, C. Nugier-Chauvin and V. Ferrières, Carbohydr. Res., 2012, 356, 44–61 CrossRef CAS PubMed.
- C. Rémond, R. Plantier-Royon, N. Aubry and M. J. O'Donohue, Carbohydr. Res., 2005, 340, 637–644 CrossRef PubMed.
- M. G. Szczepina, R. B. Zheng, G. C. Completo, T. L. Lowary and B. M. Pinto, ChemBioChem, 2009, 10, 2052–2059 CrossRef CAS PubMed.
- N. L. Rose, G. C. Completo, S.-J. Lin, M. R. McNeil, M. M. Palcic and T. L. Lowary, J. Am. Chem. Soc., 2006, 128, 6721–6729 CrossRef CAS PubMed.
- N. L. Rose, G. C. Completo, S.-J. Lin, M. McNeil, M. M. Palcic and T. L. Lowary, J. Am. Chem. Soc., 2006, 128, 6721–6729 CrossRef CAS PubMed.
- N. L. Rose, R. B. Zheng, J. Pearcey, R. Zhou, G. C. Completo and T. L. Lowary, Carbohydr. Res., 2008, 343, 2130–2139 CrossRef CAS PubMed.
- C. D. Brown, M. S. Rusek and L. L. Kiessling, J. Am. Chem. Soc., 2012, 134, 6552–6555 CrossRef CAS PubMed.
- C. Zhang, B. R. Griffith, Q. Fu, C. Albermann, X. Fu, I.-K. Lee, L. Li and J. S. Thorson, Science, 2006, 313, 1291–1294 CrossRef CAS PubMed.
- R. W. Gantt, P. Peltier-Pain, W. J. Cournoyer and J. S. Thorson, Nat. Chem. Biol., 2011, 7, 685–691 CrossRef CAS PubMed.
- I. Chlubnova, D. Filipp, V. Spiwok, H. Dvorakova, R. Daniellou, C. Nugier-Chauvin, B. Kralova and V. Ferrières, Org. Biomol. Chem., 2010, 8, 2092–2102 CAS.
- J. E. Germond, M. Delley, N. D'Amico and S. J. F. Vincent, Eur. J. Biochem., 2001, 5149–5156 CrossRef CAS.
- G. Stevenson, B. Neal, D. Liu, M. Hobbs, N. H. Packer, M. Batley, J. W. Redmond, L. Lindquist and P. Reeves, J. Bacteriol., 1994, 176, 4144–4156 CAS.
- E. Vinogradov, E. E. Egbosimba, M. B. Perry, J. S. Lam and C. W. Forsberg, Eur. J. Biochem., 2001, 3566–3576 CrossRef CAS.
- M. McNeil, M. Daffe and P. J. Brennan, J. Biol. Chem., 1990, 265, 18200–18206 CAS.
- M. Daffe, P. J. Brennan and M. McNeil, J. Biol. Chem., 1990, 265, 6734–6743 CAS.
- G. S. Besra, K. H. Khoo, M. R. McNeil, A. Dell, H. R. Morris and P. J. Brennan, Biochemistry, 1995, 34, 4257–4266 CrossRef CAS.
- D. Kaur, T. L. Lowary, V. D. Vissa, D. C. Crick and P. J. Brennan, Microbiology, 2002, 148, 3049–3057 CAS.
- M. Linnerborg, A. Weintraub and G. Widmalm, Eur. J. Biochem., 1999, 460–466 CrossRef CAS.
- A. Molinaro, V. Piscopo, R. Lanzetta and M. Parrilli, Carbohydr. Res., 2002, 340, 1707–1713 CrossRef.
- A. Prieto, J. A. Leal, A. Poveda, J. Jimenéz-Barbero, B. Gómez-Miranda, J. Domenech, O. Ahrazem and M. Bernabé, Carbohydr. Res., 1997, 304, 281–291 CrossRef CAS.
- J. P. Latgé, H. Kobayashi, J. P. Debeaupuis, M. Diaquin, J. Sarfati, J. M. Wieruszeski, E. Parra, J. P. Bouchara and B. Fournet, Infect. Immun., 1994, 62, 5424–5433 Search PubMed.
- R. M. de Lederkremer, C. Lima, M. I. Ramirez, M. A. Ferguson, S. W. Homans and J. Thomas-Oates, J. Biol. Chem., 1991, 23670–23675 CAS.
- S. B. Levery, M. S. Toledo, E. Suzuki, M. E. K. Salyan, S.-i. Hakomori, A. H. Straus and H. K. Takahashi, Biochem. Biophys. Res. Commun., 1996, 222, 639–645 CrossRef CAS.
- E. Taylor, N. Smith, J. Turkenburg, S. D'Souza, H. Gilbert and G. Davies, Biochem. J., 2006, 395, 31–37 CrossRef CAS PubMed.
- B. Hess, C. Kutzner, D. v. d. Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS.
- E. J. Taylor, N. L. Smith, J. P. Turkenburg, S. D'Souza, H. J. Gilbert and G. J. Davies, Biochem. J., 2006, 395, 31–37 CrossRef CAS PubMed ; PDB code: 32C38N.
- V. Hornak, R. Abel, A. Okur, B. Strockbine, A. Roitberg and C. Simmerling, Proteins, 2006, 65, 712–725 CrossRef CAS PubMed.
- K. N. Kirschner, A. B. Yongye, S. M. Tschampel, J. González-Outeiriño, C. R. Daniels, B. L. Foley and R. J. Woods, J. Comput. Chem., 2008, 29, 622–655 CrossRef CAS PubMed.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, J. J. A. Montgomery, T. Vreven, K. N. Kudin, J. C. Burant, J. M. Millam, S. S. Iyengar, J. Tomasi, V. Barone, B. Mennucci, M. Cossi, G. Scalmani, N. Rega, G. A. Petersson, H. Nakatsuji, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, M. Klene, X. Li, J. E. Knox, H. P. Hratchian, J. B. Cross, V. Bakken, C. Adamo, J. Jaramillo, R. Gomperts, R. E. Stratmann, O. Yazyev, A. J. Austin, R. Cammi, C. Pomelli, J. W. Ochterski, P. Y. Ayala, K. Morokuma, G. A. Voth, P. Salvador, J. J. Dannenberg, V. G. Zakrzewski, S. Dapprich, A. D. Daniels, M. C. Strain, O. Farkas, D. K. Malick, A. D. Rabuck, K. Raghavachari, J. B. Foresman, J. V. Ortiz, Q. Cui, A. G. Baboul, S. Clifford, J. Cioslowski, B. B. Stefanov, G. Liu, A. Liashenko, P. Piskorz, I. Komaromi, R. L. Martin, D. J. Fox, T. Keith, M. A. Al-Laham, C. Y. Peng, A. Nanayakkara, M. Challacombe, P. M. W. Gill, B. Johnson, W. Chen, M. W. Wong, C. Gonzalez and J. A. Pople, Gaussian 03, Revision C.02, Gaussian, Inc., Wallingford, CT, 2004 Search PubMed.
Footnote |
| † Present address: Institut de Chimie Organique et Analytique (ICOA), UMR CNRS 7311, Université d'Orléans, BP 6759, Rue de Chartres, 45067 Orléans Cedex 2, France. |
| This journal is © The Royal Society of Chemistry 2014 |