Design, synthesis and characterization of novel inhibitors against mycobacterial β-ketoacyl CoA reductase FabG4†
Deb Ranjan
Banerjee
a,
Debajyoti
Dutta
b,
Baisakhee
Saha
b,
Sudipta
Bhattacharyya
b,
Kalyan
Senapati
a,
Amit K.
Das
*b and
Amit
Basak
*a
aIIT Kharagpur, Chemistry, Kharagpur, West Bengal, India. E-mail: absk@chem.iitkgp.ernet.in
bIIT Kharagpur, Biotechnology, Kharagpur, West Bengal, India
First published on 19th September 2013
Abstract
We report the design and synthesis of triazole-polyphenol hybrid compounds 1 and 2 as inhibitors of the FabG4 (Rv0242c) enzyme of Mycobacterium tuberculosis for the first time. A major advance in this field occurred only a couple of years ago with the X-ray crystal structure of FabG4, which has helped us to design these inhibitors by the computational fragment-based drug design (FBDD) approach. Compound 1 has shown competitive inhibition with an inhibition constant (Ki) value of 3.97 ± 0.02 μM. On the other hand, compound 2 has been found to be a mixed type inhibitor with a Ki value of 0.88 ± 0.01 μM. Thermodynamic analysis using isothermal titration calorimetry (ITC) reveals that both inhibitors bind at the NADH co-factor binding domain. Their MIC values, as determined by resazurin assay against M. smegmatis, indicated their good anti-mycobacterial properties. A preliminary structure–activity relationship (SAR) study supports the design of these inhibitors. These compounds may be possible candidates as lead compounds for alternate anti-tubercular drugs. All of the reductase enzymes of the Mycobacterium family have a similar ketoacyl reductase (KAR) domain. Hence, this work may be extrapolated to find structure-based inhibitors of other reductase enzymes.
Introduction
Finding a new target to fight an existing disease has always been at the center of research in medicinal chemistry. Tuberculosis (TB) is one such existing disease which is still causing problems for mankind.1 The World Health Organization (WHO) has estimated that about one-third of the world population is infected by its causative agent Mycobacterium tuberculosis (Mtb).2 The emergence of multi drug resistant (MDR) and extremely drug resistant (XDR) Mtb strains has complicated the scenario.3 First line TB drugs such as isoniazid, rifampicin, pyraniazid, ethambutol and streptomycin have failed in recent TB cases, so it is important to find alternative drugs to fight against tuberculosis.M. tuberculosis possesses a lipid-rich cell envelope which is required for its survival within the host cell. The main component of this virtually impenetrable cell wall, mycolic acids, is produced through fatty acid synthesis (FAS). Two fatty acid synthesis pathways, FAS-I and FAS-II, have been reported in Mycobacterium.4 FAS-I, which is involved in the de novo synthesis of fatty acids, is a multi-domain enzyme. The FAS-II pathway is necessary for the synthesis of long chain fatty acids. These long-chain fatty acids are finally used in mycolic acid synthesis. Hence, the enzymes involved in FAS are attractive targets for drug designing. FabG, a ketoacyl reductase, is one of the major enzymes involved in FAS. The Mtb genome contains five FabG genes,5 but only two, FabG1 (Rv1483) and FabG4 (Rv0242c), are conserved among the mycobacterial species. FabG1 is a well-known β-ketoacyl CoA reductase6 and has been a good candidate for alternate drug discovery in the last decade.7 FabG4 is the less explored gene, which has recently been reported to be an essential and functional gene for bacterial growth, survival8 and fatty acid synthesis.9 FabG4 may also have a role in the drug resistance of mycobacterial species as it is over-expressed in sub-inhibitory concentrations of Streptomycin.10 These reports in the last three years have made FabG4 a new attractive target to fight tuberculosis.
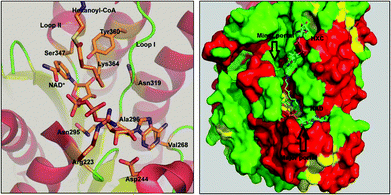
FabG4 is a high molecular weight ketoacyl reductase (HMwFabG). The crystal structure of FabG4 shows the presence of two distinct domains, domain I and II.11 Domain I is an extra N-terminal domain, and domain II is a typical ‘ketoacyl CoA reductase (KAR) domain’. FabG4 is a NADH-dependent ketoacyl reductase, whereas FabG1 is a NADPH-dependent ketoacyl reductase enzyme. The conserved catalytic tetrad Ser347, Tyr360, Lys364 and Asn319 constitute the active site residues of the KAR domain. The active site is covered by loop I and loop II. The structure of FabG4 co-crystallized with hexanoyl-CoA (HXC) and NAD+ (PDB ID: 3V1U) suggests that the active site of FabG4 could be accessed from two different sides: the major portal and minor portal.12 The major portal is wide open and is accessed by the cofactor to bind with the enzyme. The minor portal is narrower due to the helices of the dimeric interface and is accessed by the 4 phosphopantetheine-bound fatty acyl substrates. The active site is covered from one side by two conserved loops, loop I and loop II. The conserved NAG triad (Asn295, Ala296, Gly297) of loop I interacts with the pyrophosphate section of the substrate and guides the cofactor towards the active site. Asp244 and Val268 have a crucial role to hold NADH at the major portal by interacting with the adenine section of the coenzyme. Asp244 and Arg223 are the major factors behind NADH selectivity (Fig. 1).
Based on the information about the overall structure and specificity of the FabG4 enzyme, we have attempted to design novel structure-based inhibitors of FabG4. The synthesis of the designed compounds were carried out and their bio-activities were measured though biochemical assays. The drug susceptibilities of these inhibitors were tested against Mycobacterium smegmatis. A preliminary structure activity relationship (SAR) has been carried out to justify the design of the inhibitors.
Results and discussion
Design strategy
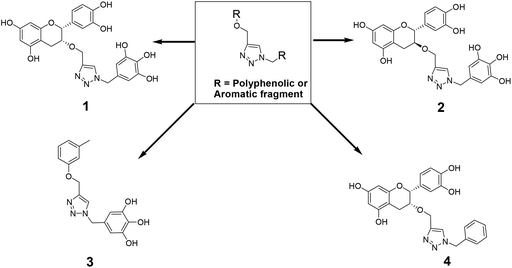
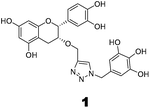
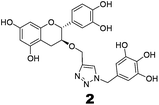
The recently-solved X-ray crystal structure of FabG4 with co-factor NADH provides the way to design the first inhibitors against this enzyme. We targeted the co-factor (NADH)-binding domain for design purposes. Inhibitors which target the NADH binding site can interact with the three subsites of the coenzyme binding domain: the nicotinamide-binding subsite (N-subsite), the adenosine-binding subsite (A-subsite) and the pyrophosphate-binding subsite (P-subsite). For structure-based design purposes, we followed the computational fragment-based drug design (FBDD) strategy.13 In this approach, fragments and linkers were selected from a literature survey and merged to form a library. The selection of the lead compounds was performed through computational docking studies. The advanced molecular grid-based docking program Autodock4.214 was used for the docking of a flexible ligand within a flexible protein.Epigallocatechin gallate and related plant polyphenols are known inhibitors of bacterial FabG.15 Hence, polyphenols were selected as one of the components of the fragment-based library. Moreover, in recent years, triazoles have received much attention as a central core in medicinal chemistry, and especially in the synthesis of anti-tubercular agents.16 The 1,4-triazole moiety can also be used as a conformationally constrained bioisosteric replacement of the pyrophosphate linker of NADH.17 With this in mind, we selected a 1,4-triazole linker for lead optimization. From the lead library, two novel 1,4-triazole linked polyphenols 1 and 2 were selected as possible inhibitors of FabG4 on the basis of their docking score (Fig. 2). The docking study suggested that the binding of the 1,4-triazole moiety occurs at the pyrophosphate binding region (P-subsite), whereas the polyphenol fragments compete for the N- and A-subsites of the NADH binding region. The detailed interactions from the docking studies are discussed later in this report. To show the importance of fragments linked by the 1,4-triazole moiety, two more hybrids, 3 and 4, were designed to draw preliminary structure–activity relationships.
Synthesis
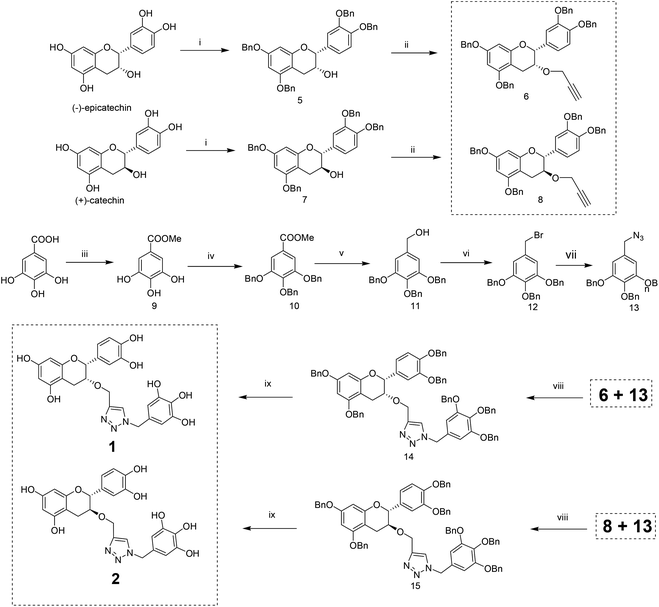
The methodology to obtain triazole derivatives was Huisgen's 1,3-dipolar cycloaddition of azides and alkynes (click protocol).18 A click reaction was performed in the presence of Cu(I), made in situ by the reduction of CuSO4 with sodium ascorbate. As expected, the regioselectivity of the click reaction was controlled by the Cu(I) catalyst, and only 1,4-regioisomers were formed.19 The respective azide and alkyne counterparts were synthesized from relatively cheaper and easily available starting materials. The benzyl ether was chosen as the protecting group of the polyphenol because of its easy deprotection under neutral conditions which will avert the possible racemization of the polyphenol at the C-2 position.For the synthesis of compounds 1 and 2, (−)-epicatechin and (+)-catechin were used as the starting materials respectively. To obtain the alkyne component for the click reaction, tetrabenzyl catechin or epicatechin was propargylated to 3-O-propargyl tetrabenzyl catechin or epicatechin. The azide component was made from gallic acid. Thus, tri-O-benzyl gallic acid was converted to the respective alcohol by NaBH4 reduction in ethanol. The SN2 replacement of the benzylic –OH by bromide, followed by azide, led to the other component for the click reaction. The click reaction was performed in the presence of Cu(I) made in situ by the reduction of CuSO4 with sodium ascorbate. Debenzylation with H2 in the presence of Pearlman’s catalyst resulted in the final compounds (Scheme 1).
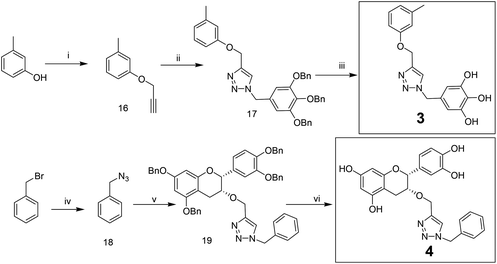
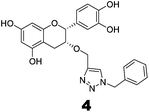
Compounds 3 and 4 were synthesized via a similar procedure using a click protocol (Scheme 2). m-Cresol was selected as the starting material to obtain compound 3. For the synthesis of compound 4, a click reaction was carried out between benzyl azide and 3-O-propargyl tetrabenzyl epicatechin. Finally, debenzylation using 20% Pd(OH)2–C at high hydrogen pressure afforded the final compounds.
All of the final compounds were purified by repeated precipitation from methanol–ether and characterized by 1H and 13C NMR, and mass spectrometry. The purity of these compounds was determined through reverse-phase analytical HPLC (see ESI†).
Inhibition kinetics
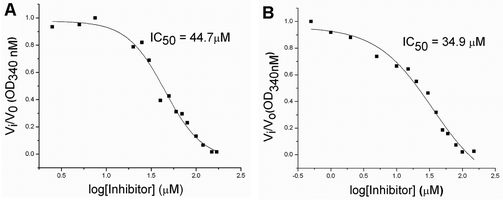
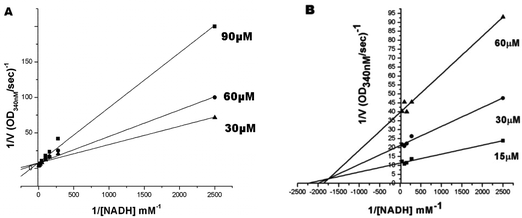
To evaluate the inhibitory effects of our compounds, the inhibition kinetics were carried out at 25 °C. The decrease in the absorbance value at 340 nM due to the conversion of NADH to NAD+ was monitored in this assay. Half maximal inhibitory concentration (IC50) values and modes of inhibition were evaluated kinetically. (−)-Epicatechin and (+)-catechin, the starting materials of compounds 1 and 2 respectively, did not inhibit FabG4 up to a 100 μM concentration. However, our designed and synthesized compounds 1 and 2 showed promising inhibition against Mtb FabG4. Both the compounds inhibited FabG4 at low micromolar concentrations. The IC50 values of compounds 1 and 2 are 44.7 μM and 34.9 μM respectively (Fig. 3). Compound 1 showed competitive inhibition with an inhibition constant (Ki) value of 3.97 ± 0.02 μM. On the other hand, compound 2 was found to be a mixed type inhibitor with a Ki value of 0.88 ± 0.01 μM (Fig. 4).Thermodynamic analysis
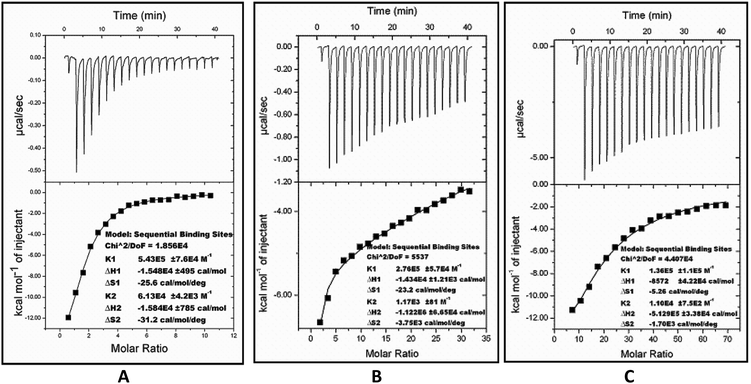
The thermodynamic parameters for inhibition were obtained from isothermal titration calorimetry (ITC). The ITC studies of FabG4 with compound 1, compound 2 and NADH were carried out at 25 °C. The FabG4–NADH titration was first performed as a control experiment because NADH was used as a substrate in the inhibition studies. The binding curve of the FabG4–NADH titration nicely fits to the ‘sequential binding’ mode, with the number of binding sites (N) equal to 2 (Fig. 5A). This sequential nature of binding is reasonable because FabG4 exists as an inseparable homo-dimer in solution. NADH binding to one monomeric unit regulates the binding of a second NADH to the second monomer, and this is co-operative in nature. This phenomenon is similar to other known FabGs where the oligomeric interface is responsible for cooperativity.20 NADH binding to both the monomers is spontaneous, as suggested by the ΔG values obtained from ITC (Table 1). The first binding constant (K1) is higher than the second binding constant (K2), which indicates negative co-operativity.| Ligand | Mode of binding | Binding constant K (mol−1) | Thermodynamic parameters | ||
|---|---|---|---|---|---|
| ΔH (kcal mol−1) | ΔS (cal mol−1 K−1) | ΔG (kcal mol−1) | |||
| NADH | Sequential binding, with number of binding sites (N) = 2 | K 1 = 5.43 × 105 | ΔH1 = −15.4 | ΔS1 = −25.6 | ΔG1 = −7.8 |
| K 2 = 6.13 × 104 | ΔH2 = −15.8 | ΔS2 = −31.2 | ΔG2 = −6.6 | ||
| Compound 1 | Sequential binding, with number of binding sites (N) = 2 | K 1 = 2.76 × 105 | ΔH1 = −14.3 | ΔS1 = −23.2 | ΔG1 = −7.4 |
| K 2 = 1.17 × 103 | ΔH2 = −1.1 × 103 | ΔS2 = −3.7 × 103 | ΔG2 = −4.5 | ||
| Compound 2 | Sequential binding, with number of binding sites (N) = 2 | K 1 = 1.36 × 105 | ΔH1 = −8.5 | ΔS1 = −5.2 | ΔG1 = −6.9 |
| K 2 = 1.10 × 104 | ΔH2 = −5.1 × 102 | ΔS2 = −1.7 × 103 | ΔG2 = −6.4 | ||
Compound 1, which has been found to be a competitive inhibitor with respect to NADH, showed a similar binding nature. The calorimetry of compound 1 with FabG4 revealed that it also binds sequentially to each monomer, as in the case of NADH (Fig. 5B). Thus, the calorimetric data supports our previous finding that compound 1 competes with NADH for the same binding sites. The binding of compound 1 is spontaneous, with a negative ΔG (Table 1). The first binding of compound 1 is strong (K1 = 2.76 × 105 M−1), and the parameters indicate interactions via good hydrogen bonding and conformational changes.21 The second binding constant is low (K2 = 1.17 × 103 M−1), which is attributed to the high negative co-operativity.
The FabG4–compound 2 titration also fits to the ‘sequential binding’ mode with two bindings (Fig. 5C). The binding of compound 2 at the NADH binding pockets of both monomers is spontaneous and comparable to compound 1 (Table 1). As evidenced from the thermodynamic parameters obtained from ITC, compound 1 binds more strongly than compound 2 at the first monomeric NADH binding site. In the case of the second binding, the extent of negative co-operativity is lower than for compound 1, as the second binding of compound 2 is relatively stronger than the second binding of compound 1.
The binding of NADH and the inhibitors to the major portal of FabG4 is mainly governed by H-bonding with Asp244, Arg223, Asn295 and the catalytic tetrad residues. Such types of hydrophilic interactions are also supported by the negative values of both ΔH and ΔS obtained from the ITC experiments (Table 1). The negative ΔH and ΔS indicates the contribution of strong H-bonding and conformational changes respectively.
Docking studies
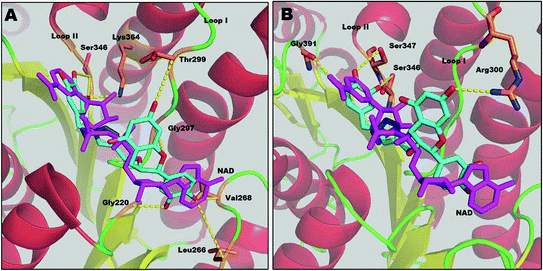
Results from docking studies support the experimental findings and provide necessary theoretical insight into the binding mode of the compounds. Docking studies can not explain the aspects of sequential binding as it was performed between one ligand and one monomer file. Here, the docking studies provides the binding interaction of one monomer with the inhibitor.Compound 1 interacts with several residues via hydrogen bonding and fully competes with the NADH binding region (major portal). The docking study shows that compound 1 interacts with Leu266, Gly220 and Val268 at the A-subsite. It also shows hydrogen bonding with Gly297 of the NAG triad and Thr299 at loop I at the P-subsite. It interferes with the nicotinamide binding region (N-subsite) of NADH by making two H-bonds with Ser346. Compound 1 also interacts with the catalytic tetrad directly via H-bonding with the active site residue Lys364. Moreover it corroborates our kinetic and thermodynamic findings that compound 1 binds at the NADH binding domain via hydrogen bonds and acts as a competitive inhibitor.
Compound 2 also binds at the major portal, though it does not compete with all of the binding sites of the NADH binding region. According to the docking study, the binding energy of compound 2 (−5.00 kcal mol−1) is lower than for compound 1 (−6.34 kcal mol−1). This supports the previous thermodynamic findings that compound 2 also binds at the NADH binding domain, but the binding is relatively weaker than for compound 1. The binding mode of compound 2 shows that it also accesses the active site through loop I via H-bonding with Ser347 of the catalytic tetrad. It interferes at the N-subsite of NADH, binding by H-bonds with Ser346 and Gly391. Compound 2 shows H-bonding with Arg300 of loop I. However, unlike compound 1, the catechin subunit of compound 2 does not show any interaction at the A-subsite or P-subsite of the NADH binding region, whereas the gallol subunit forms H-bonds with Gly391, one of the conserved residues in the α-helical sub-domain which provides the flexibility of the sub-domain to access the substrate acetoacetyl CoA.12 Thus, these findings indicate that compound 2 can interfere with the enzyme–substrate complex, as well as the free enzyme, resulting in the mixed type of inhibition (Fig. 6).
Minimum inhibitory concentration (MIC assay)
The drug MICs for Mycobacterium species were determined by an aerobic resazurin microplate reduction assay (REMA).22Mycobacterium smegmatis was used as a model organism because approximately two thousand of its proteins share homology with the pathogen M. tuberculosis. Additionally, it shares similarities with M. tuberculosis in possessing the unusual cell wall structure. The functional complementation of the essential gene, fabG1, of Mycobacterium tuberculosis by Mycobacterium smegmatis fabG is also reported.23 It is also speculated that M. smegmatis possesses multiple FabG, as both the organisms belong to the same family. Due to its fast growth and non-pathogenic nature, M. smegmatis was used in this work instead of M. tuberculosis.Both compounds have shown good anti-mycobacterial properties.
We used isoniazid (INH) as a reference compound for this assay. The standardization of the assay conditions was carried out with different concentrations of inoculum and indicator using INH. The compounds were screened within the concentration range of 0.5 μg mL−1 to 100 μg mL−1. The MICs of compounds 1 and 2 were found to be 20 μg mL−1 and 5 μg mL−1, whereas the MIC of INH was 4 μg mL−1 (see ESI†).
Structure–activity relationship (SAR)
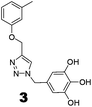
The roles of fragments linked at the 1- and 4-positions of the triazole rings in the inhibition activity have been evaluated by this preliminary SAR study. For this purpose, fragments linked to the 1- and 4-positions were substituted sequentially by groups of less interacting sites, and then their activities were compared with the original inhibitors. This strategy led to the design of compounds 3 and 4. In compound 3, the catechin/epicatechin fragment was substituted by a less interacting site, a m-cresol unit. Similarly, in compound 4, the gallol fragment linked to the 4-position of the triazole ring was replaced by a benzyl unit, a non-hydrogen bond forming counterpart (Fig. 7). Here, compound 1 was taken as a model inhibitor to compare the potential of compound 4.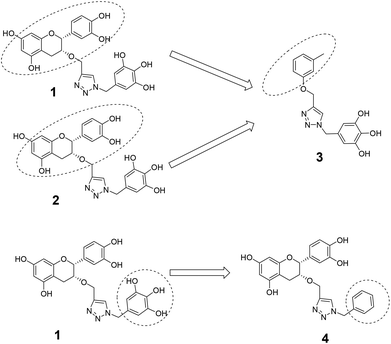 | ||
| Fig. 7 Catechin/epicatechin fragment linked to the 1-position replaced by a m-cresol unit, galloyl fragment linked at the 4-position of compound 1 replaced by a less-interacting benzyl unit. | ||
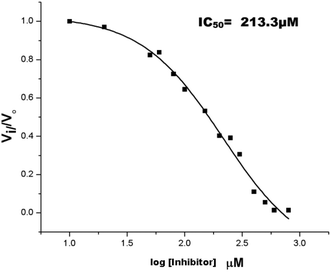
The inhibition studies of 3 and 4 were carried out under identical conditions and compared with the original inhibitors (Table 2). Compound 3 did not inhibit FabG4 up to a 400 μM concentration. Hence, the epicatechin and catechin fragments in our synthesized inhibitors play a crucial role in the inhibition potency. Compound 4 inhibited FabG4 with an IC50 value of 213.3 μM (Fig. 8). However, the IC50 value of compound 1 is 44.7 μM. This approximately five-fold decrease of the inhibition potency clearly suggests the importance of the galloyl fragment linked to the 4-position of the triazole ring for the activity of the compound.
These SAR findings are also supported by the docking studies. Compound 3 mainly floats over loop I of FabG4. The 1,4-triazole ring only makes H-bonds with Asn295 at the pyrophosphate binding subsite (P-subsite); it shows no interactions with the catalytic tetrad or NADH-binding amino acids. Therefore, the overall docking score is poor. Compound 4 binds at the major portal by making two hydrogen bonds with Asp244, which is found to play a crucial role in NADH binding. It also accesses the active site and weakly interacts with Tyr360 through the triazole ring, but it could not interact with the nicotinamide-binding subsite or loop I due to the absence of hydrogen bond acceptors/donors attached to the 4-position of the triazole ring (see ESI†). In the future, we would like to extend the SAR study by replacing the galloyl moiety with other electron-rich aromatic moieties, and also examine the role of the triazole ring by substituting with heterocyclic rings such as imidazole and pyrimidine rings, which are capable of forming H-bonds.
Conclusions
We have successfully designed and synthesized inhibitors (triazole polyphenol hybrid compounds) of the FabG4 enzyme of Mtb for the first time. The compounds have shown good inhibition at 0.88 μM and 3.97 μM respectively. Their design strategy, synthesis, inhibition potential and properties are discussed in this report. Their MIC values, as determined by a resazurin assay against M. smegmatis, indicate their good anti-mycobacterial properties. These compounds may be possible candidates as lead compounds for alternate anti-tubercular drugs. All of the reductase enzymes of the Mycobacterium family have similar ketoacyl reductase (KAR) domains. Hence, this work may be extrapolated to find structure-based inhibitors of other reductase enzymes.Experimental section
Docking details
The advanced and widely used molecular grid-based docking program Autodock4.2 was used to predict the binding modes and approximate binding free energies of all of the designed inhibitors in the lead library. The X-ray crystal coordinates of FabG4 were obtained from the Protein Data Bank (http://www.rcsb.org), PDB ID: 4FW8. The receptor structure was edited and hydrogen atoms added prior to docking. AutoDockTools was used to assign Gasteiger charges to the receptor. The catalytic tetrad and the other important amino acids in the NADH binding region of the receptor were selected as flexible residues to perform the flexible docking. The ligand structures were built up using Accelrys Discovery Studio 3.1 client. Energy minimization of the ligand structures was performed by applying a CHARMM force field. The whole receptor structure was selected for grid calculation with 0.375 Å spacing to perform blind docking. Based upon the grid maps of all of the atom types of the ligands, docking studies were carried out by using the Lamarckian genetic algorithm. Standard protocols were used, with an initial population of 150 randomly placed individuals, a maximum of 2.5 × 106 energy evaluations, a cluster tolerance of 2 Å (rms) and a maximum of 2.7 × 104 numbers of generations. After each docking execution, ten resulting conformers were clustered according to their energy. The conformer with the best binding energy was evaluated for design purposes.Chemistry details
All of the reactions were conducted with oven-dried glassware under an atmosphere of argon (Ar) or nitrogen (N2). All of the common reagents were of commercial grade and used without further purification. The solvents were dried by standard methods and purified by distillation before use. Silica gel (60–120 and 230–400 mesh) was used for column chromatography. TLC was performed on aluminum-backed plates coated with Silica gel 60 with an F254 indicator. Locally available UV-lamp chambers and I2-blowers were used as the TLC spot indicators. For solid compounds, melting points (MPs) were measured using melting point apparatus twice and reported without further calibration. The NMR spectra were recorded on Bruker 200 MHz and 400 MHz machines. The following abbreviations are used to describe the peak patterns where appropriate: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, dd = double doublet, ABq = AB quartet.General procedure for the synthesis of compounds 6 and 8
In solutions of compounds 5 and 7 (1 eq.) in dry THF, sodium hydride (4 eq.) was added at 0 °C and the mixture was stirred for 30 minutes at 0 °C. Then propargyl bromide (2 eq.), dissolved in dry THF, was added dropwise. The mixture was stirred for 12 hours at room temperature, then quenched with NH4Cl solution, and extracted with EtOAc. The organic layer was washed with brine and dried over anhydrous sodium sulphate. Evaporation under vacuum gave an oil, from which the desired compounds were isolated by column chromatography (Si-gel 60–120 mesh, PE–EA = 7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
Yield: 2.9 g (75%); state: white solid; mp: 81 °C; δH (CDCl3, 200 MHz): 7.45–7.26 (17H, m, Ar–H), 5.17 (6H, s, 3 × OCH2Ph), 3.92 (3H, s, CH3); δC (CDCl3, 50 MHz): 166.5, 152.5, 137.4, 136.6, 128.4, 128.1, 127.9, 127.9, 127.4, 125.1, 108.9, 75.0, 71.1, 52.1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
Yield: 600.5 mg (80%); state: white solid; mp: 90 °C; δH (CDCl3, 400 MHz): 7.44–7.27 (15H, m, Ar–H), 6.67 (2H, s, Ar–H), 5.10 (4H, s, 2 × –CH2Ph), 5.06 (2H, s, –CH2Ph), 4.56 (2H, s, –CH2OH); δC (CDCl3, 50 MHz): 152.8, 137.7, 137.5, 137.0, 136.6, 128.5, 128.4, 128.2, 128.1, 127.8, 127.7, 127.3, 106.2, 75.1, 65.2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
Yield: 499 mg (85%); state: yellow liquid; δH (CDCl3, 200 MHz): 7.40–7.25 (15H, m, Ar–H), 6.69 (2H, s, Ar–H), 5.09 (4H, s, 2 × CH2Ph), 5.03 (2H, s, CH2Ph), 4.39 (2H, s, CH2Br); δC (CDCl3, 50 MHz): 152.8, 137.7, 136.8, 133.1, 128.5, 128.1, 127.9, 127.8, 127.4, 108.7, 75.2, 71.2, 34.2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
Yield: 373.8 mg (90%); state: light yellow liquid; δH (CDCl3, 200 MHz): 7.46–7.30 (15H, m, Ar–H), 6.66 (2H, s, Ar–H), 5.17 (4H, s, 2 × CH2Ph), 5.11 (2H, s, CH2Ph), 4.28 (2H, s, –CH2N3); δC: 153.2, 137.9, 137.0, 131.1, 128.7, 128.3, 128.1, 128.0, 127.6, 108.0, 75.3, 71.4, 55.1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 as the eluent).
1 as the eluent).
Yield: 1.1 g (78%); state: reddish liquid; δH (CDCl3, 200 MHz): 7.19 (1H, m, Ar–H), 6.81 (3H, m, Ar–H), 4.64 (2H, d, J = 2.4 Hz, OCH2CCH), 2.49 (1H, t, J = 2.4 Hz, CCH), 2.35 (3H, s, CH3); δC (CDCl3, 50 MHz): 157.7, 139.6, 129.3, 122.5, 115.9, 111.8, 78.9, 75.5, 55.7, 21.6.
General procedure for Huisgen's 1,3-dipolar cycloaddition
Compounds 14, 15, 17 and 19 were synthesized using this procedure. The respective alkyne and azide counterparts were dissolved in acetonitrile and degasified for 15 min. Copper sulphate (1 eq.) was dissolved in an equal volume of water and degassed for 30 min. Then sodium ascorbate (2 eq.) was added, and the solution was stirred for 15 min. Then the degassed acetonitrile solution was slowly added. The mixture was stirred at room temperature until TLC indicated the disappearance of the starting materials. After that, the mixture was poured into saturated aqueous NH4Cl solution and the product was extracted four times with EtOAc. The organic layer was dried with sodium sulphate and filtered, and the solvent was removed under reduced pressure. The residue was purified by flash chromatography.General procedure of debenzylation
Debenzylation was carried out in a parr apparatus. Solutions of compounds 14, 15, 17 and 19 in a mixture of THF–MeOH–H2O (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5) were hydrogenated at 2–3 bar H2 pressure over 20% Pd (OH)2–C for 5–7 h at room temperature. The reaction mixtures were filtered through celite using 1
0.5) were hydrogenated at 2–3 bar H2 pressure over 20% Pd (OH)2–C for 5–7 h at room temperature. The reaction mixtures were filtered through celite using 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ethyl acetate–methanol and concentrated in vacuo. The gummy liquid was dissolved in a few drops of methanol and precipitation was carried out by adding diethyl ether. The compounds were purified by repeated precipitation. The pure compounds appeared as yellow gummy liquids in the absence of ether. All of the solvents were distilled prior to use and inert conditions were maintained for most of the time to avoid oxidation.
1 ethyl acetate–methanol and concentrated in vacuo. The gummy liquid was dissolved in a few drops of methanol and precipitation was carried out by adding diethyl ether. The compounds were purified by repeated precipitation. The pure compounds appeared as yellow gummy liquids in the absence of ether. All of the solvents were distilled prior to use and inert conditions were maintained for most of the time to avoid oxidation.
FabG4 inhibition assay
The inhibition studies were carried out in vitro at 25 °C by monitoring the decrease of OD340 in the kinetic mode as a function of conversion of NADH to NAD+ using an Evolution™ 300 UV-visible spectrophotometer (Thermo Fisher scientific). Protein purification was carried out as described elsewhere.24 Briefly, the recombinant protein was over-expressed in E. coli M15 cells using IPTG induction. Cells were harvested and lysed in a re-suspension buffer using ultra-sonication. The lysate was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 40 min to discard cell debris. The supernatant was loaded onto a Ni-NTA column and eluted with 300 mM imidazole solution. The eluted protein was further purified by gel filtration chromatography using a Superdex200 column. The elutant was pooled and concentrated, and extensively dialyzed against HEPES buffer (50 mM, pH 7.4) containing 50 mM NaCl prior to the experiments. The inhibitor compounds were dissolved in HEPES buffer (50 mM, pH 7.4) with 3% methanol (HPLC grade) to prepare a stock solution of 1 mM, which was further diluted repeatedly by HEPES buffer to achieve the desired concentrations. The reaction mixture contained the inhibitor solution in HEPES buffer, 0.5 mM acetoacetyl-CoA, 0.2 mM β-NADH and 1 μM of FabG4 protein in the final volume of 500 μL. A mixture of FabG4, NADH and acetoacetyl CoA was used as a positive control when no inhibitor was added. The reaction was initiated by the addition of acetoacetyl-CoA. The decrease in absorbance was recorded in 5 min intervals. The IC50 values were determined by varying the inhibitor concentrations until full inhibition occurred. To determine the mode of inhibition, we screened the enzyme activity with varying NADH concentration at three different concentrations of inhibitor. All of the experiments were repeated thrice. The IC50 values were calculated graphically from the dose–response plots. The modes of inhibition were determined from Lineweaver–Burk plots. The inhibition constants were determined from secondary plots.
000 rpm for 40 min to discard cell debris. The supernatant was loaded onto a Ni-NTA column and eluted with 300 mM imidazole solution. The eluted protein was further purified by gel filtration chromatography using a Superdex200 column. The elutant was pooled and concentrated, and extensively dialyzed against HEPES buffer (50 mM, pH 7.4) containing 50 mM NaCl prior to the experiments. The inhibitor compounds were dissolved in HEPES buffer (50 mM, pH 7.4) with 3% methanol (HPLC grade) to prepare a stock solution of 1 mM, which was further diluted repeatedly by HEPES buffer to achieve the desired concentrations. The reaction mixture contained the inhibitor solution in HEPES buffer, 0.5 mM acetoacetyl-CoA, 0.2 mM β-NADH and 1 μM of FabG4 protein in the final volume of 500 μL. A mixture of FabG4, NADH and acetoacetyl CoA was used as a positive control when no inhibitor was added. The reaction was initiated by the addition of acetoacetyl-CoA. The decrease in absorbance was recorded in 5 min intervals. The IC50 values were determined by varying the inhibitor concentrations until full inhibition occurred. To determine the mode of inhibition, we screened the enzyme activity with varying NADH concentration at three different concentrations of inhibitor. All of the experiments were repeated thrice. The IC50 values were calculated graphically from the dose–response plots. The modes of inhibition were determined from Lineweaver–Burk plots. The inhibition constants were determined from secondary plots.
ITC experiments
Isothermal titration calorimetry (ITC) was carried out using a Microcal ITC200 instrument. Titrations of FabG4 enzyme with compound 1, compound 2 and NADH were performed at 25 °C. A reference power of 5 μcal s−1, stirring speed of 800 rpm and 120 s spacing were selected. The protein solution was prepared in 50 mM HEPES buffer at pH 7.4. Protein solutions of 10 μM, 4.3 μM and 3 μM concentrations were recorded in the sample cell for the NADH (500 μM), compound 1 (1 mM) and compound 2 (1 mM) titrations respectively. The ligand solutions were prepared in the same buffer with 3% HPLC grade methanol. This extra methanol was also added to the sample cell to avoid large heat changes due to solvent mismatch (Getting Started, MicroCal ITC200). Separate buffer–ligand titrations were carried out as reference runs for each compound by recording the buffer in the sample cell. This reference value was subtracted from the protein–ligand titration to nullify the heat of dissolution. One injection of 0.4 μL, followed by nineteen injections of 2 μL of the ligand solutions, were titrated into the FabG4 solution. All of the experiments were repeated twice and the data were solved using MicroCal, LLC ITC 200 software. All of the parameters, such as tolerance limit (zero), derivative delta (0.08) and weighting method (no weighting), were default values. The data points of the first injections (0.4 μL) were neglected. The heat changes of the 19 injections (for the NADH and compound 1 titrations) and 18 injections (for the compound 2 titration) were plotted against the molar ratio of ligand.MIC study
The Mycobacterium smegmatis mc2155 strain was grown in Middlebrook 7H9 broth with 0.2% glycerol and 0.05% Tween 80 for 20 hours at 37 °C with shaking at 120 rpm, until the cells reached the mid-logarithmic phase (OD595 ∼ 0.5). The culture was diluted to OD595 = 0.0005 prior to addition into a sterile 96-well microtitre plate. Isoniazid (INH) was purchased from Aldrich and used as a reference compound for this assay. An INH stock solution of 1 mg mL−1 was prepared in distilled water and serially diluted in MB 7H9 media. 0.5 mg mL−1 stock solutions of compound 1 and 2 (in 50 mM HEPES buffer, pH 7.4, 3% HPLC grade methanol) were subjected to serial two fold dilutions in MB 7H9 media to make the desired concentrations. Resazurin sodium salt was purchased from Sisco Research Laboratories, India. 0.5% (w/v) stock solution of resazurin was prepared in distilled water, filter-sterilized and diluted to 0.02% in distilled water.One hundred microlitres of diluted culture (OD595 = 0.0005) was added to each well of the microtitre plate. The total volume was two hundred microlitres. The inoculum was omitted from the negative control (Row A), and the inhibitors were omitted from the growth control (Row B). INH, the reference compound, was used as a positive control. After 24 hours of post-drug incubation at 37 °C, 60 μL of 0.02% resazurin solution was added to each well and incubated again for 40 min at 37 °C. A color change from blue (resazurin) to pink (resorufin) indicated the growth of bacteria. The lowest concentrations of drugs which prevented such a color change were recorded as the MIC values. The assay was repeated twice and an average of both the experiments was calculated to find the MIC values.
Acknowledgements
DRB thanks the Council of Scientific and Industrial Research (CSIR) for the fellowship. Central Research Facility and Department of Chemistry, IIT Kharagpur are thanked for providing all of the instrumental facilities. AB and AKD gratefully acknowledge the support of DBT for providing the necessary research grant. AB is also grateful to DST for the JC Bose National Fellowship. BS and KS are thankful to DST for fast-track young scientist award.References
- (a) B. H. Herzog, Respiration, 1998, 65, 5 CrossRef PubMed; (b) R. P. Tripathi, N. Tewari, N. Dwivedi and V. K. Tiwari, Med. Res. Rev., 2005, 25, 93 CrossRef CAS PubMed; (c) R. Vohra, M. Gupta, R. Chaturvedi and Y. Singh, Recent Pat. Anti-Infect. Drug Discovery, 2006, 1, 95 CrossRef CAS; (d) Y. L. Janin, Bioorg. Med. Chem., 2007, 15, 2479 CrossRef CAS PubMed.
- (a) World Health Organization, World Health Forum, 1993, 14, 1 Search PubMed; (b) World Health Organization, Global tuberculosis control, 2012, WHO/HTM/TB/2012.6.
- (a) S. H. E. Kaufmann and E. Rubin, Handbook of Tuberculosis: Clinics, Diagnostics, Therapy and Epidemiology, Wiley-VCH, 2008, pp. XXIII–XXVII Search PubMed; (b) M. A. Espinal, Tuberculosis, 2003, 83, 44 CrossRef; (c) C. Lienhardt, M. Raviglione, M. Spigelman, R. Hafner, E. Jaramillo, M. Hoelscher, A. Zumla and J. Gheuens, J. Infect. Dis., 2012, 205, S241 CrossRef PubMed; (d) P. Bemer-Melchior, A. Bryskier and H. B. Drugeon, J. Antimicrob. Chemother., 2000, 46, 571 CrossRef CAS PubMed; (e) A. Jain and R. Mondal, FEMS Immunol. Med. Microbiol., 2008, 53, 145 CrossRef CAS PubMed; (f) A. S. Fauci, J. Infect. Dis., 2008, 197, 1493–1498 CrossRef PubMed; (g) G. Maartens and R. J. Wilkinson, Lancet, 2007, 370, 2030 CrossRef; (h) C. E. Barry 3rd and J. S. Blanchard, Curr. Opin. Chem. Biol., 2010, 14, 456 CrossRef PubMed.
- K. Takayama, C. Wang and G. S. Besra, Clin. Microbiol. Rev., 2005, 18, 81 CrossRef CAS PubMed.
- S. T. Cole, R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry 3rd, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, A. Krogh, J. McLean, S. Moule, L. Murphy, K. Oliver, J. Osborne, M. A. Quail, M. A. Rajandream, J. Rogers, S. Rutter, K. Seeger, J. Skelton, R. Squares, S. Squares, J. E. Sulston, K. Taylor, S. Whitehead and B. G. Barrell, Nature, 1998, 393, 537 CrossRef CAS PubMed.
- (a) H. Marrakchi, S. Ducasse, G. Labesse, E. Margeat, H. Montrozier, L. Emorine, X. Charpentier, G. Laneelle and A. Quemard, Microbiology, 2002, 148, 951 CAS; (b) M. Cohen-Gonsaud, S. Ducasse, F. Hoh, D. Zerbib, G. Labesse and A. Quemard, J. Mol. Biol., 2002, 320, 249 CrossRef CAS.
- (a) S. Ducasse-Cabanot, M. Cohen-Gonsaud, H. Marrakchi, M. Nguyen, D. Zerbib, J. Bernadou, M. Daffe, G. Labesse and A. Quemard, Antimicrob. Agents Chemother., 2004, 48, 242 CrossRef CAS; (b) A. R. Rendina and D. Cheng, Biochem. J., 2005, 388, 895 CrossRef CAS PubMed; (c) M. J. Vazquez, W. Leavens, R. Liu, B. Rodriguez, M. Read, S. Richards, D. Winegar and J. M. Dominguez, FEBS J., 2008, 275, 1556 CrossRef CAS PubMed; (d) K. Kristan, T. Bratkovic, M. Sova, S. Gobec, A. Prezelj and U. Urleb, Chem.-Biol. Interact., 2009, 178, 310 CrossRef CAS PubMed.
- D. J. Beste, M. Espasa, B. Bonde, A. M. Kierzek, G. R. Stewart and J. McFadden, PLoS One, 2009, 4, e5349 Search PubMed.
- (a) A. Gurvitz, Mol. Genet. Genomics, 2009, 282, 407 CrossRef CAS PubMed; (b) H. I. Boshoff, T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson and C. E. Barry 3rd, J. Biol. Chem., 2004, 279, 40174 CrossRef CAS PubMed.
- P. Sharma, B. Kumar, N. Singhal, V. M. Katoch, K. Venkatesan, D. S. Chauhan and D. Bisht, Indian J. Med. Res., 2010, 132, 400 CAS.
- D. Dutta, S. Bhattacharyya, S. Mukherjee, B. Saha and A. K. Das, J. Struct. Biol., 2011, 174, 147 CrossRef CAS PubMed.
- D. Dutta, S. Bhattacharyya, A. Roychowdhury, R. Biswas and A. K. Das, Biochem. J., 2013, 450, 127 CrossRef CAS PubMed.
- W. Yu, H. Xiao, J. Lin and C. Li, J. Med. Chem., 2013, 56, 4402–4412 CrossRef CAS PubMed.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785 CrossRef CAS PubMed.
- (a) Y. M. Zhang and C. O. Rock, J. Biol. Chem., 2004, 279, 30994 CrossRef CAS PubMed; (b) D. Tasdemir, G. Lack, R. Brun, P. Ruedi, L. Scapozza and R. Perozzo, J. Med. Chem., 2006, 49, 3345 CrossRef CAS PubMed; (c) S. K. Sharma, P. Parasuraman, G. Kumar, N. Surolia and A. Surolia, J. Med. Chem., 2007, 50, 765 CrossRef CAS PubMed.
- (a) M. S. Costa, N. Boechat, E. A. Rangel, F. C. Da Silva, M. T. De Souza, C. R. Rodrigues, H. C. Castro, I. N. Junior, M. C. S. Lourenco, S. M. S. V. Wardell and V. F. Ferreira, Bioorg. Med. Chem., 2006, 14, 8644 CrossRef CAS PubMed; (b) N. Boechat, V. F. Ferreira, S. B. Ferreira, M. L. G. Ferreira, F. C. Da Silva, M. M. Bastos, M. S. Costa, M. C. S. Lourenco, A. C. Pinto, A. U. Krettli, A. C. Aguiar, B. M. Teixeira, N. V. Da Silva, P. R. C. Martins, F. A. F. M. Bezerra, A. L. S. Camilo, G. P. Da Silva and C. C. P. Costa, J. Med. Chem., 2011, 54, 5988 CrossRef CAS PubMed; (c) G. R. Labadie, A. Iglesia and H. R. Morbidoni, Mol. Diversity, 2011, 15, 1017 CrossRef CAS PubMed; (d) P. Shanmugavelan, S. Nagarajan, M. Sathishkumar, A. Ponnuswamy, P. Yogeeswari and D. Sriram, Bioorg. Med. Chem. Lett., 2011, 21, 7273 CrossRef CAS PubMed; (e) C. Menendez, A. Chollet, F. Rodriguez, C. Inard, M. R. Pasca, C. Lherbet and M. Baltas, Eur. J. Med. Chem., 2012, 52, 275 CrossRef CAS PubMed.
- L. Chen, D. J. Wilson, Y. Xu, C. C. Aldrich, K. Felczak, Y. Y. Sham and K. W. Pankiewicz, J. Med. Chem., 2010, 53, 4768 CrossRef CAS PubMed.
- V. V. Rostovtsev, L. G. Green, V. V. Fokin and K. B. Sharpless, Angew. Chem., Int. Ed., 2002, 41, 2596 CrossRef CAS.
- Y. Lu and J. Gervay-Hague, Carbohydr. Res., 2007, 342, 1636 CrossRef CAS PubMed.
- D. Dutta, S. Bhattacharyya and A. K. Das, Proteins: Struct., Funct., Bioinf., 2012, 80, 1250 CrossRef CAS PubMed.
- Isothermal titration calorimetry and drug design by Microcal. http://www.philadelphia.edu.jo/academics/mbadawneh/uploadsITC-and-Drug-Design.pdf.
- (a) J. C. Palomino, A. Martin, M. Camacho, H. Guerra, J. Swings and F. Portaels, Antimicrob. Agents Chemother., 2002, 46, 2720 CrossRef CAS; (b) N. K. Taneja and J. S. Tyagi, J. Antimicrob. Chemother., 2007, 60, 288 CrossRef CAS PubMed.
- T. Parish, G. Roberts, F. Laval, M. Schaeffer, M. Daffe and K. Duncan, J. Bacteriol., 2007, 189, 3721 CrossRef CAS PubMed.
- D. Dutta, S. Bhattacharyya and A. K. Das, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2012, 68, 786 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3ob41676c |
| This journal is © The Royal Society of Chemistry 2014 |