Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Interfacial interactions between natural RBC membranes and synthetic polymeric nanoparticles†
Brian T.
Luk‡
ac,
Che-Ming
Jack Hu‡
bc,
Ronnie H.
Fang
bc,
Diana
Dehaini
bc,
Cody
Carpenter
bc,
Weiwei
Gao
bc and
Liangfang
Zhang
*bc
aDepartment of Bioengineering, University of California, San Diego, La Jolla, CA 92093, USA
bDepartment of Nanoengineering, University of California, San Diego, La Jolla, CA 92093, USA. E-mail: zhang@ucsd.edu
cMoores Cancer Center, University of California, San Diego, La Jolla, CA 92093, USA
First published on 20th December 2013
Abstract
The unique structural features and stealth properties of a recently developed red blood cell membrane-cloaked nanoparticle (RBC-NP) platform raise curiosity over the interfacial interactions between natural cellular membranes and polymeric nanoparticle substrates. Herein, several interfacial aspects of the RBC-NPs are examined, including completeness of membrane coverage, membrane sidedness upon coating, and the effects of polymeric particles' surface charge and surface curvature on the membrane cloaking process. The study shows that RBC membranes completely cover negatively charged polymeric nanoparticles in a right-side-out manner and enhance the particles' colloidal stability. The membrane cloaking process is applicable to particle substrates with a diameter ranging from 65 to 340 nm. Additionally, the study reveals that both surface glycans on RBC membranes and the substrate properties play a significant role in driving and directing the membrane–particle assembly. These findings further the understanding of the dynamics between cellular membranes and nanoscale substrates and provide valuable information toward future development and characterization of cellular membrane-cloaked nanodevices.
1. Introduction
Recent advancement in biology and materials engineering has led to surging interest in bio-inspired nanodevices with biomimetic functionalities.1–3 Exploiting the immunomodulatory self-marker proteins commonly found in cells for nanocarrier functionalization has bestowed unique anti-phagocytic properties and prolonged in vivo survival onto nanoparticles.4,5 Among bio-inspired nanocarriers, a recently developed RBC membrane-cloaked nanoparticle (RBC-NP) platform presents an intriguing system as it utilizes the RBC membrane content in its entirety for immune-evasive stealth camouflage6,7 and therapeutic purposes.8,9 Upon unilamellar membrane coating, RBC-NPs display self-marker proteins with a right-side-out orientation bias,10 which contributes to the prolonged in vivo circulation time of the platform. The unique structural features and properties of RBC-NPs raise curiosity concerning the biomembrane–particle interface that plays a significant role in enabling colloidal stability and preserving biomimetic functionalities of the platform. In this study, we dissect the RBC-NP system to shed light on the mechanisms that elegantly bridge synthetic polymeric particles with natural cellular membranes.While the development of RBC-inspired drug carriers has focused primarily on mechano-mimicry11–13 and protein functionalization,5 the present study introduces a different emphasis on surface glycans, which in fact represent the predominant moieties on cellular surfaces.14 On RBCs, the dense glycan coatings, known as glycocalyx, have significant implications in the stabilization and immune-evasive properties of the cells.15–19 These complex polysaccharides serve as a hydrophilic coating, and stabilizing strategies using analogous sugar polymers can be found in many carbohydrate-functionalized nanoformulations.20–22 The asymmetric membrane distribution of glycans, which reside exclusively on the extracellular side of RBCs, also make glycans a good indicator of membrane sidedness.23,24 In addition, the abundant, negatively charged sialyl residues at the glycan terminus bestow a charge asymmetry across cellular membranes,25 which can affect interfacial interactions between RBC membranes and synthetic polymeric particles through electrostatic interactions. Herein, we carry out a series of studies to examine several interfacial aspects of RBC-NPs, including completeness of membrane coverage, membrane sidedness on the nanoparticles, and the effects of polymeric particles' surface charge and surface curvature on the membrane cloaking process. These studies help scrutinize the RBC-NP platform from a colloidal science perspective and shed light on the implications of membrane glycans and nanoparticle properties on RBC-NP formation. The studies also provide pertinent information toward future translation of the RBC-NP platform as well as inspirations for both synthetic and naturally derived nanoparticle designs.
2. Materials and methods
2.1 Preparation and characterization of RBC-NPs
RBC membranes were derived from whole blood collected from male imprinting control region (ICR) mice (Charles River Laboratories, Wilmington, MA). Whole blood was centrifuged and subjected to hemolysis by hypotonic treatment to isolate RBC membranes. The membranes were then bath sonicated for 3 min using an FS30D bath sonicator (Fisher Scientific, Waltham, MA) at a frequency of 42 kHz and a power of 100 W and subsequently extruded through a 100 nm polycarbonate porous membrane using an Avanti mini extruder to form RBC membrane vesicles. In parallel, poly(D,L-lactic-co-glycolic acid) (PLGA) polymeric cores were prepared using a 0.67 dL g−1 carboxy-terminated 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 PLGA polymer (LACTEL Absorbable Polymers, Birmingham, AL) through a solvent displacement method. PLGA was first dissolved in acetone at a concentration of 1 mg mL−1 and added dropwise to 3 mL of water. The mixture was then stirred in open air for 2 h and filtered using an Amicon Ultra-4 Centrifugal Filter with a molecular weight cutoff (MWCO) of 10
50 PLGA polymer (LACTEL Absorbable Polymers, Birmingham, AL) through a solvent displacement method. PLGA was first dissolved in acetone at a concentration of 1 mg mL−1 and added dropwise to 3 mL of water. The mixture was then stirred in open air for 2 h and filtered using an Amicon Ultra-4 Centrifugal Filter with a molecular weight cutoff (MWCO) of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da (Millipore, Billerica, MA). To fuse the RBC membrane vesicles with the PLGA cores, 1 mg of PLGA particles was mixed with RBC membranes derived from various amounts of mouse whole blood and extruded through a 100 nm polycarbonate porous membrane using an Avanti mini extruder. The hydrodynamic diameter, polydispersity index, and surface zeta potential of the resulting RBC-NPs were determined from three repeated experiments using dynamic light scattering (DLS) on a ZEN3600 Nano Zetasizer (Malvern Instruments, UK) at 25 °C. Transmission electron microscopy (TEM) images were taken to examine the structure of the RBC-NPs. A drop of the RBC-NP solution at a concentration of 2 mg mL−1 was deposited onto a glow-discharged carbon-coated grid. After 5 min, the grid was rinsed with 10 drops of distilled water and a drop of 1% uranyl acetate stain was added for negative staining. The grid was then dried and imaged using a Tecnai G2 Sphera (FEI, Hillsboro, OR) microscope.
000 Da (Millipore, Billerica, MA). To fuse the RBC membrane vesicles with the PLGA cores, 1 mg of PLGA particles was mixed with RBC membranes derived from various amounts of mouse whole blood and extruded through a 100 nm polycarbonate porous membrane using an Avanti mini extruder. The hydrodynamic diameter, polydispersity index, and surface zeta potential of the resulting RBC-NPs were determined from three repeated experiments using dynamic light scattering (DLS) on a ZEN3600 Nano Zetasizer (Malvern Instruments, UK) at 25 °C. Transmission electron microscopy (TEM) images were taken to examine the structure of the RBC-NPs. A drop of the RBC-NP solution at a concentration of 2 mg mL−1 was deposited onto a glow-discharged carbon-coated grid. After 5 min, the grid was rinsed with 10 drops of distilled water and a drop of 1% uranyl acetate stain was added for negative staining. The grid was then dried and imaged using a Tecnai G2 Sphera (FEI, Hillsboro, OR) microscope.
2.2 Membrane coverage assay
Biotin-functionalized PLGA was prepared by conjugating amine–PEG2–biotin to 0.67 dL g−1 carboxy-terminated PLGA (Thermo Scientific, Waltham, MA). 100 mg of PLGA was dissolved in 2.5 mL chloroform and activated with 5 mg of ethyl(dimethylaminopropyl) carbodiimide (EDC) and 5.5 mg of N-hydroxysuccinimide (NHS) (Thermo Scientific, Waltham, MA). Following 60 min of activation, a 3-to-1 molar excess of amine–PEG2–biotin was added to the polymer solution along with 8 μL of N,N-diisopropylethylamine (Fisher Scientific, Waltham, MA). After overnight incubation under gentle stirring, the polymer was precipitated by adding the solution dropwise into 40 mL of −20 °C methanol. Functionalized PLGA was pelleted through centrifugation at 3500 rpm for 5 min. The pelleted polymer was then dissolved in chloroform and further purified in −20 °C methanol two more times. Following the last wash, the biotin-functionalized PLGA was dried in a vacuum and dissolved in acetone. For the preparation of biotin-functionalized PLGA particles, 1 mg mL−1 of PLGA solution containing 2% of biotin–PLGA was prepared in acetone and added dropwise to 3 mL of water. The mixture was then stirred in open air for 2 h and filtered using an Amicon Ultra-4 Centrifugal Filter with a MWCO of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da (Millipore, Billerica, MA). Biotin functionalization on nanoparticle surfaces was confirmed by incubating 1 mL of 1 mg mL−1 biotinylated nanoparticles with 20 μg of streptavidin (Invitrogen, Grand Island, NY) for 30 min. Particle aggregation induced by streptavidin–biotin interactions was monitored by DLS, using non-biotinylated PLGA particles as a negative control. The biotinylated PLGA particles were then used to examine the extent of particle surface coverage by RBC membranes. 1 mL of 1 mg mL−1 RBC-NPs prepared with different membrane-to-particle ratios were incubated with 20 μg of streptavidin for 30 min. The particle size was determined from three repeated experiments using DLS at 25 °C.
000 Da (Millipore, Billerica, MA). Biotin functionalization on nanoparticle surfaces was confirmed by incubating 1 mL of 1 mg mL−1 biotinylated nanoparticles with 20 μg of streptavidin (Invitrogen, Grand Island, NY) for 30 min. Particle aggregation induced by streptavidin–biotin interactions was monitored by DLS, using non-biotinylated PLGA particles as a negative control. The biotinylated PLGA particles were then used to examine the extent of particle surface coverage by RBC membranes. 1 mL of 1 mg mL−1 RBC-NPs prepared with different membrane-to-particle ratios were incubated with 20 μg of streptavidin for 30 min. The particle size was determined from three repeated experiments using DLS at 25 °C.
2.3 Membrane sidedness assay
RBC-NPs were first prepared with 100 nm PLGA cores and RBC membrane vesicles. For glycoprotein quantification, proteins exposed on the RBC-NP surfaces were trypsinized by incubating 1 mL of 1 mg mL−1 RBC-NPs with 5 μg of trypsin (G-Biosciences, St. Louis, MO) at room temperature for 2 h. Sialic acid removal from RBC-NPs was performed by incubating 1 mL of 1 mg mL−1 RBC-NPs in water with 100 units of sialidase (Roche Diagnostics, Indianapolis, IN) at room temperature for 2 h. The samples were then centrifuged at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 45 min using an Optima L-90K Ultracentrifuge (Beckman Coulter, Brea, CA), and the supernatant was collected and examined for glycoprotein and sialic acid content. To quantify glycoprotein, the supernatant was examined using a Glycoprotein Detection Reagent (Thermo Scientific) following the manufacturer's instructions. To quantify sialic acid, the supernatant was examined using a Sialic Acid Quantification Kit (Sigma-Aldrich, St. Louis, MO) following the manufacturer's instructions. Equivalent amounts of RBC ghosts and bare PLGA cores were used as a positive and a negative control, respectively.
000×g for 45 min using an Optima L-90K Ultracentrifuge (Beckman Coulter, Brea, CA), and the supernatant was collected and examined for glycoprotein and sialic acid content. To quantify glycoprotein, the supernatant was examined using a Glycoprotein Detection Reagent (Thermo Scientific) following the manufacturer's instructions. To quantify sialic acid, the supernatant was examined using a Sialic Acid Quantification Kit (Sigma-Aldrich, St. Louis, MO) following the manufacturer's instructions. Equivalent amounts of RBC ghosts and bare PLGA cores were used as a positive and a negative control, respectively.
2.4 Stability study of RBC-NPs
A series of RBC-NP formulations were prepared by coating 1 mg of PLGA cores with RBC membranes collected from 200 μL, 100 μL, 75 μL, 50 μL, 25 μL or 0 μL of mouse blood. Each formulation was then adjusted to 1× PBS buffer (pH = 7.4) and sonicated for 5 min using an FS30D bath sonicator (Fisher Scientific, Waltham, MA) at a frequency of 42 kHz and a power of 100 W to facilitate the aggregation process. After sonication, the hydrodynamic diameter of the particles was determined using DLS. Trypsinized RBC-NPs were prepared by incubating a stable RBC-NP formulation (100 μL blood per mg PLGA core) with 50 μg mL−1 trypsin (G-Biosciences, St. Louis, MO). Two hours following the trypsinization, the particle size was measured by DLS.2.5 Preparation of RBC-NPs with positively charged polymeric cores
To create positively charged polymeric cores, 100 nm PLGA nanoparticles prepared using the aforementioned protocol were mixed with 5% (w/w) polyethylenimine (PEI, molecular weight = 1800 Da) and bath sonicated for 5 min. The resulting nanoparticle solution was filtered using an Amicon Filter with a MWCO of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da. The hydrodynamic size, polydispersity index, and surface zeta potential of the PEI-coated PLGA particles were characterized by DLS. 1 mg of the resulting positively charged nanoparticles was then mixed with RBC membrane vesicles derived from 100 μL of whole blood and extruded through a 200 nm, 400 nm or 1000 nm polycarbonate porous membrane using an Avanti mini extruder. The particle size and surface zeta potential before and after RBC membrane coating were measured by DLS. The structure of the particles was examined using TEM after negative staining with uranyl acetate.
000 Da. The hydrodynamic size, polydispersity index, and surface zeta potential of the PEI-coated PLGA particles were characterized by DLS. 1 mg of the resulting positively charged nanoparticles was then mixed with RBC membrane vesicles derived from 100 μL of whole blood and extruded through a 200 nm, 400 nm or 1000 nm polycarbonate porous membrane using an Avanti mini extruder. The particle size and surface zeta potential before and after RBC membrane coating were measured by DLS. The structure of the particles was examined using TEM after negative staining with uranyl acetate.
2.6 Preparation of RBC-NPs with differently sized polymeric cores
Differently sized PLGA cores between 65 and 340 nm in diameter were prepared by adjusting the polymer concentration, the solvent-to-water ratio, and the ionic content in the aqueous phase during the nanoprecipitation process. Specifically, to prepare 65 nm PLGA cores, 1 mL of 5 mg mL−1 PLGA dissolved in acetone was pipetted into 3 mL of water. 120 nm cores were made by pipetting 1 mL of 10 mg mL−1 PLGA dissolved in acetone into 3 mL of water. 200 nm cores were prepared by pipetting 1 mL of 10 mg mL−1 PLGA dissolved in acetone into 4 mL of 2.5× PBS. 340 nm cores were prepared by adding water dropwise into 1 mL of 3 mg mL−1 PLGA dissolved in acetone until cloudiness was observed in the solution. All formulations were then evaporated in a vacuum chamber overnight to remove excess acetone. The prepared cores were washed 3 times using Amicon Filters with a MWCO of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da prior to coating with RBC membranes. A 10% excess of RBC membranes was added to each core type based on the theoretical membrane-to-polymer ratio for complete membrane coverage (ESI†). For 65 nm cores, RBC membranes derived from 165 μL of blood were added and the mixture was extruded through a 100 nm porous membrane. For 120 nm cores, RBC membranes derived from 75 μL of blood were added and the mixture was extruded through a 200 nm porous membrane. For 200 nm cores, RBC membranes derived from 40 μL of blood were added and the mixture was extruded through a 400 nm porous membrane. For 340 nm cores, RBC membranes derived from 23 μL of blood were added and the mixture was extruded through a 400 nm porous membrane. The particle size and surface zeta potential before and after RBC membrane coating were measured by DLS. The core–shell structure of the particles was examined using TEM after negative staining with uranyl acetate.
000 Da prior to coating with RBC membranes. A 10% excess of RBC membranes was added to each core type based on the theoretical membrane-to-polymer ratio for complete membrane coverage (ESI†). For 65 nm cores, RBC membranes derived from 165 μL of blood were added and the mixture was extruded through a 100 nm porous membrane. For 120 nm cores, RBC membranes derived from 75 μL of blood were added and the mixture was extruded through a 200 nm porous membrane. For 200 nm cores, RBC membranes derived from 40 μL of blood were added and the mixture was extruded through a 400 nm porous membrane. For 340 nm cores, RBC membranes derived from 23 μL of blood were added and the mixture was extruded through a 400 nm porous membrane. The particle size and surface zeta potential before and after RBC membrane coating were measured by DLS. The core–shell structure of the particles was examined using TEM after negative staining with uranyl acetate.
3. Results
3.1 Completeness of RBC membrane coverage
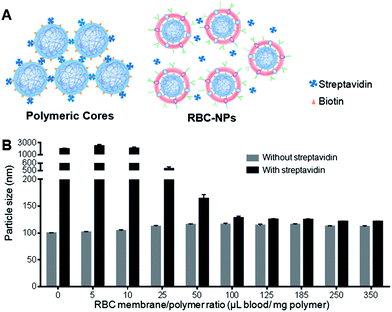
RBC-NPs were first examined for the completeness of the membrane coverage. An aggregation assay based on streptavidin–biotin cross-linking chemistry was implemented to examine whether polymeric surfaces were exposed. In the study, biotinylated PLGA nanoparticles were prepared, which readily aggregate upon direct exposure to free streptavidin in solution (Fig. 1A). Carboxylated PLGA polymers were first functionalized with amine–PEG2–biotin using EDC/NHS chemistry, and the resulting biotinylated PLGA polymers were mixed with carboxylated PLGA at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio to form biotinylated polymeric nanoparticles. The biotin-conjugated polymers showed a negligible effect on the physicochemical properties of the PLGA cores (ESI, Fig. S1 and S2†), as both biotinylated and non-biotinylated particles were ∼100 nm in size and −45 mV in surface zeta potential. Upon mixing with free streptavidin, the biotinylated cores aggregated to approximately 2000 nm, whereas the non-biotinylated cores remained similar in size, demonstrating a particle bridging mechanism that was specific to the strong streptavidin–biotin interaction. A slight size increase by ∼10 nm was observed in the non-biotinylated cores upon mixing with streptavidin, which was likely due to the non-specific protein absorption on the particle surface.
50 ratio to form biotinylated polymeric nanoparticles. The biotin-conjugated polymers showed a negligible effect on the physicochemical properties of the PLGA cores (ESI, Fig. S1 and S2†), as both biotinylated and non-biotinylated particles were ∼100 nm in size and −45 mV in surface zeta potential. Upon mixing with free streptavidin, the biotinylated cores aggregated to approximately 2000 nm, whereas the non-biotinylated cores remained similar in size, demonstrating a particle bridging mechanism that was specific to the strong streptavidin–biotin interaction. A slight size increase by ∼10 nm was observed in the non-biotinylated cores upon mixing with streptavidin, which was likely due to the non-specific protein absorption on the particle surface.
Given that bilayer membranes can preclude surface-attached biotin from interacting with the 60 kDa streptavidin, the completeness of RBC membrane coating was then evaluated using the biotinylated polymeric cores. Using a previously described extrusion process,6 the cores were coated with increasing amounts of RBC membrane content ranging from 0 to 350 μL of mouse blood per mg of polymer. The coated cores were then mixed with streptavidin and monitored for particle size change. At low membrane-to-polymer ratios (below 25 μL mg−1), significant aggregation was observed. However, as the membrane-to-polymer ratio increased to 100 μL mg−1 or higher, the addition of streptavidin failed to induce any considerable size increase among the biotinylated cores (Fig. 1B). The preclusion of streptavidin-induced aggregation suggests that the particle surfaces were completely shielded, and 100 μL mg−1 was sufficient to fully coat all the nanoparticles present. Based on the particle surface area, RBC surface area, and RBC concentration in mouse blood, it was estimated that ∼85 μL of mouse blood would be needed to derive enough RBC membrane material to completely coat 1 mg of 100 nm PLGA nanoparticles (ESI†). The close match between the theoretical and the experimental membrane-to-polymer ratio for full coverage suggests a high membrane coating efficiency, which is consistent with a previous study that showed a high protein translocation yield through the membrane coating approach.10
3.2 Sidedness of RBC membranes upon coating
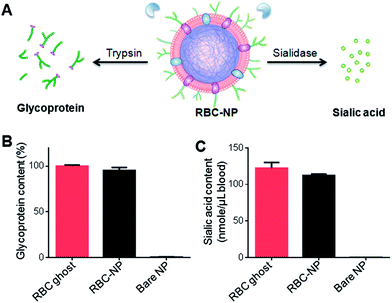
Owing to the asymmetric distribution of glycans on the extracellular side of cellular membranes, these glycans can be used as an indicator to quantitatively analyze the membrane sidedness on RBC-NPs. A trypsinization method was herein applied to examine the glycoprotein content on the outer surface of RBC-NPs.26 In the study, RBC-NPs were first prepared with excess polymeric cores to ensure that all RBC membranes were occupied. RBC ghosts containing equivalent amounts of membrane content and bare polymeric cores were prepared as a positive control and a negative control, respectively. The samples were then trypsinized for 2 h and subjected to ultracentrifugation at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 45 min. Since RBC bilayer membranes are impermeable to trypsin, membrane sidedness can be assessed by measuring the enzymatically removed glycoprotein content.26 Detached glycoproteins were collected from the sample supernatant and quantified using a periodate-based glycoprotein detection assay (Fig. 2A). Relative to the glycoprotein content in trypsinized RBC ghosts, trypsinized RBC-NPs yielded ∼95% of the glycoprotein content in the sample supernatant (Fig. 2B). This result indicates that the majority of surface glycoproteins on RBC-NPs were exposed to the trypsin treatment, corroborating a right-side-out membrane orientation that was previously observed by immunogold staining of surface proteins.10
000×g for 45 min. Since RBC bilayer membranes are impermeable to trypsin, membrane sidedness can be assessed by measuring the enzymatically removed glycoprotein content.26 Detached glycoproteins were collected from the sample supernatant and quantified using a periodate-based glycoprotein detection assay (Fig. 2A). Relative to the glycoprotein content in trypsinized RBC ghosts, trypsinized RBC-NPs yielded ∼95% of the glycoprotein content in the sample supernatant (Fig. 2B). This result indicates that the majority of surface glycoproteins on RBC-NPs were exposed to the trypsin treatment, corroborating a right-side-out membrane orientation that was previously observed by immunogold staining of surface proteins.10
To further verify the membrane sidedness of RBC-NPs, a secondary assay was applied to quantify sialic acid, a characteristic carbohydrate terminus on RBC glycans. A sialidase enzyme was used to remove the terminal sialyl groups on the RBC-NPs (Fig. 2A). Unlike the complete glycoprotein removal by trypsin, sialidase treatment only removed the terminal carbohydrate and did not affect the size of the RBC-NPs (ESI Fig. S3A†). The enzymatic activity, however, could be observed through surface zeta potential measurements as the removal of negatively charged sialic acid altered the surface charge of the RBC-NPs. Following 2 h of sialidase treatment, the zeta potential of RBC-NPs shifted from −23 mV to −0.6 mV, whereas that for bare nanoparticles remained largely unchanged (ESI, Fig. S3B†), verifying the enzymatic removal of sialic acid. Following sialidase treatment and ultracentrifugation at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g, the sample supernatants were collected and assayed using a sialic acid quantification kit. It was found that per RBC membrane content derived from 1 mL of blood, 122 ± 8 and 112 ± 2 nmol of sialic acid was recovered from RBC ghosts and RBC-NPs, respectively (Fig. 2C). The values are consistent with the reported sialic acid density on erythrocytes,27 and the close match in sialic acid recovery further supports the right-side-out-membrane orientation of the RBC-NPs. Based on the quantification result, it can also be estimated that at full coverage each RBC-NP with 100 nm in core diameter possesses ∼43
000×g, the sample supernatants were collected and assayed using a sialic acid quantification kit. It was found that per RBC membrane content derived from 1 mL of blood, 122 ± 8 and 112 ± 2 nmol of sialic acid was recovered from RBC ghosts and RBC-NPs, respectively (Fig. 2C). The values are consistent with the reported sialic acid density on erythrocytes,27 and the close match in sialic acid recovery further supports the right-side-out-membrane orientation of the RBC-NPs. Based on the quantification result, it can also be estimated that at full coverage each RBC-NP with 100 nm in core diameter possesses ∼43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 sialyl moieties (ESI†). This dense population of surface sialyl moieties has strong implications on the RBC-NPs' properties as they are a key modulator of cellular interaction and immune activation and contribute to erythrocytes' in vivo survival.18,19,28
500 sialyl moieties (ESI†). This dense population of surface sialyl moieties has strong implications on the RBC-NPs' properties as they are a key modulator of cellular interaction and immune activation and contribute to erythrocytes' in vivo survival.18,19,28
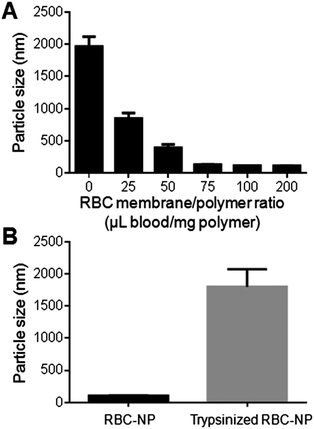
3.3 RBC membrane cloaks stabilizing polymeric cores
Upon verification that the RBC membrane can completely envelop PLGA nanoparticles with a right-side-out membrane orientation, the stabilizing effect of the membrane coating was then evaluated. Carboxylated PLGA was used to prepare polymeric cores that were ∼100 nm in diameter with a surface zeta potential of −45 mV. The cores were then extruded with various amounts of RBC membrane to form RBC-NPs. To test the stabilizing effect of RBC membranes, the particle solution was adjusted to 1× PBS (pH = 7.4), in which non-stabilized nanoparticles are known to aggregate owing to the charge screening effect by the ionic environment. Following brief sonication to agitate the sample solutions, the nanoparticle sizes were monitored using DLS. For the formulations containing 0, 25, 50, 75, 100, and 200 μL of RBC membrane per mg of PLGA core, the final particle sizes following sonication in PBS were 1965 ± 152 nm, 850 ± 84 nm, 395 ± 51 nm, 135 ± 2 nm, 117 ± 1 nm, and 116 ± 2 nm respectively (Fig. 3A). The results reflect the increasing particle stability with higher RBC membrane content, and above 100 μL mg−1 of membrane-to-particle ratio, which is above the theoretical value for full particle coverage, the nanoparticles showed negligible size increase following sonication.The stabilizing effect of RBC membranes can be attributed to the copious surface glycans present, which are highly hydrophilic and contribute to the steric stabilization of cells.15,17 To examine the role of these glycans in enabling colloidal stability, stabilized RBC-NPs (membrane-to-polymer ratio: 100 μL blood per mg polymer) were treated with trypsin, which enzymatically cleaves glycoproteins that serve to anchor the majority of RBC surface glycans. 2 h following trypsinization, significant RBC-NP aggregation was observed (Fig. 3B). The loss of colloidal stability following trypsinization suggests that the phospholipid membrane bilayer alone is not sufficient to stabilize the particles, and that steric stabilization was enabled by the polysaccharides on RBC membranes. The stabilizing capability of hydrophilic glycans is not surprising, as synthetic polysaccharides have been commonly applied for nanoparticle stabilization.20–22 In the RBC-NP platform, polymeric nanoparticles are functionalized by naturally derived surface glycans. This glycan stabilization mechanism also provides an explanation to the efficient membrane cloaking process, in which unilamellar membrane coating readily occurs and yet multilamellar coating is not observed despite excess membrane materials (ESI, Fig. S4†); the non-stabilized polymeric surfaces with higher surface energy would readily interact with glycan-stabilized membranes to minimize the overall energy, whereas glycan-stabilized RBC-NP surfaces preclude further membrane interactions.
3.4 Surface charge of polymeric cores affecting RBC membrane coating
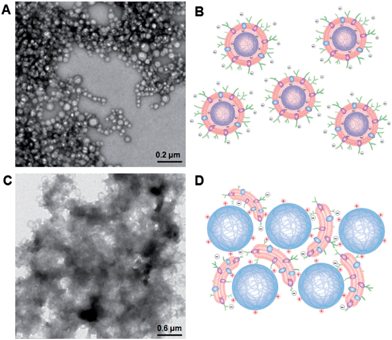
To further advance the understanding and development of cell membrane-cloaked nanoparticles, we examined effects of the nanoparticle core properties on the formation of the RBC-NPs. Since the electrostatic effect has shown a strong influence on lipid membrane interactions with a nanoparticle substrate,29 we investigated the use of positively and negatively charged polymeric cores for RBC-NP preparations. The negatively charged cores were prepared from carboxyl-terminated PLGA, and the positively charged cores were prepared by modifying carboxyl-terminated PLGA particles with polyethyleneimine (PEI). Upon mixing PLGA–COOH cores with 5% PEI solution, a change in the surface charge from −45 mV to +27 mV was observed with a negligible effect on the nanoparticle size (ESI, Fig. S5†). The two oppositely charged cores were then used to prepare RBC-NPs using the extrusion method.Unlike the negatively charged cores, which were readily extruded with RBC membrane vesicles to form RBC-NPs, the positively charged cores formed observable aggregates upon mixture with the vesicles. Extruding the positively charged cores with RBC membrane vesicles through 200, 400, and 1000 nm porous polycarbonate membranes resulted in significant membrane clogging that impeded the extrusion process. To better examine the membrane–particle interactions, the particle–membrane mixtures were visualized using transmission electron microscopy (TEM). The negatively charged cores formed consistent spherical particles with a core–shell structure (Fig. 4A). The absence of aggregates in membranes mixed with negatively charged cores suggests a more subtle membrane–particle interaction that is less disruptive to the membrane structure and fluidity (Fig. 4B). In contrast, the positively charged cores coalesced into large polydisperse aggregates with the membranes (Fig. 4C). The aggregate formation can likely be attributed to the strong electrostatic interactions between positively charged cores and the negatively charged RBC membranes. Owing to the dense, negatively charged sialyl moieties on the extracellular membrane side, the strong affinity to positively charged nanoparticles could collapse the fluidic lipid bilayer and impede the local arrangement necessary for lipid coverage.30 Bridging between cores and collapsed lipid membranes with opposite charges can therefore account for the observed aggregate formation (Fig. 4D). While understanding the dynamics between the negatively charged cores and RBC membranes demands in-depth molecular analysis beyond the scope of the present study, we demonstrate that the negative surface charge is a major factor in enabling proper RBC-NP formation. The electrostatic repulsion between negatively charged particle surfaces and asymmetrically charged RBC membranes also provides a driving force that favors the right-side-out membrane orientation on the RBC-NPs.
3.5 Surface curvature of polymeric cores affecting RBC membrane coating
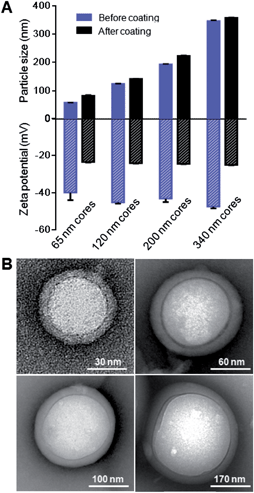
As lipid bilayer coating on nanoscale substrates can be influenced by the substrates' radius of curvature,31 or, alternatively speaking, the cores' size, we demonstrate the applicability of the RBC membrane cloaking process over a range of particle sizes that are relevant for nanomedicine applications. We prepared carboxyl-terminated PLGA cores of different diameters using variations of the solvent displacement method,32 creating unimodal nanoparticles that ranged from 65 nm to 340 nm. Each of these PLGA nanoparticle cores was extruded with the corresponding amount of RBC membrane vesicles (ESI†) through a polycarbonate membrane with appropriate pore sizes to form RBC-NPs. For all the differently sized particles, RBC membrane coating resulted in an increase in size by 10–20 nm. The cloaking process also shifted the zeta potential of the particles from approximately −45 mV to −24 mV, which corresponds closely to the zeta potential of RBC membrane vesicles (Fig. 5A and ESI, S6†). TEM visualization of particle cores 65, 120, 200, and 340 nm in diameter showed that uniform RBC membrane cloaks were broadly applied (Fig. 5B). The flexibility of the membrane coating technique offers the freedom to functionalize a broad range of nanodevices with different sizes and geometries. The size of RBC-NPs can also be fine-tuned to match the need of specific medical applications.4. Discussion
By examining multiple interfacial aspects of RBC-NPs, the present study provides a better understanding of the platform. The particles were shown to be completely enveloped, suggesting a continuous membrane coating on the particle surfaces. The completeness of membrane coverage has significant implications as it can shield the particles from external exposure, thereby minimizing the risk of complement activation and immunological responses typically associated with foreign materials. The RBC-NPs were also found to possess right-side-out oriented membrane coating, which exposes dense surface glycan contents to the aqueous environment and ensures the proper protein presentation on the particle surface.10 The hydrophilic glycan layer also enhances the colloidal stability of RBC-NPs under ionic conditions, and its enzymatic removal by trypsin led to rapid particle aggregation. Taken together with previous findings that demonstrate the membrane and surface protein retention6 and cargo encapsulation by the particles,33 the RBC-NPs can be considered a multifunctional nanoparticle system that integrates the steric stabilization of polysaccharides, the biological immunoevasive activities of membrane proteins, the cargo loading capacity of polymeric cores, and the anchoring and insulating functions of the lipid membranes, into a single, unified package. The present study also demonstrates RBC-NPs' size-tunability and enzyme-triggered particle destabilization. These features provide the platform with the opportunity to be tailored for specific delivery goals.In addition to elucidating structural composition of RBC-NPs, the present study also extends the understanding of cell membrane–nanoparticle interaction that drives the membrane cloaking process. Both the surface glycans on RBC membranes and the physicochemical properties of nanoparticle cores were found to play a significant role in RBC-NP formation. With carboxy-terminated PLGA cores as nanoparticle substrates, the membrane cloaking process was highly efficient. Achieving complete particle shielding and particle stabilization requires a membrane-to-polymer ratio closely matched to the theoretical value for complete particle coverage. This efficient cloaking process can be attributed to the glycan-driven colloidal stabilization of the polymeric particles, which renders the system energetically favorable. The asymmetrically distributed sialic acids, which bestow a negative charge to the extracellular membrane side, can also affect the membrane–particle interaction electrostatically. Rapid aggregation occurred upon the mixture of positively charged polymeric cores with RBC membranes, as strong electrostatic attractions resulted in membrane–particle bridging. In contrast, negatively charged cores readily form RBC-NPs with right-side-out membrane coating, which is likely aided by the repulsion between particle and extracellular membrane surfaces. As surface sialic acids and their associated membrane charge asymmetry are broadly present in biology,34,35 such a membrane–particle interaction should be considered broadly in the development of cell membrane-functionalized nanodevices.
5 Conclusions
RBC-NPs present a unique nanocarrier platform that combines the immunomodulatory properties of natural cellular components with the cargo carrying capacity of polymeric nanoparticles. In the present study, we examined the platform's interfacial features and assessed the roles of RBC surface glycans and nanoparticle properties in RBC-NP formation. RBC-NPs are completely shielded by lipid membranes and are stabilized by surface glycans. The hydrophilic glycans and the negatively charged sialic acid residues contribute to the structural organization of RBC-NPs, which possess a unilamellar, right-side-out membrane cloak. Owing to the inherent electrostatic properties of RBC membranes, positively charged nanoparticles did not yield proper RBC-NP formation. The membrane cloaking approach was successfully applied to nanoparticle substrates between 65 and 340 nm in diameter (the size range tested in this study), demonstrating the versatility of the camouflaging technique toward a variety of nanocarriers.Acknowledgements
This work is supported by the National Science Foundation Grant DMR-1216461 and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number R43AI100398. B.L. is supported by a National Institutes of Health R25CA153915 training grant from the National Cancer Institute.Notes and references
- J. Yoo, D. Irvine, D. Discher and S. Mitragotri, Nat. Rev. Drug Discovery, 2011, 10, 521 CrossRef CAS PubMed.
- S. Balmert and S. Little, Adv. Mater., 2012, 24, 3757 CrossRef CAS PubMed.
- C. Hu, R. Fang, B. Luk and L. Zhang, Nanoscale, 2014, 6, 65 RSC.
- R. Tsai, P. Rodriguez and D. Discher, Blood Cells, Mol. Dis., 2010, 45, 67 CrossRef CAS PubMed.
- P. Rodriguez, T. Harada, D. Christian, D. Pantano, R. Tsai and D. Discher, Science, 2013, 339, 971 CrossRef CAS PubMed.
- C. Hu, L. Zhang, S. Aryal, C. Cheung and R. Fang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10985 Search PubMed.
- W. Gao, C. Hu, R. Fang, B. Luk, J. Su and L. Zhang, Adv. Mater., 2013, 25, 3549 CrossRef CAS PubMed.
- C. Hu, R. Fang, J. Copp, B. Luk and L. Zhang, Nat. Nanotechnol., 2013, 8, 336 CrossRef CAS PubMed.
- R. Fang, C. Hu, K. Chen, B. Luk, C. Carpenter, W. Gao, S. Li, D. Zhang, W. Lu and L. Zhang, Nanoscale, 2013, 5, 8884 RSC.
- C. Hu, R. Fang, B. Luk, K. Chen, C. Carpenter, W. Gao, S. Li, D. Zhang, W. Lu and L. Zhang, Nanoscale, 2013, 5, 2664 RSC.
- N. Doshi, S. Zahr, S. Bhaskar, J. Lahann and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 21495 CrossRef CAS PubMed.
- T. Merkel, S. Jones, K. Herlihy, F. Kersey, A. Shields, M. Napier, J. Luft, H. Wu, W. Zamboni, A. Wang, J. Bear and J. DeSimone, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 586 CrossRef CAS PubMed.
- C. Hu, R. Fang and L. Zhang, Adv. Healthcare Mater., 2012, 1, 537 CrossRef CAS PubMed.
- M. Mager, V. LaPointe and M. Stevens, Nat. Chem., 2011, 3, 582 CrossRef CAS PubMed.
- W. Evans and J. Graham, Membrane Structure and Function, Oxford Univ. Press, New York, 1991, pp. 1–86 Search PubMed.
- E. Eylar, M. Madoff, O. Brody and J. Oncley, J. Biol. Chem., 1962, 237, 1992 CAS.
- M. Fukuda, Molecular Glycobiology, Oxford Univ. Press, New York, 1994, pp. 1–52 Search PubMed.
- R. Schauer, Curr. Opin. Struct. Biol., 2009, 19, 507 CrossRef CAS PubMed.
- S. Kelm and R. Schauer, Int. Rev. Cytol., 1997, 175, 137 CAS.
- P. Raveendran, J. Fu and S. Wallen, J. Am. Chem. Soc., 2003, 125, 13940 CrossRef CAS PubMed.
- C. Cho, J. Jeong, T. Ishihara, R. Takei, J. Park, K. Park, A. Maruyama and T. Akaike, Biomaterials, 1997, 18, 323 CrossRef CAS.
- C. Lemarchand, R. Gref and P. Couvreur, Eur. J. Pharm. Biopharm., 2004, 58, 327 CrossRef CAS PubMed.
- J. Rothman and J. Lenard, Science, 1977, 195, 743 CAS.
- T. Steck and G. Dawson, J. Biol. Chem., 1974, 249, 2135 CAS.
- R. Winzler, Red Cell Membrane, Lippincott, Philadelphia, 1969, p. 157 Search PubMed.
- H. Heidrich and G. Leutner, Eur. J. Biochem., 1974, 41, 37 CrossRef CAS.
- A. Miller, J. Sullivan and J. Katz, Cancer Res., 1963, 23, 485 CAS.
- J. Durocher, R. Payne and M. Conrad, Blood, 1975, 45, 11 CAS.
- S. Mornet, O. Lambert, E. Duguet and A. Brisson, Nano Lett., 2005, 5, 281 CrossRef CAS PubMed.
- M. Fischlechner, M. Zaulig, S. Meyer, I. Estrela-Lopis, L. Cuellar, J. Irigoyen, P. Pescador, M. Brumen, P. Messner, S. Moya and E. Donath, Soft Matter, 2008, 4, 2245 RSC.
- Y. Roiter, M. Ornatska, A. Rammohan, J. Balakrishnan, D. Heine and S. Minko, Nano Lett., 2008, 8, 941 CrossRef CAS PubMed.
- J. Chan, L. Zhang, K. Yuet, G. Liao, J. Rhee, R. Langer and O. Farokhzad, Biomaterials, 2009, 30, 1627 CrossRef CAS PubMed.
- S. Aryal, C. Hu, R. Fang, D. Dehaini, C. Carpenter, D. Zhang and L. Zhang, Nanomedicine, 2013, 8, 1271 CrossRef CAS PubMed.
- R. Schauer, Adv. Carbohydr. Chem. Biochem., 1982, 40, 131 CrossRef CAS.
- M. Glick, C. Comstock, M. Cohen and L. Warren, Biochim. Biophys. Acta, 1971, 233, 247 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Theoretical calculations and supporting figures. See DOI: 10.1039/c3nr06371b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |