Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Mass spectrometry imaging of plant metabolites – principles and possibilities
Nanna
Bjarnholt
a,
Bin
Li
b,
Janina
D'Alvise
b and
Christian
Janfelt
*b
aDepartment of Plant and Environmental Sciences, University of Copenhagen, Bülowsvej 17, 1870 Frederiksberg C, Copenhagen, Denmark
bDepartment of Pharmacy, University of Copenhagen, Universitetsparken 2, 2100 Copenhagen, Denmark. E-mail: christian.janfelt@sund.ku.dk; Fax: +45 35 33 60 30; Tel: +45 35 33 65 57
First published on 23rd January 2014
Abstract
Covering: up to the end of 2013
New mass spectrometry imaging (MSI) techniques are gaining importance in the analysis of plant metabolite distributions, and significant technological improvements have been introduced in the past decade. This review provides an introduction to the different MSI techniques and their applications in plant science. The most common methods for sample preparation are described, and the review also features a comprehensive table of published studies in MSI of plant material. A number of significant works are highlighted for their contributions to advance the understanding of plant biology through applications of plant metabolite imaging. Particular attention is given to the possibility for imaging of surface metabolites since this is highly dependent on the methods and techniques which are applied in imaging studies.
1 Introduction
Higher plants are versatile organisms, consisting of many different and highly specialized tissue types with very diverse metabolic profiles, of which many undergo dramatic changes in metabolism during development. In addition, plants are amongst Nature's most skilled chemists, biosynthesizing an estimated 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 different general and specialized metabolites1 to attract pollinators, to protect themselves against a multitude of biotic and abiotic stresses and to communicate with their surroundings. This overwhelming number of highly diverse small molecules poses a great challenge for plant metabolomics in general, and to the analysis of metabolite spatial distributions in particular. Mass spectrometry imaging (MSI) has been established as a valuable tool for imaging metabolite distributions in mammalian tissues, and is at the same time the only approach versatile enough to cover a broad range of plant natural products. Consequently, the research into plant MSI has expanded rapidly during the last decade as the basic techniques have become well documented and instruments have become commercially available. This review describes the current state of MSI, covering the most abundant methods and their particular advantages as well as the pitfalls within plant tissue analysis; it is written with the aim of helping the scientific community choose the right method for a given application. Special attention will be given to the challenge presented by the diverse nature of plant tissue physical characteristics, as these may be key factors in determining the preferred method. Specifically, plant cuticles and epicuticular waxes form a critical barrier for distribution analyses of internal metabolites, which may be more efficiently overcome by some methods than others. The different potentials of the different methods rely in part on sample preparation which will be described in detail. Finally, as plant metabolite MSI is still in its infancy, the number of actual biological results obtained with these methods is still limited. The most important ones will be described in relation to the perspectives for future plant research.
000 different general and specialized metabolites1 to attract pollinators, to protect themselves against a multitude of biotic and abiotic stresses and to communicate with their surroundings. This overwhelming number of highly diverse small molecules poses a great challenge for plant metabolomics in general, and to the analysis of metabolite spatial distributions in particular. Mass spectrometry imaging (MSI) has been established as a valuable tool for imaging metabolite distributions in mammalian tissues, and is at the same time the only approach versatile enough to cover a broad range of plant natural products. Consequently, the research into plant MSI has expanded rapidly during the last decade as the basic techniques have become well documented and instruments have become commercially available. This review describes the current state of MSI, covering the most abundant methods and their particular advantages as well as the pitfalls within plant tissue analysis; it is written with the aim of helping the scientific community choose the right method for a given application. Special attention will be given to the challenge presented by the diverse nature of plant tissue physical characteristics, as these may be key factors in determining the preferred method. Specifically, plant cuticles and epicuticular waxes form a critical barrier for distribution analyses of internal metabolites, which may be more efficiently overcome by some methods than others. The different potentials of the different methods rely in part on sample preparation which will be described in detail. Finally, as plant metabolite MSI is still in its infancy, the number of actual biological results obtained with these methods is still limited. The most important ones will be described in relation to the perspectives for future plant research.
The high specialization in plant tissue metabolism leads to very diverse metabolite distributions, for example the pollinator attracting flower pigmentation patterns2 or the accumulation of defense compounds in sensitive tissues.3 Traditionally, such discoveries have been based on extraction of whole tissue for chemical analyses, in some cases guided by specific molecular characteristics of the analytes, for example the UV fluorescence of hydroxycoumarins accumulated during deterioration of cassava (Manihot esculenta) tubers4 or stilbenes produced as a response to downy mildew infection or UV light in grapevine (Vitis vinifera).5 However, in the chemical analysis of bulk tissue, the finer details of the distributions are lost, whereas on the other hand, imaging based on functional group histochemistry normally does not distinguish between individual compounds. Some compounds can be stained by reactions with exogenously applied chemicals and visualized under the microscope, for example alkaloids, terpenoids, tannins6 and free sugars.7 These methods also lack specificity for individual compounds, and moreover they rely on chemical treatment of tissues, which may in itself cause dislocation or other distortions of localization. On the cellular level, defense compounds, such as terpenoids, alkaloids and glucosinolates, are known to accumulate in single or few-cell structures, including, but not limited to, trichomes, laticifers or specialized cells scattered between other cells in a given tissue.8–11 In barley (Hordeum vulgare), the defense compounds hydroxynitrile glucosides are exclusively found in the single cell layer of the epidermis,12 and from other plants it is known that α-hydroxynitrile glucosides (commonly known as cyanogenic glucosides) are stored in the vacuoles of cells,13i.e. a subcellular compartmentalization of metabolites. These results have mainly been obtained by relatively cumbersome and lengthy procedures to isolate a given tissue, cell or compartment for chemical analysis of contents. Moreover, in the case of, for example, glucosinolates, the initial identification of relevant cells relied on molecular characteristics, namely the high sulfur content of these specialized metabolites.11 With MSI it is possible in one analysis to get a snapshot of the metabolic status in the tissue investigated, and of course to obtain information on the majority of all the above mentioned compounds and more, without prior knowledge or assumptions based on functional group characteristics.
Amongst biosynthetic pathways, several are known to be compartmentalized on the cellular or subcellular level. For example, in general metabolism the carbon assimilation pathway of C4 plants is divided between mesophyll and bundle-sheath (or similar) cells, and the amino acid biosynthesis is specifically localized to the chloroplast. In production of specialized metabolites, the terpenoid and glucosinolate biosynthetic pathways are divided between cytosol and chloroplasts (or even more compartments),14,15 and the complex biosynthetic pathway of monoterpenoid indole alkaloids in Madagascar periwinkle (Catharanthus roseus) is separated between at least three different cell types and five different subcellular compartments.16 Such discoveries are almost exclusively based on studies of localization and in vitro activities of the involved enzymes. Proteins can be tagged by e.g. antibody labeling or fluorescent markers, such as GFP (Green Fluorescent Protein), and studied with high resolution microscopy. These and other methods for studying protein localization are becoming common procedures, which in many cases allow determination of the subcellular localization of enzymes. Thus, one major bottleneck in further understanding of details in plant metabolism has become the determination of subcellular localization of metabolites, i.e. substrates and products of the enzymes present in various cell compartments. This review will therefore also deal with the issue of spatial resolution; as it will be revealed, the spatial resolution that can be obtained by MS imaging has improved considerably over the past few years, but there are still several inherent limitations to be addressed before the ultimate goal of MS imaging of plant metabolites with subcellular resolution is reached.
2 Mass spectrometry imaging
Mass spectrometry is one of the most widely used analytical techniques and extensively used within fields such as pharmaceutical and environmental analysis, food analysis and throughout the biological and medical sciences, e.g. in microbiology, plant science, clinical diagnostics, forensics and protein research. The reason for the success of mass spectrometry in all these fields is the sensitivity provided by the technique (extremely low amounts of e.g. pollutants or metabolites can be detected), the selectivity (even compounds with highly similar structures can be distinguished, in contrast to what many spectroscopic techniques are capable of) and the identification capabilities (structural information for confirmation of known compounds and identification of new compounds) of the technique. The goal of mass spectrometry imaging is to provide an imaging technique which, in contrast to all optical imaging techniques, has exactly these qualities with regards to the chemical information that is an inherent part of mass spectrometry images.The vast majority of mass spectrometric analyses today are performed in combination with separation techniques, such as gas chromatography (GC) and liquid chromatography (LC) in the well-known combinations GC-MS and LC-MS. For analyses of plant or animal tissues, these traditional approaches to mass spectrometry require preceding extractions, bringing the analytes to a solution that can be injected into a GC-MS or LC-MS system. Such an approach may very well be used to show that a particular compound was present in the sample, but all the information about its distribution in the sample is lost in the extraction process.
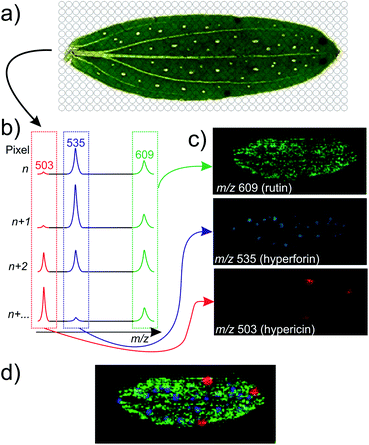
The spatial information in the mass spectrometric analysis can be retained if the extraction process is avoided or performed very locally in the sample. This is possible with many of the new ambient ionization techniques, such as Desorption Electrospray Ionization (DESI).17 DESI and certain other ionization techniques18 are particular in the way that they ionize and desorb analytes directly from a surface, such that traditional liquid extractions are avoided. One can thus move the sample below the ionization spot while looking at the mass spectra being recorded and thereby see exactly where in the sample a given compound is detected. By automating the procedure, images can be generated showing the spatial distributions of all the detected compounds (Fig. 1). In MSI the mass spectrometer is typically set to operate in full-scan mode, and in this way from one experiment of a few hours duration, images of a large number of different compounds can be generated.
What differs between the different MS imaging techniques is the way ions are generated, e.g. by ion bombardment as in Secondary Ion Mass Spectrometry (SIMS), by laser illumination with or without the use of matrix molecules as in (Matrix Assisted) Laser Desorption Ionization (LDI/MALDI), or as secondary ions generated by an electrospray of charged solvent droplets as in Desorption Electrospray Ionization (DESI). More recently, a number of alternative techniques based on the ablation19 or extraction20,21 of compounds from a surface followed by ionization have been introduced, increasing the number of MS imaging techniques to choose from.
Over the years, a number of review papers have been written about MS imaging.22–29 While most of them are focusing on the imaging of animal tissue – which is still the preferred type of sample in the MS imaging community – the principles in MS imaging of plant material are the same, and more recently a number of review papers dedicated to MS imaging of plant material30–34 and natural products35 have also been published, some of them focusing mainly on MALDI imaging. The present review is an attempt to give an overview of the possibilities with several of these techniques and the knowledge which has been obtained in such imaging studies of plant material. More detailed information about the fundamentals of MS imaging may be found in the above mentioned reviews.
3 Laser Desorption Ionization (LDI) and Matrix Assisted Laser Desorption Ionization (MALDI)
Laser Desorption Ionization (LDI) is a widely used group of ionization techniques in mass spectrometry, which use laser light (UV or IR) for the direct desorption and ionization of compounds from a surface. In Matrix Assisted Laser Desorption Ionization (MALDI) mass spectrometry, which is by far the most commonly used form of LDI, the analyte is co-crystallized with a chemical matrix, which absorbs the laser energy and releases the analytes into the gas-phase in a process leading to ionization. The exact mechanism of the desorption/ionization in MALDI is far from fully understood and is still being debated.36,37 The addition of the matrix has a number of advantages, in particular that it allows ionization of compounds which do not absorb light themselves. Moreover, since the energy of the laser light is absorbed by the matrix and not directly by the analytes, MALDI is a soft ionization technique, showing little fragmentation in the mass spectra.The focused ultraviolet (typical wavelengths 337 nm, 355 nm or 266 nm) or infrared (2.94 μm or 10.6 μm) laser beam is irradiated directly onto a metal plate with the sample deposited or mounted on the surface. In traditional non-imaging MALDI, as introduced by Karas and Hillenkamp in 1987,38 the dissolved sample is mixed with a matrix compound and deposited on a metal plate where the solution evaporates to dryness prior to the analysis. The sample plate is placed in the ion source in the vicinity of the ion entrance of a mass spectrometer where it is illuminated by a pulsed laser beam with pulse widths of 2–100 nanoseconds; several laser shots may be used for one mass spectrum. In MALDI imaging, as introduced by Spengler et al.39 and Caprioli et al.40 in the mid 1990's, the sample is already mounted on a plate and the matrix is subsequently deposited. The choice and deposition of the MALDI matrix, which are discussed in the sample preparation section below, are of paramount importance in MALDI imaging.
Commercial instruments, mainly for UV-MALDI, are available from many vendors, offering spatial resolutions around 20 μm. Although in a few cases higher resolution (around 10 μm) has been presented on commercial instruments,41 spatial resolution of a few microns has so far only been presented on prototype instruments from the research groups of Spengler26,42 and Caprioli,43 recently allowing for MALDI imaging of single cells.43,44 The problem in attempting MALDI imaging at higher resolution is not focusing the laser to a smaller diameter, but the fact that the ion yield (and thus the sensitivity) drops significantly with laser spot sizes smaller than 1 μm. Moreover, the application of the matrix becomes extremely critical when high resolution imaging is attempted,45 since the size of matrix crystals incorporated with analytes should be smaller than the required lateral resolution of the imaging experiment.
The majority of the commercially available MALDI instruments are based on a TOF (Time-of-flight) mass analyzer and has the sample enclosed in a vacuum chamber during the analysis. An alternative approach, although not nearly as commercially abundant, is atmospheric pressure MALDI (AP-MALDI).46 Among the advantages of AP-MALDI is the possibility of using liquid matrixes, as well as interchangeability with other atmospheric ionization sources (e.g. ESI and APCI) on the same mass spectrometer.47 The possibility for AP-MALDI imaging can thus be purchased as an add-on to an existing standard electrospray ionization mass spectrometer. In addition, another advantage with AP-MALDI imaging is a lower degree of fragmentation compared to vacuum MALDI due to the rapid collisional cooling that occurs at atmospheric pressure.48 In return, however, AP-MALDI mass spectra are also characterized by a presence of analyte–matrix cluster ions,49 which sometimes hinder the detection of some targeted analytes more prone to cluster with matrix ions. The signals from analyte–matrix cluster ions can be recognized and identified in subsequent MS/MS experiments. The concern about the lower sensitivity in AP-MALDI in early times has been largely eliminated with the advancement of the technique.
The main challenge of MALDI analysis is the interference between the high matrix ion background in the low mass range (m/z < 1000) and the signals from small metabolites. This problem can be reduced with the use of high resolution mass spectrometry or MS/MS, but completely eliminated with the use of matrix-free LDI. Direct UV or IR laser desorption/ionization for detection and imaging of plant metabolites is in some cases advantageous due to simplified sample pretreatment, minimized interferences with matrix ions and reduced risk of metabolite relocation. It does, however, rely on strong UV light absorption of the analytes, and the absence of matrix causes a higher degree of molecular fragmentations due to the direct exposure to laser pulse. In principle, any metabolites with condensed aromatic rings could absorb UV light and be desorbed/ionized with UV-LDI, such as plant pigments (flavonoids, flavanes, anthocyanins) or indolic alkaloids and related structures.41 However, the results of such metabolite imaging tend to be biased, as many analytes do not absorb enough UV light to be ionized.
Compared to UV-LDI, infrared LDI enables the direct detection and imaging of a wide range of primary and secondary plant metabolites without the addition of matrix, since the IR laser beam simply uses the endogenous water as matrix. The O–H band stretch vibrations of tissue water or other cell constituents match the 3 μm wavelength of an IR laser well. A few reports have described the utilization of IR lasers for the analysis or imaging of metabolites directly from plant tissues.50–52 The disadvantage of IR-LDI compared to the traditional UV-LDI is that infrared laser radiation is much more difficult to focus than UV. While UV laser radiation can easily be focused to a 1 μm spot size, what has been obtained with IR lasers is a spot size around 30 μm.53 Thus, IR-LDI imaging of mouse brain with a spatial resolution of 30 μm has recently been presented,54 but no such results have yet been reported for plants. The use of physiological water as the matrix is indeed the real advantage of IR-LDI imaging, making LDI imaging of samples in their natural state much more direct, but also a point of concern, since the water tends to evaporate during the hour-long acquisition on an image, causing a severe reduction in the ion yield. Therefore, the sample must be kept cold during the entire imaging experiment to avoid premature evaporation of the physiological water. Furthermore, the use of physiological water as matrix requires a fairly homogenous distribution of water in the sample, a requirement which is not necessarily met in all samples.
MALDI imaging is today by far the most widely applied MS imaging technique for animal tissue as well as for plant tissue. The reason for this is that MALDI routinely provides resolutions down to 20 μm and a wide selection of instruments is commercially available at prices comparable to other mass spectrometers. In fact, most MALDI instruments sold today, e.g. for protein research, are capable of imaging with regards to the hardware and the accompanying software.
As seen in Table 1, MALDI has been used for imaging of a wide range of analytes in many different types of plant material. The advantage of MALDI is seen for example when MALDI is applied for imaging of lipids in Camelina sativa55 and barley56 seeds with a spatial resolution of 15–25 μm. This resolution is not readily obtained with DESI, and the relatively large analytes are only poorly detected with SIMS. What is particular for MALDI is the capability of imaging biomacromolecules, such as proteins, which cannot be imaged by DESI and SIMS. While MALDI imaging is frequently applied on proteins in animal tissue samples, e.g. as biomarkers for cancer, protein imaging on plant material has been demonstrated to be possible,57,58 but also much more complicated than imaging of smaller metabolites.33
| Year published | Species | Sample type | Imaging technique and image resolution | Imaged analytes | Sample preparation | Reference |
|---|---|---|---|---|---|---|
| 2005 | Soya | Leaf, stem | MALDI | Mesotrione and azoxystrobin (pesticides) | Freeze-drying, mounting with conductive tape or blotting onto and acetone wetted cellulose membrane. CHCA matrix | 133 |
| 2007 | Wheat (Triticum aestivum) | Seed | MALDI 100 μm | Metabolites, amino acids, carbohydrates | Cryosectioning CHCA or 9-aminoacridine matrix | 134 |
| 2007 | Wheat | Stem | MALDI 200 μm | Oligosaccharides | Cryosectioning (50 μm) CHCA matrix | 135 |
| 2007 | Strawberry | Fruit skin | AP IR-MALDI 200 μm | Saccharides, citric acid | Sectioned (0.2–0.5 mm) at room temperatures with knife. Fresh samples mounted to steel surface without use of adhesive | 136 |
| 2008 | White lily (Lilium candidum) | Petal | AP IR-MALDI 200 μm | GABA, L-glutamine, saccharides | Mounting of sample on stage. No matrix applied | 129 |
| 2008 | Thale cress/Arabidopsis (Arabidopsis thaliana) | Leaf | MALDI 200 μm | Glucosinolates | Mounting of samples using double-sided tape. 9-AA matrix | 93 |
| 2009 | Peach | Fruit | MALDI 400 μm | Lipid transfer protein | Cryosectioning (250 μm). Thaw mounting on conductive microscope slides. Sinapinic acid matrix | 57 |
| 2009 | Sunflower | Stem | MALDI 200 μm | Nicosulfuron (pesticide) | Cryosectioning CHCA matrix | 137 |
| 2010 | Egg plant (Solanum melongena) | Fruit | MALDI 200 and 25 μm | GABA, amino acids, carbohydrates | Cryosectioning (14 μm) DHB matrix | 138 |
| 2010 | Juveline poplar (Populus deltoids) | Stem | MALDI ≈ 20 μm | Cellulose compounds | Cryosectioning (50 μm) DHB matrix | 139 |
| 2010 | Rice (Oryza sativa) | Seed | MALDI100 μm | Lipids and other metabolites | Cryosectioning (8 μm) DHB matrix | 140 |
| 2010 | Thale cress/Arabidopsis, date palm (Phoenix sp.) | Leaf | MALDI 200 μm | Cuticular lipids | Mounting of samples using double-sided tape. DHB matrix | 117 |
| 2011 | Eastern cottonwood (Populus deltoides) | Stem | MALDI 50 μm | Cellulose | Sectioning on vibratome (50 μm). DHB matrix | 141 |
| 2012 | Barley, tobacco (Nicotiana tabacum) | Grain, root | MALDI 15–35 μm | Lipids | Cryosectioning (20–55 μm). Drying. DHB and HCCA matrix | 56 |
| 2012 | Apple (Malus domestica) | Fruit | MALDI 75–150 μm | Glycosylated flavonols and dihydrochalcones | Manual slicing with razor blade CHCA matrix | 142 |
| 2012 | Rabbiteye blueberry (Vaccinium ashei) | Fruit | MALDI 100 μm | Anthocyanins | Cryosectioning (50 μm) DHB matrix | 91 |
| 2012 | Potato (Solanum tuberosum) | Tuber | MALDI200 μm | Glycoalkaloids | Cryosectioning (6 μm) DHB matrix | 143 |
| 2012 | Black rice (Oryza sativa L. Japonica) | Seed | MALDI 50 μm | Anthocyanins, lipids | Embedding in 2% CMC and cryosectioning (10 μm) DHB matrix | 108 |
| 2012 | Cotton (Gossypium hirsutum) | Seed | MALDI 50 μm | Lipids | Cryosectioning (30 μm) DHB matrix | 106 |
| 2012 | Thale cress/Arabidopsis | Flower bud, sepal, silique | MALDI 50 μm | Glucosinolates | Mounting with conductive tape. 9-AA matrix | 144 |
| 2012 | Petunia (Petunia × hybrid) | Leaf | MALDI 100 μm | Cyclotides | Cryosectioning (15 μm) CHCA matrix | 105 |
| 2012 | Capsicum annuum | Fruit | MALDI 250 μm | Capsaicin | Cryosectioning (70 μm) CHCA matrix | 145 |
| 2013 | Populus nigra, Ambrosia trifida, Artemisia absinthium, Hibiscus syriacus | Pollen grains | MALDI 50–150 μm | Unidentified, characteristic high-mass compounds | Embedding in resin layer on ITO-coated glass slide. CHCA matrix | 58 |
| 2013 | Medicago (Medicago truncatula) | Root, root nodule | MALDI 50 μm | Amino acids, saccharides, lipids, flavonoids, organic acids | Embedding in gelatin and cryosectioning (12 μm). DMAN and DHB matrix | 122 |
| 2013 | Avocado (Persea americana) | Mesocarp | MALDI 300 μm | Lipids: triacylglycerols | Imprinting onto nitrocellulose membrane. DHB matrix | 110 |
| 2013 | Camelina sativa | Seed | MALDI 25 μm | Lipids: triacylglycerols and phosphatidylcholines | Embedding in gelatin and cryosectioning (30–50 μm). DHB Matrix | 55 |
| 2007 | Apple, strawberry | Fruit | GALDI 100 μm | Organic acids, flavonoids and oligosaccharides | Cryosectioning (15 μm), mounting on stainless steel plate | 146 |
| 2008 | Thale cress/Arabidopsis | Leaf, flower, stem | GALDI 50–150 μm | Lipids, cerebrosides, saccharides, flavonoids and other secondary metabolites | Leaves and petals: either no sample pretreatment or dipping in chloroform. Stem: cryosectioning (50–80 μm). Coated with colloidal graphite solution | 130 |
| 2009 | Thale cress/Arabidopsis | Flower, leaf | LDI 50–100 μm | Epicuticular wax metabolites | Fixing to stainless steel plate with conductive doublesided tape, drying. Coated with colloidal silver solution | 102 |
| 2009 | Thale cress/Arabidopsis, St. John's wort (H. reflexum and H. perforatum) | Stamen, petal, leaves, placenta, pollen | LDI 10 μm | Secondary metabolites | Laser microdissection (stigma), cryosectioning (placenta, 60 μm). Mounting with carbon conductive adhesive tape | 88 |
| 2009 | Ginger | Rhizome | AP-LDI 10–20 μm | Gingerol, terpenoids, saccharides | Slicing using razor blade, mounting on ITO-coated glass slides using double-sided conductive tape | 147 |
| 2010 | Grape vine (Vitis vinifera) | Leaf | LDI 25 μm | Stilbenoids | Fixing to MALDI plate with aluminized tape | 5 |
| 2010 | Thale cress/Arabidopsis | Flower, stem, root | LDI 12–50 μm | Epicuticular lipids | Mounting of samples using conductive double-sided tape. Coating with colloidal silver | 73 |
| 2011 | Chamberbitter (Phyllanthus urinaria) | Leaf | LDI 90 μm | Antifungal and antibacterial metabolites | Washing with water, drying with nitrogen | 148 |
| 2012 | Thale cress/Arabidopsis | Flower, petal | LDI 50 μm | Flavonoid metabolites | Mounting on stainless steel sample plates using conductive double-sided tape. Coated with colloidal graphite | 128 |
| 2013 | Wild tomato (Solanum habrochaites) | Leaf, trichome | LDI 25 μm | Acyl sugars, alka-loids, flavonoids, terpenoid acids | Imprinting onto graphite-coated glass slide. Carbon coating | 149 |
| 2010 | Switchgrass (Miscanthus × giganteus) | Stem | LDI/MALDI, 100 μm SIMS, ≈ 2 μm, 22 keV Au1 + beam | Saccharides | Cryosectioning (50 μm) LDI: Thaw mounting on glass slides. No matrix, CHB or CHCA matrix. Coating with gold. SIMS: Thaw mounting on Si wafer, drying, coating with gold | 68,69 |
| 2005 | Sugi (Cryptomeria japonica) | Wood tissue | SIMS 15 keV Ga+ beam | Ferruginol (a diterpene phenol) | Microtome slicing (30 μm) | 150 |
| 2008 | Hinoki cypress (Chamaecyparis obtuse) | Wood tissue | SIMS 2 μm spot diameter, 22 keV Au1+ beam | Hinokiresinol, hinokione, hinokiol, hinokinin | Microtome slicing (100 μm). Drying at room temperature | 151 |
| 2011 | Poplar (Populus trichocarpa) | Wood tissue | SIMS 300 nm spot diameter 25 keV Bi3+ beam | Guaiacyl and syringyl lignin units | Dehydration, incubation in wax, microtome slicing, incubation in wax, dewaxing and drying | 152 |
| 2012 | Maple (A. micranthum) | Wood tissue | SIMS 1–2 μm spot diameter, 22 keV Au1+ beam | Guaiacyl and syringyl lignin units | Microtome slicing (100 μm) | 70 |
| 2008 | Zebra plant (Aphelandra squarrose) | Leaf | LAESI 350 μm | Primary and secondary metabolites | Mounting of sample on glass slides | 153 |
| 2009 | Peace lily (Spathiphyllum lynise) and zebra plant (Aphelandra squarrose) | Leaf | 3D LEASI 300 μm lateral 30–40 μm depth | Secondary metabolites | Mounting of sample on glass slides | 63 |
| 2009 | Red macroalga (C. serratus) | Blade | DESI 200 μm | Bromophycolides | Mounting on PTFE substrate | 131 |
| 2011 | St. John's wort (H. perforatum), thorn apple (Datura stramonium), opium poppy (Papaver somniferum) | Leaf, petal, capsule | DESI 100–125 μm | Phloroglucinols, flavonoids, naphthodianthrones, saccharides, alkaloids | Imprinting onto porous PTFE | 89 |
| 2011 | Barley | Leaf | DESI 100–200 μm | Hydroxynitrile glucosides | Stripping of epidermis or imprinting onto porous PTFE | 132 |
| 2011 | Katsura tree (C. japonicum) and American sweetgum (Liquidambar styraciflua) | Leaf | DESI 130–310 μm | Chlorophyll catabolites | Imprinting onto porous PTFE | 127 |
| 2011 | Myristica malabarica | Seed | DESI 250 μm | Alkaloid | Cross-sectioning and imprinting onto printer paper | 154 |
| 2012 | Red alga (Phacelocarpus neurymeniodides) | Blade | DESI 180 μm | Antibacterial metabolite neurymenolide A | Mounting to glass slides with glue, followed by direct DESI imaging | 101 |
| 2013 | Japanese birdsfoot trefoil (L. japonicus), cassava (Manihot esculenta), sorghum (Sorghum bicolor) | Leaf, tuber, seedling | DESI 150–200 μm | Hydroxynitrile glucosides, saccharides | Imprinting onto porous PTFE. Cryosectioning (50 μm) | 94 |
| 2013 | St. John's wort (H. perforatum) | Leaf, petal | DESI 100–150 μm | Phloroglucinols, flavonoids, naphthodianthrones | Washing with chloroform. Mounting on double sided tape | 90 |
| 2013 | Potato, ginko (Gingko biloba), strawberry (Fragaria × ananassa) | Sprout, leaf, fruit | DESI 150–200 μm | Glycoalkaloids, flavonoids, glucosides, saccharides | Imprinting onto TLC plates | 155 |
| 2013 | Madagascar periwinkle, thale cress/Arabidopsis, potato, tomato. | Leaves and petals | DESI | Secondary metabolites | Imprinting onto TLC plates | 126 |
| 2012 | St. John's wort (H. perforatum) | Leaf | EASI 100 μm | Phloroglucinols, flavonoids, naphthodianthrones | Imprinting onto porous PTFE | 61 |
| 2009 | Sage (Salvia sp.) | Leaf | DAPPI 500 μm | Tocopherol, methyl carnosic acid | Mounting of sample (dry plant material) on stage | 62 |
| 2013 | Sunflower (Helianthus annuus) | Leaf | LA-ICP-MS 22 μm (x-axis) 300 μm (y-axis) | Selenium and sulfur | Drying, mounting with double-sided adhesive tape | 156 |
4 Desorption Electrospray Ionization (DESI)
Desorption Electrospray Ionization (DESI) was first presented as an ionization technique in 2004,17 and first applied for MS imaging in 2006.59 The ionization technique utilizes an electrospray generated by a low flow (0.5–10 μL min−1) of solvent and the application of a nebulizer gas and a high voltage potential (typically 4–5 kV), resulting in a beam of charged micro-droplets. When the spray is directed towards a surface, the surface will be wetted and simultaneous extraction, desorption and ionization will occur from the sample spot below the spray. The ionization principles of DESI are very similar to those of the normal electrospray technique, making the technique applicable to polar compounds in positive or negative ionization mode.As in all microprobe imaging techniques, the limiting factor for the spatial resolution is the size of the ionization spot. In a DESI experiment the spray cannot be focused the same way as the laser is in a MALDI experiment. The size of the ionization spot is typically around 200 μm, strongly depending on the composition of the spray solution and the flow rate, and the typical spatial resolution that can be obtained with DESI is around 100 μm, although a resolution of 35 μm was recently obtained by careful optimization of several experimental parameters.60 The limited spatial resolution offered by DESI is therefore the obvious limitation of the technique compared to the two other principal MS imaging techniques, MALDI and SIMS. The advantages of the technique are that the analysis takes place under ambient conditions and that the sample is analyzed directly without any application of matrix compounds, thus keeping the sample preparation to a minimum, avoiding matrix interferences and enabling analysis of wet surfaces as well. While MALDI analysis traditionally is performed preferably in the positive ion mode, DESI works equally well in the negative and positive ion mode. Furthermore, the DESI imaging experiment is performed on a normal electrospray ionization mass spectrometer like the ones present in thousands of research labs worldwide. They just need to be equipped with a DESI imaging ion source, which is commercially available for most mass spectrometers on the market. The simplicity of the DESI technique compared to SIMS or MALDI makes the technology quite affordable and even makes it possible for an engineering workshop to construct a DESI imaging source of their own design. This means that for many laboratories, the cost of entering the field of MS imaging with DESI is much lower than with SIMS and MALDI, which both require the purchase of a new mass spectrometer dedicated to the respective ionization technique (with the exception of AP-MALDI).
In situations where the application of high voltage under ambient conditions is undesirable, quite similar imaging may be performed with EASI (Easy Ambient Sonic spray Ionization), whose operating principles are highly similar to those of DESI but only relies on the pressure of the nebulizer gas in the creation of ions. EASI imaging was applied in imaging of secondary metabolites in the St. John's wort (Hypericum perforatum),61 but is not widely used in plant metabolite imaging. Likewise, a photo ionization analogue of DESI has been constructed, Desorption Atmospheric Pressure Photoionization (DAPPI), which utilizes UV light and heat for desorption and ionization of analytes, thus enabling ionization also of non-polar molecules. DAPPI has been applied in plant metabolite imaging once, but unfortunately the application of high temperature has a deteriorating impact on the spatial resolution that can be obtained, which is thus around 500 μm.62
DESI has been applied in imaging of a variety of different plant samples (see Table 1 and sections about applications of mass spectrometry imaging in plant science), in particular leaves and petals, which are often imaged indirectly via an imprint on a PTFE surface, as discussed in the section about sample preparation. DESI imaging may also be applied on cryo-sections of e.g. tubers or seeds, although the technique is of limited usefulness in imaging of very small seeds where the spatial resolution of 100 μm only provides very few pixels in an image.
5 Laser ablation based imaging
While the laser in a MALDI experiment is responsible for both desorption and ionization of analytes, the two events may be decoupled such that the desorption taking the analyte from the surface to the gas phase is performed by a laser and the subsequent ionization by some post-ionization technique, e.g. electrospray ionization. The principal technique using this approach is Laser Ablation Electrospray Ionization (LAESI), which under ambient conditions utilizes a mid-infrared laser to evaporate tiny portions of the sample, due to absorption of the laser light by native water molecules in the sample.19 The evaporated sample plume is extracted into an electrospray, thereby ionizing molecules from the ablated spot on the sample.LAESI has been applied in a number of imaging studies of plant material (see Table 1) and shows one distinct advantage in this context, namely that the ablation by the IR laser is an efficient way to overcome the problems of penetrating through the cuticle, as discussed later in this text. The sample preparation in imaging of e.g. leaves with LAESI is very simple since no application of matrix is needed, and the analytes, also in deeper parts of the tissue, are readily available for analysis. The laser can be applied not only for ablation of the sample to be ionized, but also as a means to probe several layers in the sample, i.e. depth profiling and acquisition of three dimensional images, since the laser gradually removes tissue as part of the ablation process.63 LAESI is commercially available as an imaging ion source to fit on the atmospheric interface of mass spectrometers from selected vendors, though at a significantly higher price than the cost of a DESI imaging ion source. The typical spatial resolution in a LAESI experiment is 200–300 μm, although ablation spots of 30 μm size have been obtained, in some cases making in situ analysis (but not imaging) of single plant cells possible, as demonstrated on onion epidermis cells.53
Laser ablation is also applied in combination with inductively coupled plasma mass spectrometry (LA-ICP-MS), which enables spatially resolved elemental analysis of solid samples, typically with a spatial resolution of 10–200 μm.64 This technique provides information about the distribution of elements (e.g. metals) in a sample, but not about the chemical context they are appearing in. LA-ICP-MS may be achieved by coupling a commercially available laser ablation source to an existing ICP-MS, but is also possible on dedicated commercially available LA-ICP-MS instruments.
6 Secondary Ion Mass Spectrometry (SIMS)
Although Secondary Ion Mass Spectrometry (SIMS) was the first of the MS imaging techniques to be presented (introduced for imaging in the 1980's65), its use for imaging of plant material is very limited. SIMS, which operates under high vacuum and uses high-energetic (5–40 keV) primary ions for ionization, has the advantage over all other MS imaging techniques that very high spatial resolution (better than 100 nm) can be obtained. The major disadvantage is that a very high degree of fragmentation is related to this very high spatial resolution. The situation has improved with the introduction of new cluster ion beams which provide a much higher ion yield, in particular in the higher mass range, thus enabling detection of intact lipids. However, the ion intensities in the m/z 800 region are still orders of magnitude lower than around m/z 100,66 and the sensitivity for intact lipids is therefore much lower compared to other MS imaging techniques. The higher ion yield of the cluster ion sources is an effect of a higher sputter yield from the surface, which thus limits the resolution in imaging of such larger molecules to around 1 μm. Due to the high degree of fragmentation in SIMS experiments, characteristic fragments of analytes are often imaged rather than the intact analytes.67SIMS does not require much sample preparation (no matrix is needed), but the fact that the analysis takes place under high vacuum, means the samples are often freeze dried before introduction into the vacuum, and the use of vacuum also implies the risk that volatile analytes evaporate before they are detected. SIMS instrumentation has been commercially available for many years, but the cost of a SIMS instrument is significantly higher than for any of the other MS imaging techniques and the abundance of the equipment in laboratories around the world is correspondingly lower.
In a plant imaging context, SIMS has been applied for imaging of stem68,69 and wood tissue70 for the distributions of saccharides and lignin units, thus benefiting from the inherent fragmentation of large polymers into smaller units as a consequence of the hard ionization.
7 Instrumental considerations in the imaging experiment
Sensitivity, selectivity and identification power were mentioned as key advantages of mass spectrometry, but the degree of those virtues in an MS imaging experiment is very much subject to how the experiment is performed and to which type of mass spectrometer is used.Most MS imaging experiments are performed in full-scan mode, which means that an entire mass spectrum is recorded in each pixel of the image. The advantage of this approach is that all the compounds which are ionized are detected and can therefore be imaged simultaneously. After such an experiment it is therefore possible to browse through the entire list of masses and observe which ions show interesting distributions in the sample. This is indeed a non-targeted approach, which often leads to interesting and unforeseen observations, as no specific knowledge about the sample is required prior to such an experiment. One must of course be aware that normally the imaging only takes place in one ion polarity at a time; that means that if images are desired of all compounds that ionize in the positive or the negative ion mode, two full scan experiments must be conducted, one in each polarity. However, methods have been presented on how to combine this in one single experiment.71,72 A problem in full scan mode can be spectral interferences between compounds having similar masses (isobars) or even identical masses (isomers). The problem with isobars is commonly occurring in imaging of lipids in animal tissue, but has also been demonstrated to exist in plant imaging, e.g. for epicuticular lipid metabolites from Arabidopsis (Arabidopsis thaliana) where the silver adducts of the C28 aldehyde (m/z 515.3376) and the C29 alkane (m/z 515.3728) have the same nominal mass73 and may thus appear as one compound in the images. Interferences with unknown molecules in the background (and with the matrix in MALDI) are also commonly occurring. Such interferences can be resolved on high-resolution mass spectrometers (spectral resolution, not to be confused with spatial resolution), such as TOF, Orbitrap and ion cyclotron resonance (ICR) instruments, but not on low resolution mass spectrometers, such as ion traps. The incidence of isomers is frequently occurring with plant metabolites, e.g. the well-known alkaloids cocaine and scopolamine (which both have the formula C17H21NO4 and appear at m/z 304.1548 when protonized), apigenin and genistein (C15H10O5) or nicotine and anabasine (C10H14N2). Such interferences cannot be resolved in a full scan experiment, but can be distinguished in an MS/MS experiment where the ion of interest is fragmented and the imaging is based on the fragment ions. In the example with cocaine and scopolamine, one would thus choose m/z 304 as precursor ion and use the m/z 182 fragment of cocaine and the m/z 156 fragment of scopolamine for much more selective imaging. The improved selectivity in MS/MS mode is typically accompanied by an improved sensitivity as a result of the increased signal-to-noise ratio obtained in an MS/MS experiment; in animal studies, this is often the preferred approach in the imaging of drugs and metabolites where maximum sensitivity is needed.74,75 MS/MS imaging is thus a targeted approach where the analytes and their fragmentation patterns are known prior to the imaging experiment. The disadvantage is that an MS/MS imaging experiment takes as long as a full-scan experiment, but only yields the image of one single compound. On some MALDI instruments and with the displaced dual-mode imaging method in DESI,72 it is possible to perform simultaneous full-scan and MS/MS imaging, however, with a correspondingly longer total acquisition time. MS/MS imaging is not common in SIMS, nor is it available on TOF instruments. It is found on triple quadrupoles (rarely used for MS imaging; though inferior in full-scan mode they are excellent for MS/MS imaging75), ion traps, TOF-TOFs, Q-TOFs, ICRs and most Orbitraps.
The high spectral resolution on some instruments makes it possible to determine the mass with very high accuracy.76 As an example, when an analyte is determined to have the mass of m/z 286.1438, the only molecular formula which can provide this exact mass is C17H20NO3, which in this example corresponds to protonated morphine. This feature, implemented in software for data analysis, is of great importance in the confirmation of detected compounds as well as in the identification of new compounds, and particularly useful in MS imaging experiments, which often provide a vast number of interesting distributions of unknown compounds.
In the planning of an imaging experiment, it is important to have realistic expectations of the combination of desired spatial resolution and the physical dimensions of the sample to be imaged. Together they determine the number of pixels in the images and thus the time it takes to acquire one image. In MALDI and DESI a typical acquisition time per pixel is around 1 s, depending on instrument and settings. That means that a 200 μm image of a 15 mm × 10 mm sample requires 3750 pixels, giving an approximate total acquisition time around 1 h. If 100 μm resolution is desired, the number of pixels grows by a factor of 22 = 4, giving an acquisition time around 4 h. Therefore, in high resolution MALDI MSI only quite small sample sections are imaged, and often the imaging experiment is set to run overnight. Work is in progress on the enhancement of the image acquisition times with MALDI MSI.77
8 Data analysis and image generation
The data acquisition is in many ways similar to conventional LC-MS. For every pixel the ion intensities of all detected compounds are recorded and from this, extracted ion chromatograms (EIC) are generated for ions of interest. In LC-MS the intensity of a compound is shown as a function of time, whereas in an MS image the intensity is shown as a function of position with the relative intensities typically indicated by a color scale in a 2-dimensional heat map (Fig. 1). Data acquisition and image generation is normally performed in two separate steps. To account for differences in the matrix distribution, the ionization efficiency, ion suppression effects, but also to be able to compare data obtained on different days, normalization and post-acquisition data analysis can be included in the data processing.78–81For most commercial imaging ion sources software is provided by the manufacturer, supporting only the data format of that particular instrument. Another approach, in particular used in publication from the early years of MS imaging, is to handle the imaging data manually, without the use of dedicated software for image generation, e.g. by importing the raw data into MATLAB.82 This requires some technical skills, but also gives some extra possibilities. Once the raw data exist as a matrix in MATLAB they can be processed and scaled in various ways, including normalization as part of a quantitative or semi-quantitative approach, as discussed below. What may make this complicated is of course the amount of data which must be handled. While the total amount of raw data from an MS imaging experiment on a low-resolution instrument (e.g. an ion trap) has the size of some hundred megabytes, for an image on a high-resolution instrument (e.g. an Orbitrap) the total amount of data from one image may be several gigabytes.
Recently, a common data format, imzML, has been developed in order to facilitate the exchange and comparison of MSI data across different MS vendors.83 ImzML allows loading of data from different instrument types and ionization techniques and creates files that can be read by freely available image generation software (available at http://www.maldi-msi.org), such as BioMap (Novartis) or the DataCube Explorer (AMOLF). Other software solutions, which combine image creation, analysis and data processing e.g. Mirion,84 are currently under development.
Often the resulting chemical image is matched to the optical image, to compare physiological structures with the distribution of the selected compounds.85 This represents the easiest way of validating the measured distribution of secondary metabolites, especially if the metabolite distribution is known in advance. St. John's wort and other Hypericum species accumulate hyperforin and other phloroglucinols in translucent glands, and naphthodianthrones, such as hypericin, in dark nodules of petals and leaves.86,87 The already known and easily identified localization of secondary metabolites has been used for proof-of-concept studies with LDI,88 indirect89 and direct DESI90 and EASI.61 Optical validation has also been used in distribution studies of anthocyanins in the exocarp and endocarp of blueberry (Vaccinium ashei).91 In MS imaging studies of drug distributions, other optical methods, such as fluorescence microscopy, have been used as an alternative, allowing the direct localization of the drug.92
Another approach is the use of selective extraction in combination with other analytical methods, such as separation techniques, often combined with mass spectrometry. This is especially useful for the validation of quantitative and semi-quantitative results. Shroff et al. used dissection and HPLC-UV analysis to validate the distribution and abundance of glucosinolates in Arabidopsis leaves.93 Hydrolyzed glucosinolate products are defense compounds against lepidopteran larvae, requiring the enzyme myrosinase in direct proximity to the glucosinolates for the reaction. For this reason another validation of the distribution of glucosinolates in the leaves was possible by measuring the myrosinase activity. Also the distribution of hydroxynitrile glucosides in Lotus japonicus leaves and cassava tubers was validated by LC-MS analysis.94
With mass spectrometry imaging experiments today primarily qualitative information is obtained. Quantitative approaches are currently developed in several laboratories, especially for the detection of drugs and metabolites in animal tissue.75,95,96 Mainly two different approaches have been used. In the first approach, ion suppression effects are calculated by the uniform application of an internal standard to the tissue section.97 For the second approach, the tissue section is covered with an isotope labeled internal standard for normalization. A standard curve is then established on a control tissue section, which does not contain the analyte, in order to obtain the same matrix effects as on the sample of interest.98 If no isotope labeled standard is available other normalization methods, such as normalization to another standard or the TIC (total ion current), have been performed instead.98,99 In biological applications it is of course not always possible to obtain a standard curve on a control sample that does not contain the metabolite of interest. This has been solved by Pirman and Yost by using a standard addition curve directly on the tissue.100 Using DESI imaging Andras et al. estimated the concentration of neurymenolide A, an allelochemical produced by the red algae Phacelocarpus neurymenioides that induces coral bleaching, by comparing averaged signal intensities on the algae surface to the signal intensities observed from a standard deposited on PTFE.101 However, this approach suffers from the lack of identical matrix backgrounds, and the result could only be considered semi-quantitative. As experimental validation of quantitative data is often complicated and labor intensive it is, as in the above mentioned study, not always performed. In addition, difficulties for plant surface analysis can be expected due to the heterogeneous distribution of metabolites in the different layers of the plant material. For example, Cha et al. found differences in the relative signal intensities for cuticular wax metabolites of Arabidopsis leaves measured with LDI, compared to the abundance of waxes determined in GC-MS analysis of the plant extracts.102 The authors suggest that the differences can be explained by the shallower penetration depths of LDI than for the GC-MS analysis and by differences in ionization efficiency for the different compounds.
9 Sample preparation
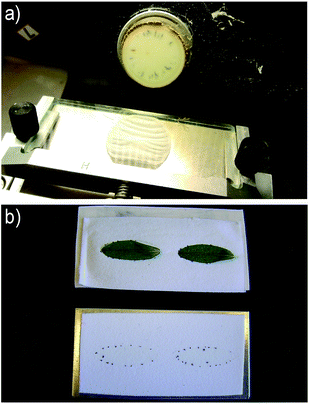
Development of sample preparation methods and protocols of MSI is an active area in MSI research that demands in-depth studies based on individual and community characteristics of plant samples. The issues of tissue sectioning, matrix application in MALDI, improvement of signal-to-noise ratios for small molecules, peptides and proteins are reviewed in some publications.30,103,104 Most of the methodologies of sample preparation in MSI are developed for animal tissues and cannot always be applied directly to plant samples due to the differences in composition and structure between plant and animal tissues. Plant tissue has some specific features compared to animal tissue, such as phloem and xylem tissue, large vacuoles with cell sap, presence of oil cells and in general many different cell types which behave differently, e.g. in the application of a MALDI matrix. All of these factors generate serious problems for sample preparation of plant tissues and need to be sorted out for the development of more specific sample protocols.Cryo-sectioning and imprinting of plant tissue are the two dominating methods applied in the first step of MSI sample preparation. The critical part in cryo-sectioning is to make a high-quality tissue section while avoiding contamination and relocation of metabolites. The sample size and shape are factors that need to be considered in cryo-sectioning. Root, stem and fruit tissue with the appropriate size not exceeding the imaging area available for the sample stage and regular shape can successfully be transformed to tissue sections that represent the histological features of tissue. Leaves and petals, however, which are generally very thin and fragile tissues, are a challenge to cut in sections, although paradermal sectioning (parallel to the surface) of petunia (Petunia × hybrida) leaves has been performed in an MS imaging study.105 With the exception of wood samples, which are typically cut at room temperature using a sliding microtome, most samples of plant material are cut at low temperature (around −20 °C) using a cryo-microtome. These instruments are traditionally used in histology for slicing of samples prior to microscopy studies. The sample may be embedded in an appropriate material, making the sample easier to cut, and attached to a sample holder by some adhesive medium. The sample is then passed by a knife in sequential circular motions such that the sample is moved forwards towards the knife in small, well-defined increments of some micrometers for each turn. In this way sections are produced in thicknesses from a few μm up to around 100 μm, as seen in Fig. 2a.
When cryo-sectioning is applied in the context of MS imaging, one should avoid the use or contamination from the optimum cutting temperature (OCT) polymer. The OCT is widely used in traditional histology for embedding and adhesion, but it causes a severe degree of ion suppression which often renders subsequent MS analysis impossible. Paraformaldehyde (4% solution) has been applied for embedding of seeds, but with a subsequent washing step to remove the excess fixative to minimize the interference from it.106 Pure water, gelatin107 and 2% CMC (carboxymethyl cellulose) solution108 have been reported as appropriate embedding media with minimized tissue tearing and no significant background interference. In addition, cryo-sectioning with the assistance of adhesive film or tape may be applied in sectioning of very hard and dry samples in order to keep the original sample structure.109 For hard tissues the normal cryo-microtome blades should be exchanged for the stronger tungsten carbide blades.109
Regarding the section thickness, there is no uniform standard. It ranges from a few to 100 μm with typical thicknesses around 20–50 μm. Typically, the thickness applied is determined by the physical properties of the sample; with some samples, the thinner sections tend to disintegrate and the thickness must be increased. The freshly cut tissue sections must be quickly transferred onto a sample slide by thaw-mounting. In many MALDI experiments, conductive indium tin oxide (ITO) coated glass slides are applied, but in DESI experiments and with some AP-MALDI instruments26 ordinary glass slides may be applied. It should be noted that for DESI measurements on some cryo-sections, mounting on double-sided adhesive tape prefixed on the glass slide is a good way to keep the sample from blowing away under the high pressure of the nitrogen gas. The prepared tissue sections can be stored in a −80 °C freezer or analyzed immediately after drying in a vacuum desiccator. In the case of MALDI the sections must be covered by matrix before analysis.
As discussed in a later section, in imaging of leaves and petals the presence of cuticular waxes on the surface can cause problems in the detection of metabolites. This problem can be circumvented by producing an imprint and then performing the MS imaging experiment on the imprint rather than on the original plant material.89 The method, mainly used in DESI imaging with porous PTFE as imprinting surface, was first developed for the visualization of the plant metabolites, especially for the soft and uneven tissues (e.g. leaf and petal) coated by layers of cuticular waxes. In the imprinting process, the cell contents of the fresh plant material are transferred to the pores in the PTFE surface while maintaining the spatial distribution from the original sample (Fig. 2b). Subsequently, the analytes are easily desorbed and ionized in a subsequent DESI experiment, which benefits from the very good properties of the porous PTFE surface for DESI analysis. The method may also be applied on samples which are difficult to cryo-section due to their shape, e.g. as shown in DESI imaging of the longitudinal distribution of dhurrin in 2–6 days etiolated sorghum (Sorghum bicolor) seedlings, which were too long (>6 cm) and tortuous for cryo-sectioning to be feasible.94 Finally, the imprinting method can be used to obtain a snapshot of liable components that are readily degraded when the plant tissue is disrupted, a process which is stopped when the sample is dried immediately after imprinting.94 The apparent limitation of the imprinting method is the possible dislocation of metabolites when the sample is squeezed and the flattening of the 3D structure onto a 2D surface. Regarding the risk of sample relocation, with a typical porosity of the imprint surface of around 10 μm, the potential sample relocation is normally negligible compared to the 100 μm spatial resolution in a DESI imaging experiment, but considerable in a high-resolution MALDI experiment. The imprinting method has been applied for MALDI imaging of plant material as well,110 but not in high-resolution experiments.
10 Matrix application
In MALDI imaging, the correct choice and application of the chemical matrix is a crucial step, which has direct consequences for the spatial resolution, potential artificial relocation and ionization efficiency of metabolites.45,111 The interaction between matrix and analytes, the matrix solvent, as well as the chemical properties of the metabolites (e.g. polarity, molecular weight and mobility in solution) are all parameters that need to be considered due to their influence on the authenticity of imaging results. The search for better matrices in general or for specific applications of the existing ones still remain mostly a trial-and-error process, and the most widely applied matrices in UV-MALDI imaging are the 2,5-dihydroxybenzoic acid (2,5-DHB),106 sinapinic acid (SA)112 or α-cyano-4-hydroxycinnamic acid (CHCA).113 Other matrices for specific targeting of metabolites are also reported in MALDI imaging, for instance 9-aminoacridine and 3-aminoquinoline applied to Arabidopsis leaves for the negatively charged molecules, such as glucosinolates and acids,114 colloidal graphite and colloidal silver for low-molecular-weight metabolites e.g. epicuticular wax, flavonol aglycones and glycosides in Arabidopsis115,116 or lithium 2,5-dihydroxybenzoate for imaging of very highly apolar metabolites, such as wax esters located on Phoenix sp. and Arabidopsis leaves.117 Generally, both positive and negative ion modes can be used for MALDI imaging, but the latter is not extensively applied due to lower ionization efficiency with most commonly used matrices, such as DHB. In the positive ion mode and especially in low mass range, adducts with sodium and potassium ions are present in the spectra to a large extent. The presence of adducts makes the mass spectra more complicated, and their different fragmentation patterns compared to the protonated ions is a challenge when searching in databases of MS/MS spectra of [M + H]+ ions as part of a structure elucidation. p-Nitroaniline (PNA) is one of the commonly used matrices for lipids analysis in the negative mode but it is toxic and favors the ionization of phosphatidylcholines.118 Ionic matrices have been demonstrated as a good way to increase the signal intensity specifically in the negative ion mode for lipid imaging.119,120 Furthermore, 1,8-bis(dimethyl-amino)naphthalene (DMAN), classified as a “proton sponge”, has proven to be a very useful matrix in the negative ion mode; the main advantage of DMAN is the absence of matrix-related interfering signals in the mass spectra, also in the low-mass region.121,122The deposition of the matrix can be performed in a number of different ways, with the aim of obtaining a layer of matrix crystals on the sample surface that are homogenous in size and distribution. The matrix crystallizes as the solvent evaporates to form a bulk of small crystals that range in size from a few to a few hundred micrometers, depending on the matrix properties and the deposition method. The size of matrix crystals incorporated with analytes should be smaller than the required lateral resolution of the imaging measurement. Various matrix application methods have been described.123 The most common devices available for the application of matrix are aerosol sprayers or airbrushes by which the pixel size of imaging is usually limited to 50–100 μm.123 Bouschen et al. described an alternative matrix deposition method termed pneumatic spray and vapour deposition/recrystallization specifically for high spatial resolution scanning microprobe MALDI-MSI, enabling a lateral analytical resolution of 2–10 μm.111 Recently, sublimation has been introduced as a method for matrix deposition, reducing the risk of analyte relocation because of the direct phase transitions between the solid–vapor–solid phases. The application time, temperature, and pressure settings should be finely controlled for the repeatability of such matrix applications.124 A number of different matrix deposition systems based on different techniques are commercially available.
11 Application of mass spectrometry imaging in plant science
Table 1 is a summary of most of the plant metabolite MSI studies performed in the last 8 years where the technique has begun to be extensively explored. Most of the works in the table are “proof-of-concept” where the emphasis is on the method development and/or validation. However, a few have taken one step further and applied MSI of plant metabolites to obtain biological insight, sometimes still published as a technical paper involving much method optimization and validation. The breakthrough came when Shroff et al. used MALDI-MSI to reveal a non-uniform distribution of glucosinolates across leaves of Arabidopsis, and were able to correlate that with the feeding pattern of a caterpillar.93 This significant result demonstrated the great potential of plant metabolite MSI for answering biological questions. In addition, the distribution was verified by conventional methods of dissection and extractions, thus substantiating the validity of MALDI-MS for imaging plant metabolites. Others have since repeated and confirmed the distribution analyses and been able to partly correlate it with the expression of certain transcription factors.125 However, in this study the distribution analyses of wild type and transgenic plants were indeed done using conventional methods, and it would have benefited greatly from the faster and less laborious MALDI-MSI analysis.Since the breakthrough of Shroff et al.,93 much of the effort has been focused partly on developing alternative MSI methods for plants, which may in some cases have advantages over MALDI-MS, and partly on increasing the resolution of MALDI-MSI to take this method to a level in detail where conventional methods cannot be used. However, a few more studies have provided some biological insight, further demonstrating the usefulness of the techniques for plant research. A number of studies have been able to correlate the distributions of metabolites found with MSI with already existing knowledge on other physiological phenomena. Thus, grapevine leaves produce stilbenes as a response to infection by downy mildew or UV irradiation, and using LDI-TOF-MS it could be shown that the accumulations of individual stilbenes were differentially correlated with the exposure sites, and with the different types of stress.5 Also, age-dependent and/or presumably stress-induced changes in metabolite distributions in various leaves or seedlings were visualized with imprint DESI-MSI by Hemalatha and Pradeep126 and Li et al.94 Utilizing the same technique Müller et al. found the abundance of chlorophyll degradation products to be correlated with the visually detectable loss of green color in senescing leaves.127 Finally, with DESI-MSI we were able to show that distributions of hydroxynitrile glucosides in L. japonicus is related to the expression of a key biosynthetic enzyme in a non-trivial age-dependent manner.94 This demonstrates that RNA and protein localization analyses or plant metabolite MSI cannot stand alone in explaining the complex dynamics of living tissues.
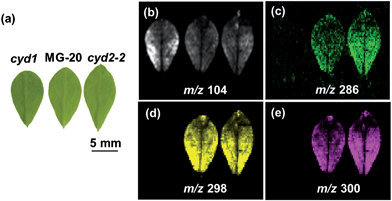
In a proof-of-concept paper, a mutant of Arabidopsis defective in a specific enzyme in the biosynthesis of certain flavonoids was employed to demonstrate the utility of LDI-MSI in functional genomics, i.e. the assignment of gene function.128 Apart from the absence of the flavonoids in question, the petals of the mutant plant also showed different distributions of flavonoids originating from another branch of flavonoid biosynthesis compared to the wild type petals. This new distribution could be explained by the mutated enzyme being a branch point between the two metabolic pathways and indicated that the expression pattern of the gene in question is responsible for the wild type distribution of the different flavonoids. Unfortunately, confirming this was beyond the scope of the study, and seen in the light of our findings mentioned above, the relation between gene expression and metabolite distributions are not always trivial. Nevertheless, metabolite distribution data of various origins has and will be a useful tool for directing the search for genes and enzymes of interest, especially in non-model plant species where the available genetic information is limited. Another important role of MSI in this respect is the potential for discovery of novel metabolites in untargeted approaches utilizing high resolution and/or tandem MS equipment for compound identification. Thus, our identification of a possible turnover product of cyanogenic glucosides in cassava tubers and the discovery of its specific localization to certain tissues of the tuber94 have prompted us to initiate the search for the responsible enzymes (work in progress).
Other studies which deserve mentioning include the impressive number of big and small, general and specialized, metabolites recently found to be differentially localized in and around nodules of the nitrogen fixing plant medicago (Medicago truncatula)122 (Fig. 3). Although the findings were only directly related to the distributions in and between the physiological structures of root and nodules, this study is an example of how untargeted metabolite imaging may provide the starting point for formulating biological questions which would otherwise not have been asked. The authors made use of mutant strains of plant and symbiotic bacteria to identify metabolites specifically absent in non-functional nodules compared to functional ones, confirming the usefulness of such approaches. Likewise, we demonstrated that mutant plants defective in specific enzymes can be used in combination with MSI to visualize and confirm the wild type enzymatic activity in situ.94 Such approaches will ease the assignment of gene and enzyme function by providing the missing link between transcript/protein localization data and substrate/product distributions in planta. However, as pointed out above, the relation is not always straight forward and many more biological questions can be expected to arise when it becomes standard to combine RNA/enzyme and metabolite localization data.
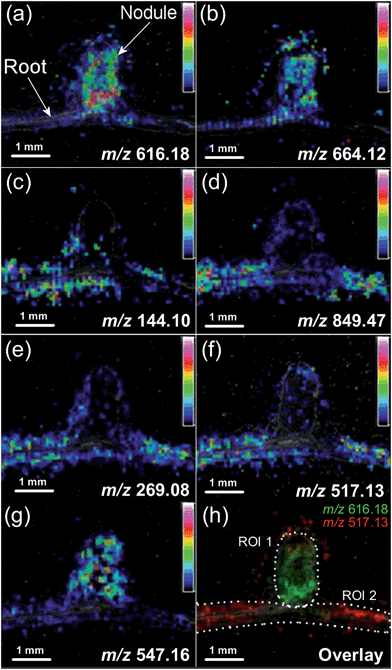 | ||
| Fig. 3 Distributions of selected metabolites in nitrogen fixing nodules of medicago and the roots around them, as imaged by MALDI-MSI. For identifications of individual metabolites and general explanations of the experiment, please refer to Ye et al.122 Adapted from ref. 122 with permission from the authors and the publisher. | ||
11.1 Imaging surface metabolites of intact tissues
Interestingly, although many researchers have performed MSI of intact tissues, such as leaves or petals, only a few have investigated how deeply the chosen methods penetrate into the tissue. From a biological point of view, however, this question is potentially important. As mentioned, at this stage in development many studies are technical reports imaging the distribution of compounds already known to be present in certain physiological structures, e.g. the lipids of epicuticular waxes on a leaf surface. However, with the ultimate goal of unbiased analyses of the distribution of “all” small molecules to bring us new insights on the detailed metabolism of plants, it is necessary to know that the chosen mode of analysis is indeed sampling exclusively e.g. the wax layer, and not the underlying epidermal cells. Using MALDI, Shroff et al. reported that the number of metabolites detected in the mass spectra of the imaging analyses increased with the number of matrix applications performed.93 They were essentially demonstrating a sequential extraction of the leaf material by the matrix, but it has not been investigated whether this could be exploited to deliberately and reproducibly limit the MALDI-MSI analysis to the upper layers. With LAESI-MSI it is possible to perform a sequential analysis of layers of increasing depth and thus construct a 3D-image of metabolite distributions. Unfortunately, due to the resolution constraints of LAESI, one image layer consists of several cell layers.63 Using scanning electron microscopy Li et al. examined the depth of the crater left by a single pulse of their IR laser used for AP IR-MALDI-MS analysis of various plant tissues.129 They concluded that the laser had successfully desorbed the wax and cuticle of a peace lily (Spathifyllum sp.) petal and a few, but not all, upper epidermal cells. Several consecutive pulses led to penetration into the mesophyll layer. Consequently, both methods lack the ability to distinguish reproducibly between individual cell layers or leaf surface and leaf epidermis.In a number of studies using less invasive techniques than the infrared laser, it is stated that the method of choice samples the tissue surface, but whether this includes the contents of the upper epidermal cells plus the cuticular wax layer or only the latter, is not always clear. In two cases it has been shown that chloroform extraction of leaves to remove epicuticular waxes was necessary to make flavonoids and other specialized metabolites available for imaging , respectively, GALDI-MS130 and direct DESI-MS.90 In both cases, the small molecules were readily available for imaging in petals without prior chloroform treatment. Likewise, Jun et al. showed that two “rounds” of colloidal graphite application followed by MS imaging of stem samples, rendered flavonoids available for imaging in the “second layer”.73 For LDI-MSI Hamm et al. simply reported that “high laser energy” was necessary to desorb stilbenes from grapevine leaves,5 and Hölscher et al. successfully imaged flavonoids in petals of St. John's wort and also naphtodianthrones in the dark glands of H. reflexum leaves with no pretreatment.88 The latter compounds were also available for direct DESI-MSI in St. John's wort without prior chloroform extraction, most likely owing to the physical nature of the dark glands.90 On the other hand, the compounds found in translucent glands of St. John's wort were only accessible to direct DESI-MSI after chloroform extraction.90 All of these results most likely reflect that the layer of cuticular waxes (and/or cuticle) on petals is more permeable to laser or solvent spray than that of leaves or stems, and that the flavonoids and other specialized metabolites in question are found in the epidermal cells (or special structures protruding from the leaf surface, such as dark glands). They also reflect that the some specific structures of intact leaves are more easily penetrated than others, and that it is difficult to determine exactly which physical structure is being interrogated in the MSI analysis. Hopefully, with further development these softer methods may become tools for investigating metabolite distributions in specific layers, i.e. the cuticle/wax layer separately from the epidermal cells.
As mentioned, direct DESI-MSI has also been used to visualize the presence of anti-microbial and allelopathic substances on surfaces of macroalgae.101,131 Although they are not strictly plants and therefore do not harbor a proper cuticle, they have characteristics in common with plants, and the experiments are interesting because attempts were made to investigate whether the DESI-MS experiment caused rupture of the epidermal cells.131 Light microscopy did not reveal any obvious differences before and after DESI-MSI, and thus it appears that the compounds in question were indeed present on the surface of the algal blade. If this result can be extrapolated to land plants, it indicates that with direct DESI-MSI it is possible to sample only the actual surface of the leaf and leave the epidermal cells intact. For direct DESI-MSI of plant leaves however, it was necessary to optimize the solvent spray composition to obtain signals stable enough for imaging,90 and as mentioned above this spray appears to be able to image the contents inside the various glands in H. perforatum leaves, i.e. not leaving these cells intact. With this spray it is also possible to directly image the distribution of hydroxynitrile glucosides in/on leaves of L. japonicus (Fig. 4) and obtain a result similar to that previously obtained with imprint DESI-MSI,94i.e. representing the sum of internal and external metabolites. To complicate matters further, the developed ternary solvent spray for direct DESI-MSI does not work in the same way for all leaves. As mentioned, the hydroxynitrile glucosides of barley are located in the leaf epidermis, and we have successfully imaged these compounds using both indirect DESI-MSI of whole leaves and direct DESI-MSI of epidermis stripped from the barley leaf.132 However, no signals from these compounds are obtained in direct DESI-MSI of barley leaves using the ternary spray, with or without prior chloroform treatment (data not shown). Whether this is a barley specific issue or it is general for monocots, and whether the other methods suffer from the same restrictions remain to be elucidated. Finally, the influence of surface topology on MSI results is an overlooked, but very important issue to consider when applying methods of direct imaging on whole leaves and petals. A homogeneous surface topology is important for uniform application of matrix for MALDI-MSI as well as for the stable and homogeneous desorption of analytes in DESI-MSI. As the above described examples with the dark glands of Hypericum species illustrate, plant leaf surfaces are far from homogeneous. Furthermore, the translucent glands of these leaves were best sampled from the abaxial side because they were more protuberant here.90 Likewise, trichomes and general surface inhomogeneity can be expected to influence the imaging results. In Fig. 4 it is evident that there was less signal from all shown metabolites around the leaf midvein. This is in contrast to our previously reported DESI-MS images obtained by imprinting where only the control compound γ-aminobutyric acid (Fig. 4b) showed decreased concentration in the midvein area.94 Thus the decreased abundance of hydroxynitrile glucosides in the midvein in Fig. 4c–e may be a “false result” caused by less accessibility to the solvent spray. All in all, it can be concluded that even when applying the same method for “surface metabolite imaging” great variations in the results can be expected depending on the physical nature of the sample material. As the desire in such experiments often is to image distributions on an intact surface, this problem may not be very likely to be solved in the near future.
 | ||
| Fig. 4 Direct DESI images (200 μm pixel size) of major metabolites from L. japonicus leaves. (a) Optical images of leaves from left to right are cyd1 (cyanogenesis deficient1, completely lacking hydroxynitrile glucosides), MG-20 (L. japonicus cv. Miyakojima, wild type) and cyd2-2 (cyanogenesis deficient 2–2, lacking β-glycosidases responsible for hydrolysis of cyanogenic glucosides). (b–e) ion images of m/z 104 = [γ-aminobutyric acid + K]+, m/z 286 = [linamarin + K]+, m/z 298 = [rhodiocyanoside A/D/E + K]+, m/z 300 = [lotaustralin + K]+. See Li et al.94 for description of mutant plants and ion identifications and Li et al.90 for experimental procedures of direct DESI-MSI. | ||
12 Future directions and concluding remarks
The major focus in the future development of MS imaging is above all related to the spatial resolution that can be obtained. As mentioned above, commercial and quite user-friendly MALDI imaging instruments routinely provide spatial resolution of around 20 μm, but MALDI imaging with resolutions of 1–3 μm has so far only been presented on custom built instruments from university laboratories. It is just a matter of time before similar solutions are commercially available, but as explained making images at such high resolution is not just a matter of purchasing the right instrument, but also to a high extent a matter of extremely careful sample preparation. Care must be taken that analyte molecules are not relocated during sample preparation and that the matrix is evenly distributed in very small crystals, effects which become much more crucial in high-resolution imaging.While an impressive increase in the performance of particularly MALDI imaging has been observed, it is also clear that there is a limit to how much more the resolution can be improved if meaningful results for other than the very most abundant compounds are expected. The challenge of performing MSI at cellular and subcellular resolution is that when the ionization spot is focused to sub-micron dimensions the number of available molecules of a given species simply becomes very low, and the number of ionized molecules is thus very often below the limit of detection of the applied mass spectrometer. Hopefully, the gradual technical improvement of mass spectrometers will help push the sensitivity and thus the obtainable spatial resolution even further.
It is therefore important to remember than even if imaging is not available, other spatially resolved techniques may be applied to answer the biological questions at hand, and alongside the development of MS imaging a significant development in the field of single cell analysis by mass spectrometry is taking place.157 It has, for example, recently been described how a well-established technique such as static nanospray can be used to probe the contents of a single plant (leaf, stem or petal) cell.158 With the sample present below a microscope, a nanospray tip with a 1 μm top diameter is introduced into a plant cell by means of a micromanipulator and the content of the cell is sucked into the nanospray capillary. The capillary is subsequently mounted in the nanospray ion source of the mass spectrometer, and a steady signal with the possibility of MS/MS and MS3 identification of the detected compounds is obtained. Likewise, in the already mentioned LAESI analysis of single onion epidermal cells, the contents of the cell were probed individually and each cell targeted manually below a microscope.53 The current limitation of the technique is the 30 μm ablation spot, which is not in all cases low enough to target single cells, but this may be within reach with future improvements of the LAESI technology.
A current challenge in MS imaging of plant material, which might be addressed in future research, is the sample stability and dynamics in certain types of plant material. Plants are full of very labile compounds that are either oxidized during sample handling or subject to various enzymatic reactions of much shorter timescales than the hour-long acquisition of MS images. The issue can be approached from the side of the image acquisition time as with the presentation of a MALDI instrument, which records a 100 μm resolution image of a rat brain in just 10 minutes,77 or it can be dealt with by introducing new procedures for sample preparation and storage conditions during image acquisition, for example with regards to the temperature and the atmosphere the sample is kept in during the imaging experiment.
Another concern is of course the type of analytes that can be imaged. The limitation with SIMS is obviously that larger molecules are severely fragmented during the ionization. DESI is capable of ionizing roughly the same analytes as normal electrospray, i.e. more or less polar but not directly non-polar molecules. For MALDI, the type of molecules which are ionized is highly matrix dependent, but generally the technique is most successful in the analysis of polar compounds as the ionization most often takes place by proton transfer as in ESI. With the advent of new matrix compounds, the applicability of MALDI is expanded, as with the recent introduction of 9,10-diphenylantracene for analysis of non-polar compounds.159
A different laser-based approach to the analysis of non-polar compounds is the recently presented Laser Ablation Atmospheric Pressure Photoionization (LAAPI).160 LAAPI is similar to LAESI, but the ESI post-ionization has been replaced by APPI, which also ionizes non-polar compounds. Although LAAPI has not been demonstrated for imaging yet, it represents a potential technique for imaging of polar and non-polar compounds with a spatial resolution around 300 μm, which may also be subject to improvement in the future.
Mass spectrometry imaging has come a long way during the past ten years, and it is obvious from the referenced examples in Table 1 that its application in studies of plant material has become significantly more widespread over the past few years. With sufficient focus on the points mentioned above, MS imaging will become even more relevant to the plant research community. While many of the first papers are written by analytical chemists with a new tool looking for a problem to use it on, much of the future work will hopefully be driven by plant scientists with a problem at hand for which MS imaging is the natural tool to use. In particular the technological improvements in spatial resolution towards the cellular level will be important, with MS imaging established as the “small molecule” approach to biological imaging, complementing the well-established and efficient tools for studying localization of proteins and enzymes in plant samples.
13 References
- O. Fiehn, Plant Mol. Biol., 2002, 48, 155–171 CrossRef CAS.
- T. R. Martins, J. J. Berg, S. Blinka, M. D. Rausher and D. A. Baum, New Phytol., 2013, 197, 958–969 CrossRef CAS PubMed.
- B. A. Halkier and B. L. Møller, Plant Physiol., 1989, 90, 1552–1559 CrossRef CAS PubMed.
- H. Buschmann, M. X. Rodriguez, J. Tohme and J. R. Beeching, Ann. Bot., 2000, 86, 1153–1160 CrossRef CAS.
- G. Hamm, V. Carré, A. Poutaraud, B. Maunit, G. Frache, D. Merdinoglu and J. F. Muller, Rapid Commun. Mass Spectrom., 2010, 24, 335–342 CrossRef CAS PubMed.
- R. Satyan, N. Aveek, P. Eganathan and A. Parida, Biotech. Histochem., 2010, 85, 285–293 CrossRef CAS.
- R. Sánchez-Pérez, K. Jørgensen, M. S. Motawia, F. Dicenta and B. L. Møller, Plant J., 2009, 60, 894–906 CrossRef PubMed.
- M. Weid, J. Ziegler and T. M. Kutchan, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 13957–13962 CrossRef CAS PubMed.
- J. Q. Goodger, A. M. Heskes, M. C. Mitchell, D. J. King, E. H. Neilson and I. E. Woodrow, Plant Methods, 2010, 6, 20 CrossRef PubMed.
- E. Werker, E. Putievsky, U. Ravid, N. Dudai and I. Katzir, Ann. Bot., 1993, 71, 43–50 CrossRef CAS.
- O. A. Koroleva, T. M. Gibson, R. Cramer and C. Stain, Plant J., 2010, 64, 456–469 CrossRef CAS PubMed.
- K. A. Nielsen, C. E. Olsen, K. Pontoppidan and B. L. Møller, Plant Physiol., 2002, 129, 1066–1075 CrossRef CAS PubMed.
- C. Gruhnert, B. Biehl and D. Selmar, Planta, 1994, 195, 36–42 CrossRef CAS.
- I. E. Sønderby, F. Geu-Flores and B. A. Halkier, Trends Plant Sci., 2010, 15, 283–290 CrossRef PubMed.
- N. Dudareva, A. Klempien, J. K. Muhlemann and I. Kaplan, New Phytol., 2013, 198, 16–32 CrossRef CAS PubMed.
- P. J. Facchini and B. St-Pierre, Curr. Opin. Plant Biol., 2005, 8, 657–666 CrossRef CAS PubMed.
- Z. Takats, J. M. Wiseman, B. Gologan and R. G. Cooks, Science, 2004, 306, 471–473 CrossRef CAS PubMed.
- M. E. Monge, G. A. Harris, P. Dwivedi and F. M. Fernández, Chem. Rev., 2013, 113, 2269–2308 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, Anal. Chem., 2007, 79, 8098–8106 CrossRef CAS PubMed.
- V. Kertesz and G. J. Van Berkel, J. Mass Spectrom., 2010, 45, 252–260 CrossRef CAS PubMed.
- P. J. Roach, J. Laskin and A. Laskin, Analyst, 2010, 135, 2233–2236 RSC.
- E. R. Amstalden van Hove, D. F. Smith and R. M. A. Heeren, J. Chromatogr., A, 2010, 1217, 3946–3954 CrossRef CAS PubMed.
- L. A. McDonnell and R. M. A. Heeren, Mass Spectrom. Rev., 2007, 26, 606–643 CrossRef CAS PubMed.
- P. M. Angel and R. M. Caprioli, Biochemistry, 2013, 52, 3818–3828 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, TrAC, Trends Anal. Chem., 2012, 34, 22–34 CrossRef CAS PubMed.
- A. Römpp and B. Spengler, Histochem. Cell Biol., 2013, 139, 759–783 CrossRef PubMed.
- A. Svatoš, Trends Biotechnol., 2010, 28, 425–434 CrossRef PubMed.
- P. Chaurand, J. Proteomics, 2012, 75, 4883–4892 CrossRef CAS PubMed.
- T. Greer, R. Sturm and L. Li, J. Proteomics, 2011, 74, 2617–2631 CrossRef CAS PubMed.
- J. Grassl, N. L. Taylor and A. H. Millar, Plant Methods, 2011, 7, 21–21 CrossRef CAS PubMed.
- S. Kaspar, M. Peukert, A. Svatos, A. Matros and H.-P. Mock, Proteomics, 2011, 11, 1840–1850 CrossRef CAS PubMed.
- Y. J. Lee, D. C. Perdian, Z. Song, E. S. Yeung and B. J. Nikolau, Plant J., 2012, 70, 81–95 CrossRef CAS PubMed.
- A. Matros and H.-P. Mock, Front. Plant Sci., 2013, 4, 1–7 CrossRef PubMed.
- A. Svatoš and H. P. Mock, The Handbook of Plant Metabolomics, pp. 93–110 Search PubMed.
- E. Esquenazi, Y. L. Yang, J. Watrous, W. H. Gerwick and P. C. Dorrestein, Nat. Prod. Rep., 2009, 26, 1521–1534 RSC.
- K. Dreisewerd, Chem. Rev., 2003, 103, 395–426 CrossRef CAS PubMed.
- M. Karas, M. Glückmann and J. Schäfer, J. Mass Spectrom., 2000, 35, 1–12 CrossRef CAS.
- M. Karas, D. Bachmann, U. Bahr and F. Hillenkamp, Int. J. Mass Spectrom. Ion Processes, 1987, 78, 53–68 CrossRef CAS.
- B. Spengler, M. Hubert and R. Kaufmann, 42nd Annual Conference on Mass Spectrometry and Allied Topics, Illinois, Chicago, 1994 Search PubMed.
- R. M. Caprioli, T. B. Farmer and J. Gile, Anal. Chem., 1997, 69, 4751–4760 CrossRef CAS.
- D. Hölscher, R. Shroff, K. Knop, M. Gottschaldt, A. Crecelius, B. Schneider, D. G. Heckel, U. S. Schubert and A. Svatoš, Plant J., 2009, 60, 907–918 CrossRef PubMed.
- A. Rompp, S. Guenther, Y. Schober, O. Schulz, Z. Takats, W. Kummer and B. Spengler, Angew. Chem., Int. Ed., 2010, 49, 3834–3838 CrossRef PubMed.
- A. Zavalin, E. M. Todd, P. D. Rawhouser, J. Yang, J. L. Norris and R. M. Caprioli, J. Mass Spectrom., 2012, 47, 1473–1481 CrossRef PubMed.
- Y. Schober, S. Guenther, B. Spengler and A. Römpp, Anal. Chem., 2012, 84, 6293–6297 CrossRef CAS PubMed.
- W. Bouschen and B. Spengler, Int. J. Mass Spectrom., 2007, 266, 129–137 CrossRef CAS PubMed.
- V. V. Laiko, S. C. Moyer and R. J. Cotter, Anal. Chem., 2000, 72, 5239–5243 CrossRef CAS.
- C. S. Creaser and L. Ratcliffe, Curr. Anal. Chem., 2006, 2, 9–15 CrossRef CAS.
- G. J. Van Berkel, S. P. Pasilis and O. Ovchinnikova, J. Mass Spectrom., 2008, 43, 1161–1180 CrossRef CAS PubMed.
- C. S. Creaser, J. C. Reynolds, A. J. Hoteling, W. F. Nichols and K. G. Owens, Eur. J. Mass Spectrom., 2003, 9, 33 CrossRef CAS PubMed.
- A. Ibanez, J. Scharte, P. Bones, A. Pirkl, S. Meldau, I. Baldwin, F. Hillenkamp, E. Weis and K. Dreisewerd, Plant Methods, 2010, 6, 14 CrossRef PubMed.
- K. Dreisewerd, F. Draude, S. Kruppe, A. Rohlfing, S. Berkenkamp and G. Pohlentz, Anal. Chem., 2007, 79, 4514–4520 CrossRef CAS PubMed.
- Y. Li, B. Shrestha and A. Vertes, Anal. Chem., 2007, 79, 523–532 CrossRef CAS PubMed.
- B. Shrestha, J. M. Patt and A. Vertes, Anal. Chem., 2011, 83, 2947–2955 CrossRef CAS PubMed.
- A. Römpp, K. Schäfer, S. Guenther, Z. Wang, M. Köstler, A. Leisner, C. Paschke, T. Schramm and B. Spengler, Anal. Bioanal. Chem., 2013, 405, 6959–6968 CrossRef PubMed.
- P. J. Horn, J. E. Silva, D. Anderson, J. Fuchs, L. Borisjuk, T. J. Nazarenus, V. Shulaev, E. B. Cahoon and K. D. Chapman, Plant J., 2013, 76, 138–150 CAS.
- M. Peukert, A. Matros, G. Lattanzio, S. Kaspar, J. Abadía and H.-P. Mock, New Phytol., 2012, 193, 806–815 CrossRef CAS PubMed.
- V. Cavatorta, S. Sforza, G. Mastrobuoni, G. Pieraccini, S. Francese, G. Moneti, A. Dossena, E. A. Pastorello and R. Marchelli, J. Mass Spectrom., 2009, 44, 891–897 CrossRef CAS PubMed.
- S. Weidner, R. D. Schultze and B. Enthaler, Rapid Commun. Mass Spectrom., 2013, 27, 896–903 CrossRef CAS PubMed.
- J. M. Wiseman, D. R. Ifa, Q. Y. Song and R. G. Cooks, Angew. Chem., Int. Ed., 2006, 45, 7188–7192 CrossRef CAS PubMed.
- D. Campbell, C. Ferreira, L. Eberlin and R. G. Cooks, Anal. Bioanal. Chem., 2012, 404, 389–398 CrossRef CAS PubMed.
- C. Janfelt and A. W. Nørgaard, J. Am. Soc. Mass Spectrom., 2012, 23, 1670–1678 CrossRef CAS PubMed.
- J. Pol, V. Vidova, G. Kruppa, V. Kobliha, P. Novak, K. Lemr, T. Kotiaho, R. Kostiainen, V. Havlicek and M. Volny, Anal. Chem., 2009, 81, 8479–8487 CrossRef CAS PubMed.
- P. Nemes, A. A. Barton and A. Vertes, Anal. Chem., 2009, 81, 6668–6675 CrossRef CAS PubMed.
- J. S. Becker, M. Zoriy, J. S. Becker, J. Dobrowolska and A. Matusch, J. Anal. At. Spectrom., 2007, 22, 736–744 RSC.
- D. Briggs, Surf. Interface Anal., 1983, 5, 113–118 CrossRef CAS.
- D. Touboul, F. Halgand, A. Brunelle, R. Kersting, E. Tallarek, B. Hagenhoff and O. Laprévote, Anal. Chem., 2004, 76, 1550–1559 CrossRef CAS PubMed.
- J. C. Vickerman, Analyst, 2011, 136, 2199–2217 RSC.
- Z. Li, P. W. Bohn and J. V. Sweedler, Bioresour. Technol., 2010, 101, 5578–5585 CrossRef CAS PubMed.
- Z. Li, L.-Q. Chu, J. V. Sweedler and P. W. Bohn, Anal. Chem., 2010, 82, 2608–2611 CrossRef CAS PubMed.
- K. Saito, Y. Watanabe, M. Shirakawa, Y. Matsushita, T. Imai, T. Koike, Y. Sano, R. Funada, K. Fukazawa and K. Fukushima, Plant J., 2012, 69, 542–552 CrossRef CAS PubMed.
- A. Thomas, J. L. Charbonneau, E. Fournaise and P. Chaurand, Anal. Chem., 2012, 84, 2048–2054 CrossRef CAS PubMed.
- C. Janfelt, N. Wellner, H. S. Hansen and S. H. Hansen, J. Mass Spectrom., 2013, 48, 361–366 CrossRef CAS PubMed.
- J. H. Jun, Z. Song, Z. Liu, B. J. Nikolau, E. S. Yeung and Y. J. Lee, Anal. Chem., 2010, 82, 3255–3265 CrossRef CAS PubMed.
- V. Kertesz, G. J. Van Berkel, M. Vavrek, K. A. Koeplinger, B. B. Schneider and T. R. Covey, Anal. Chem., 2008, 80, 5168–5177 CrossRef CAS PubMed.
- B. Prideaux and M. Stoeckli, J. Proteomics, 2012, 75, 4999–5013 CrossRef CAS PubMed.
- H.-K. Lim, J. Chen, C. Sensenhauser, K. Cook and V. Subrahmanyam, Rapid Commun. Mass Spectrom., 2007, 21, 1821–1832 CrossRef CAS PubMed.
- J. M. Spraggins and R. M. Caprioli, J. Am. Soc. Mass Spectrom., 2011, 22, 1022–1031 CrossRef CAS PubMed.
- S.-O. Deininger, D. S. Cornett, R. Paape, M. Becker, C. Pineau, S. Rauser, A. Walch and E. Wolski, Anal. Bioanal. Chem., 2011, 401, 167–181 CrossRef CAS PubMed.
- L. A. McDonnell, A. van Remoortere, R. J. van Zeijl and A. M. Deelder, J. Proteome Res., 2008, 7, 3619–3627 CrossRef CAS PubMed.
- S. L. Luxembourg, L. A. McDonnell, M. C. Duursma, X. Guo and R. M. Heeren, Anal. Chem., 2003, 75, 2333–2341 CrossRef CAS.
- J. L. Norris, D. S. Cornett, J. A. Mobley, M. Andersson, E. H. Seeley, P. Chaurand and R. M. Caprioli, Int. J. Mass Spectrom., 2007, 260, 212–221 CrossRef CAS PubMed.
- P. J. Trim, M.-C. Djidja, T. Muharib, L. M. Cole, B. Flinders, V. A. Carolan, S. Francese and M. R. Clench, J. Proteomics, 2012, 75, 4931–4940 CrossRef CAS PubMed.
- T. Schramm, A. Hester, I. Klinkert, J.-P. Both, R. M. A. Heeren, A. Brunelle, O. Laprévote, N. Desbenoit, M.-F. Robbe, M. Stoeckli, B. Spengler and A. Römpp, J. Proteomics, 2012, 75, 5106–5110 CrossRef CAS PubMed.
- C. Paschke, A. Leisner, A. Hester, K. Maass, S. Guenther, W. Bouschen and B. Spengler, J. Am. Soc. Mass Spectrom., 2013, 1–11 Search PubMed.
- P. Chaurand, S. A. Schwartz, D. Billheimer, B. J. Xu, A. Crecelius and R. M. Caprioli, Anal. Chem., 2004, 76, 1145–1155 CrossRef CAS PubMed.
- J. Soelberg, L. B. Jorgensen and A. K. Jager, Ann. Bot., 2007, 99, 1097–1100 CrossRef CAS PubMed.
- S. M. A. Zobayed, F. Afreen, E. Goto and T. Kozai, Ann. Bot., 2006, 98, 793–804 CrossRef CAS PubMed.
- D. Hölscher, R. Shroff, K. Knop, M. Gottschaldt, A. Crecelius, B. Schneider, D. G. Heckel, U. S. Schubert and A. Svatos, Plant J., 2009, 60, 907–918 CrossRef PubMed.
- J. Thunig, S. H. Hansen and C. Janfelt, Anal. Chem., 2011, 83, 3256–3259 CrossRef CAS PubMed.
- B. Li, S. H. Hansen and C. Janfelt, Int. J. Mass Spectrom., 2013, 348, 15–22 CrossRef CAS PubMed.
- Y. Yoshimura, H. Enomoto, T. Moriyama, Y. Kawamura, M. Setou and N. Zaima, Anal. Bioanal. Chem., 2012, 403, 1885–1895 CrossRef CAS PubMed.
- B. Enthaler, J. Pruns, S. Wessel, C. Rapp, M. Fischer and K.-P. Wittern, Anal. Bioanal. Chem., 2012, 402, 1159–1167 CrossRef CAS PubMed.
- R. Shroff, F. Vergara, A. Muck, A. Svatos and J. Gershenzon, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 6196–6201 CrossRef CAS PubMed.
- B. Li, C. Knudsen, N. K. Hansen, K. Jørgensen, R. Kannangara, S. Bak, A. Takos, F. Rook, S. H. Hansen, B. L. Møller, C. Janfelt and N. Bjarnholt, Plant J., 2013, 74, 1059–1071 CrossRef CAS PubMed.
- B. Prideaux, V. Dartois, D. Staab, D. M. Weiner, A. Goh, L. E. Via, C. E. Barry III and M. Stoeckli, Anal. Chem., 2011, 83, 2112–2118 CrossRef CAS PubMed.
- N. Takai, Y. Tanaka, K. Inazawa and H. Saji, Rapid Commun. Mass Spectrom., 2012, 26, 1549–1556 CrossRef CAS PubMed.
- G. Hamm, D. Bonnel, R. Legouffe, F. Pamelard, J.-M. Delbos, F. Bouzom and J. Stauber, J. Proteomics, 2012, 75, 4952–4961 CrossRef CAS PubMed.
- P. Källback, M. Shariatgorji, A. Nilsson and P. E. Andrén, J. Proteomics, 2012, 75, 4941–4951 CrossRef PubMed.
- A. Nilsson, T. E. Fehniger, L. Gustavsson, M. Andersson, K. Kenne, G. Marko-Varga and P. E. Andrén, PLoS One, 2010, 5, e11411 Search PubMed.
- D. A. Pirman and R. A. Yost, Anal. Chem., 2011, 83, 8575–8581 CrossRef CAS PubMed.
- T. D. Andras, T. S. Alexander, A. Gahlena, R. M. Parry, F. M. Fernandez, J. Kubanek, M. D. Wang and M. E. Hay, J. Chem. Ecol., 2012, 38, 1203–1214 CrossRef CAS PubMed.
- S. Cha, Z. Song, B. J. Nikolau and E. S. Yeung, Anal. Chem., 2009, 81, 2991–3000 CrossRef CAS PubMed.
- B. K. Kaletaş, I. M. van der Wiel, J. Stauber, J. D. Lennard, C. Güzel, J. M. Kros, T. M. Luider and R. M. A. Heeren, Proteomics, 2009, 9, 2622–2633 CrossRef PubMed.
- S. Kaspar, M. Peukert, A. Svatos, A. Matros and H. P. Mock, Proteomics, 2011, 11, 1840–1850 CrossRef CAS PubMed.
- A. G. Poth, J. S. Mylne, J. Grassl, R. E. Lyons, A. H. Millar, M. L. Colgrave and D. J. Craik, J. Biol. Chem., 2012, 287, 27033–27046 CrossRef CAS PubMed.
- P. J. Horn, A. R. Korte, P. B. Neogi, E. Love, J. Fuchs, K. Strupat, L. Borisjuk, V. Shulaev, Y.-J. Lee and K. D. Chapman, Plant Cell, 2012, 24, 622–636 CrossRef CAS PubMed.
- H. Ye, E. Gemperline, M. Venkateshwaran, R. Chen, P.-M. Delaux, M. Howes-Podoll, J.-M. Ané and L. Li, Plant J., 2013, 75, 130–145 CrossRef CAS PubMed.
- Y. Yoshimura, N. Zaima, T. Moriyama and Y. Kawamura, PLoS One, 2012, 7, e31285 CAS.
- T. Kawamoto, Arch. Histol. Cytol., 2003, 66, 123–143 CrossRef.
- P. J. Horn, C. N. James, S. K. Gidda, A. Kilaru, J. M. Dyer, R. T. Mullen, J. B. Ohlrogge and K. D. Chapman, Plant Physiol., 2013, 162, 1926–1936 CrossRef CAS PubMed.
- W. Bouschen, O. Schulz, D. Eikel and B. Spengler, Rapid Commun. Mass Spectrom., 2010, 24, 355–364 CrossRef CAS PubMed.
- A. K. Mullen, M. R. Clench, S. Crosland and K. R. Sharples, Rapid Commun. Mass Spectrom., 2005, 19, 2507–2516 CrossRef CAS PubMed.
- D. M. G. Anderson, V. A. Carolan, S. Crosland, K. R. Sharples and M. R. Clench, Rapid Commun. Mass Spectrom., 2009, 23, 1321–1327 CrossRef CAS PubMed.
- R. Shroff, F. Vergara, A. Muck, A. Svatos and J. Gershenzon, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 6196–6201 CrossRef CAS PubMed.
- S. Cha, H. Zhang, H. I. Ilarslan, E. S. Wurtele, L. Brachova, B. J. Nikolau and E. S. Yeung, Plant J., 2008, 55, 348–360 CrossRef CAS PubMed.
- S. Cha, Z. Song, B. J. Nikolau and E. S. Yeung, Anal. Chem., 2009, 81, 2991–3000 CrossRef CAS PubMed.
- V. Vrkoslav, A. Muck, J. Cvačka and A. Svatoš, J. Am. Soc. Mass Spectrom., 2010, 21, 220–231 CrossRef CAS PubMed.
- R. Estrada and M. C. Yappert, J. Mass Spectrom., 2004, 39, 412–422 CrossRef CAS PubMed.
- R. Lemaire, J. C. Tabet, P. Ducoroy, J. B. Hendra, M. Salzet and I. Fournier, Anal. Chem., 2006, 78, 809–819 CrossRef CAS PubMed.
- C. Meriaux, J. Franck, M. Wisztorski, M. Salzet and I. Fournier, J. Proteomics, 2010, 73, 1204–1218 CrossRef CAS PubMed.
- R. Shroff and A. Svatoš, Anal. Chem., 2009, 81, 7954–7959 CrossRef CAS PubMed.
- H. Ye, E. Gemperline, M. Venkateshwaran, R. Chen, P.-M. Delaux, M. Howes-Podoll, J.-M. Ané and L. Li, Plant J., 2013, 75, 130–145 CrossRef CAS PubMed.
- D. S. Cornett, M. L. Reyzer, P. Chaurand and R. M. Caprioli, Nat. Methods, 2007, 4, 828–833 CrossRef CAS PubMed.
- J. A. Hankin, R. M. Barkley and R. C. Murphy, J. Am. Soc. Mass Spectrom., 2007, 18, 1646–1652 CrossRef CAS PubMed.
- I. E. Sønderby, M. Burow, H. C. Rowe, D. J. Kliebenstein and B. A. Halkier, Plant Physiol., 2010, 153, 348–363 CrossRef PubMed.
- R. G. Hemalatha and T. Pradeep, J. Agric. Food Chem., 2013, 61, 7477–7487 CrossRef CAS PubMed.
- T. Müller, S. Oradu, D. R. Ifa, R. G. Cooks and B. Kräutler, Anal. Chem., 2011, 83, 5754–5761 CrossRef PubMed.
- A. R. Korte, Z. Song, B. J. Nikolau and Y. J. Lee, Anal. Methods, 2012, 4, 474–481 RSC.
- Y. Li, B. Shrestha and A. Vertes, Anal. Chem., 2008, 80, 407–420 CrossRef CAS PubMed.
- S. W. Cha, H. Zhang, H. I. Ilarslan, E. S. Wurtele, L. Brachova, B. J. Nikolau and E. S. Yeung, Plant J., 2008, 55, 348–360 CrossRef CAS PubMed.
- A. L. Lane, L. Nyadong, A. S. Galhena, T. L. Shearer, E. P. Stout, R. M. Parry, M. Kwasnik, M. D. Wang, M. E. Hay, F. M. Fernandez and J. Kubanek, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7314–7319 CrossRef CAS PubMed.
- B. Li, N. Bjarnholt, S. H. Hansen and C. Janfelt, J. Mass Spectrom., 2011, 46, 1241–1246 CrossRef CAS PubMed.
- A. K. Mullen, M. R. Clench, S. Crosland and K. R. Sharples, Rapid Commun. Mass Spectrom., 2005, 19, 2507–2516 CrossRef CAS PubMed.
- M. Burrell, C. Earnshaw and M. Clench, J. Exp. Bot., 2007, 58, 757–763 CrossRef CAS PubMed.
- S. Robinson, K. Warburton, M. Seymour, M. Clench and J. Thomas-Oates, New Phytol., 2007, 173, 438–444 CrossRef CAS PubMed.
- Y. Li, B. Shrestha and A. Vertes, Anal. Chem., 2007, 79, 523–532 CrossRef CAS PubMed.
- D. M. Anderson, V. A. Carolan, S. Crosland, K. R. Sharples and M. R. Clench, Rapid Commun. Mass Spectrom., 2009, 23, 1321–1327 CrossRef CAS PubMed.
- N. Goto-Inoue, M. Setou and N. Zaima, Anal. Sci., 2010, 26, 821–825 CrossRef CAS.
- S. Jung, Y. Chen, M. C. Sullards and A. J. Ragauskas, Rapid Commun. Mass Spectrom., 2010, 24, 3230–3236 CrossRef CAS PubMed.
- N. Zaima, N. Goto-Inoue, T. Hayasaka and M. Setou, Rapid Commun. Mass Spectrom., 2010, 24, 2723–2729 CrossRef CAS PubMed.
- K. A. Lunsford, G. F. Peter and R. A. Yost, Anal. Chem., 2011, 83, 6722–6730 CrossRef CAS PubMed.
- P. Franceschi, Y. Dong, K. Strupat, U. Vrhovsek and F. Mattivi, J. Exp. Bot., 2012, 63, 1123–1133 CrossRef CAS PubMed.
- M. Ha, J. H. Kwak, Y. Kim and O. P. Zee, Food Chem., 2012, 133, 1155–1162 CrossRef CAS PubMed.
- J. Sarsby, M. W. Towers, C. Stain, R. Cramer and O. A. Koroleva, Phytochemistry, 2012, 77, 110–118 CrossRef CAS PubMed.
- S. Taira, S. Shimma, I. Osaka, D. Kaneko, Y. Ichiyanagi, R. Ikeda, Y. Konishi-Kawamura, S. Zhu, K. Tsuneyama and K. Komatsu, Int. J. Biotechnol. Wellness Ind., 2012, 1, 61–65 CAS.
- H. Zhang, S. W. Cha and E. S. Yeung, Anal. Chem., 2007, 79, 6575–6584 CrossRef CAS PubMed.
- T. Harada, A. Yuba-Kubo, Y. Sugiura, N. Zaima, T. Hayasaka, N. Goto-Inoue, M. Wakui, M. Suematsu, K. Takeshita and K. Ogawa, Anal. Chem., 2009, 81, 9153–9157 CrossRef CAS PubMed.
- C. H. Hsiao, C. W. Hong, B. H. Liu, C. W. Chen, C. C. Wu and Y. S. Wang, Rapid Commun. Mass Spectrom., 2011, 25, 834–842 CrossRef CAS PubMed.
- C. Li, Z. Wang and A. D. Jones, Anal. Bioanal. Chem., 2013, 1–12 Search PubMed , In press.
- T. Imai, K. Tanabe, T. Kato and K. Fukushima, Planta, 2005, 221, 549–556 CrossRef CAS PubMed.
- K. Saito, T. Mitsutani, T. Imai, Y. Matsushita and K. Fukushima, Anal. Chem., 2008, 80, 1552–1557 CrossRef CAS PubMed.
- C. Zhou, Q. Li, V. L. Chiang, L. A. Lucia and D. P. Griffis, Anal. Chem., 2011, 83, 7020–7026 CrossRef CAS PubMed.
- P. Nemes, A. A. Barton, Y. Li and A. Vertes, Anal. Chem., 2008, 80, 4575–4582 CrossRef CAS PubMed.
- D. R. Ifa, A. Srimany, L. S. Eberlin, H. R. Naik, V. Bhat, R. G. Cooks and T. Pradeep, Anal. Methods, 2011, 3, 1910–1912 RSC.
- E. Cabral, M. Mirabelli, C. Perez and D. Ifa, J. Am. Soc. Mass Spectrom., 2013, 24, 956–965 CrossRef CAS PubMed.
- M. A. O. da Silva and M. A. Z. Arruda, Metallomics, 2013, 5, 62–67 RSC.
- A. Oikawa and K. Saito, Plant J., 2012, 70, 30–38 CrossRef CAS PubMed.
- M. n. Lorenzo Tejedor, H. Mizuno, N. Tsuyama, T. Harada and T. Masujima, Anal. Chem., 2012, 84, 5221–5228 CrossRef CAS PubMed.
- M. Nazim Boutaghou and R. B. Cole, J. Mass Spectrom., 2012, 47, 995–1003 CrossRef PubMed.
- A. Vaikkinen, B. Shrestha, T. J. Kauppila, A. Vertes and R. Kostiainen, Anal. Chem., 2012, 84, 1630–1636 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |