Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The zinc repository of Cupriavidus metallidurans†
Martin
Herzberg
a,
Dirk
Dobritzsch
b,
Stefan
Helm
b,
Sacha
Baginsky
b and
Dietrich H.
Nies
*a
aMolecular Microbiology, Institute for Biology/Microbiology, Martin-Luther-University Halle-Wittenberg, Kurt-Mothes-Str. 3, 06120 Halle/Saale, Germany. E-mail: d.nies@mikrobiologie.uni-halle.de
bPlant Biochemistry, Institute for Biochemistry, Martin-Luther-University Halle-Wittenberg, Kurt-Mothes-Str. 3, 06120 Halle/Saale, Germany
First published on 24th September 2014
Abstract
Zinc is a central player in the metalloproteomes of prokaryotes and eukaryotes. We used a bottom-up quantitative proteomic approach to reveal the repository of the zinc pools in the proteobacterium Cupriavidus metallidurans. About 60% of the theoretical proteome of C. metallidurans was identified, quantified, and the defect in zinc allocation was compared between a ΔzupT mutant and its parent strain. In both strains, the number of zinc-binding proteins and their binding sites exceeded that of the zinc ions per cell, indicating that the totality of the zinc proteome provides empty binding sites for the incoming zinc ions. This zinc repository plays a central role in zinc homeostasis in C. metallidurans and probably also in other organisms.
Introduction
Similar to other transition metal cations, zinc is an important trace element, but is nevertheless toxic at high concentrations.1,2 Zn(II) is redox-inactive, useful to tie flexible polypeptide chains into a more rigid confirmation around a tetrahedral Zn(II) complex, and allow catalysis as a Lewis-acid.3–5 Zinc is therefore the second most important transition metal in most prokaryotes, and up to 6% of the total number of proteins in bacteria and archaea, respectively, are zinc-binding proteins. In eukaryotes, this number even goes up to 8.8%.6We applied a systems biology approach, connecting and integrating cellular pathways and processes involving transition metal ions, to understand bacterial zinc homeostasis.7 The beta-proteobacterium Cupriavidus metallidurans is adapted to high concentrations of transition metals, and is able to maintain its zinc homeostasis even under adverse conditions.8,9 This is accomplished by a battery of highly redundant metal cation uptake systems with only minimal cation selectivity. These operate in combination with heavy metal efflux systems that export surplus cations10 and transport e.g. Zn(II) from the cytoplasm or periplasm to the outside.11–13 The only known import system with some specificity for zinc in C. metallidurans is ZupT of the ZIP protein family {TC#1.A.35; TC, Transporter classification14,15}, which is a protein family ubiquitous from bacteria to humans.
In C. metallidurans ZupT is needed to provide zinc under conditions of low availability.10 While the C. metallidurans parent strain keeps its zinc content at 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) per cell in growth media with nM zinc concentrations, ΔzupT mutant cells are only able to accumulate 20
000 Zn(II) per cell in growth media with nM zinc concentrations, ΔzupT mutant cells are only able to accumulate 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) per cell.5 Other unspecific uptake systems may provide Zn(II) to C. metallidurans cells at moderate zinc availability,10,16 filling up the cellular zinc pool to about 125
000 Zn(II) per cell.5 Other unspecific uptake systems may provide Zn(II) to C. metallidurans cells at moderate zinc availability,10,16 filling up the cellular zinc pool to about 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 atoms per cell in the ΔzupT mutant and its parent strain.5 Although the number of zinc atoms per cell is similar in both strains, the ΔzupT mutant nevertheless suffers from several defects resulting from disturbed zinc homeostasis,5 indicating that ZupT is also required for efficient zinc allocation at higher zinc concentrations. Cells of a quadruple mutant devoid of the most important zinc efflux systems accumulate 250
000 atoms per cell in the ΔzupT mutant and its parent strain.5 Although the number of zinc atoms per cell is similar in both strains, the ΔzupT mutant nevertheless suffers from several defects resulting from disturbed zinc homeostasis,5 indicating that ZupT is also required for efficient zinc allocation at higher zinc concentrations. Cells of a quadruple mutant devoid of the most important zinc efflux systems accumulate 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) per cell when 10 μM Zn(II) is added to the growth medium. When more zinc is provided, these cells were not able to grow.5 Thus, 250
000 Zn(II) per cell when 10 μM Zn(II) is added to the growth medium. When more zinc is provided, these cells were not able to grow.5 Thus, 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) per cell might be the maximum zinc content of C. metallidurans, 125
000 Zn(II) per cell might be the maximum zinc content of C. metallidurans, 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) the optimum adjusted by efflux systems, and 20
000 Zn(II) the optimum adjusted by efflux systems, and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Zn(II) the minimum.
000 Zn(II) the minimum.
Since the molecular entities responsible for adjusting the zinc concentrations to the above-mentioned number should be different in the ΔzupT mutant and its parent strain, we used a bottom-up quantitative approach to compare the proteomes of both strains. We identified the zinc repository in C. metallidurans, the container of its cytoplasmic zinc pool. This zinc repository buffers the incoming zinc and other transition metal ions, thereby mollifying the differences between the mutant and the wild type. Moreover, as the tip of the iceberg, the zinc depository is also involved in regulation of synthesis of the nickel-dependent hydrogenase in the ΔzupT mutant strain.
Results and discussion
Quantitative proteomic analysis of C. metallidurans strain AE104 and its ΔzupT mutant
Since ΔzupT deletion is incompatible with the presence of the plasmid-encoded CzcCBA efflux system in C. metallidurans,5 the plasmid-free C. metallidurans strain AE104 and its ΔzupT deletion strain were compared (three biological repeats each). For reference, C. metallidurans CH34(pMOL28, pMOL30) wild type was included in one experiment. The proteome of the soluble and solubilized membrane protein fractions was analyzed using a SYNAPT G2-S (Waters, Eschborn, Germany). Protein in-solution digestions, Nano-LC separation HD-MSE and data processing were performed as described earlier.17Out of the 5804 proteins in the theoretical proteome of C. metallidurans, 3469 (60%) were found in at least one experiment (Table S1, ESI†). No lower detection limit was evident although the percent deviation increased with the decrease in the number of individual proteins per cell (Fig. S1, ESI†). The quantities of each protein in the supernatant and solubilized ultracentrifugation sediment were summed up and the resulting sum was normalized to an overall number of 1.86 million proteins per cell, which was derived from the experimentally determined average protein content. This number was lower than 2.6 million proteins per E. coli cell, although E. coli has a 1.5-fold lower cell volume and should have lower dry mass.18–21 Alternatively, the number of proteins per cell was also calculated from the dry mass of 0.6 pg of the C. metallidurans cell derived from the cellular dimensions,20 50% protein in the dry mass, an estimated mean molecular mass of 40 kDa per average protein, all together resulting in 7.5 amol protein per cell. Multiplication with the Avogadro number gave 4.5 million proteins per cell. The actual in vivo number per C. metallidurans cell could thus be higher than the 1.86 million proteins per cell used for normalization. Out of the 1.86 million proteins per cell assumed, only 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 were encoded by the plasmids in the comparison between the CH34 wild type strain and its plasmid-free derivative AE104 (8000 pMOL28 and 12
000 were encoded by the plasmids in the comparison between the CH34 wild type strain and its plasmid-free derivative AE104 (8000 pMOL28 and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pMOL30, 1.07% of the proteome, data not shown).
000 pMOL30, 1.07% of the proteome, data not shown).
Evaluation of the protein numbers per cell
Before the individual number of all identified proteins in the ΔzupT mutant and its parent strain AE104 were compared, the numbers of the constituents of three different protein complexes with known numbers per cell, the F1F0 ATPase (Table 1), the RNA polymerase (Table 2) and the ribosome (Fig. 1), were analyzed to evaluate the quality of the data obtained.| Name | Complex−1 | WT, cell−1 | Mut., cell−1 | Da | Description |
|---|---|---|---|---|---|
| a The number of proteins per AE104 cell (WT) or ΔzupT cell (Mut.) is shown plus the number of monomers per F1F0 complex (unknown for the c subunit, 10 is assumed, also unknown for AtpI) and the predicted molecular mass is in Da. “NeF”, never found in the proteome analysis. The numbers in italic font indicate down-regulation and in bold font up-regulation. | |||||
| Rmet_3493, AtpC | 1 | 2930 ± 246 | 2976 ± 271 | 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 958 958 |
Q1LHL1 ATP synthase epsilon chain |
| Rmet_3494, AtpD | 3 | 7664 ± 709 | 6955 ± 667 | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120 120 |
Q1LHL0 ATP synthase subunit beta |
| Rmet_3495, AtpG | 1 | 2406 ± 139 | 2347 ± 487 | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 320 320 |
Q1LHK9 ATP synthase gamma chain |
| Rmet_3496, AtpA | 3 | 5312 ± 439 | 4656 ± 229 | 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 410 410 |
Q1LHK8 ATP synthase subunit alpha |
| Rmet_3497, AtpH | 1 | 1892 ± 978 | 1624 ± 453 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 753 753 |
Q1LHK7 ATP synthase delta subunit |
| Rmet_3498, AtpF | 2 | 1934 ± 177 | 1021 ± 354 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 384 |
Q1LHK6 ATP synthase b chain |
| Rmet_3499, AtpE | 10? | 601 ± 87 | 104 | 9676 | Q1LHK5 ATP synthase c subunit |
| Rmet_3500, AtpB | 1 | 778 ± 37 | 274 ± 171 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 968 968 |
Q1LHK4 ATP synthase a chain |
| Rmet_3501, AtpI | ? | NeF | NeF | 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 707 707 |
Q1LHK3 ATP synthase I chain |
| Name | WT, cell−1 | Mut., cell−1 | Description |
|---|---|---|---|
| a The number of proteins per AE104 cell (WT) or ΔzupT cell (Mut.) is shown; “NeF”, never found in the proteome analysis; “NF”, not found in this strain. The numbers in italic font indicate down-regulation and in bold font up-regulation of strain AE104 compared to CH34 and the AE104ΔzupT mutant compared to AE104. | |||
| Rmet_3291, RpoA | 7174 ± 1044 | 7959 ± 1314 | Q1LI63 DNA-directed RNA polymerase subunit alpha |
| Rmet_3334, RpoB | 4908 ± 340 | 5411 ± 962 | Q1LI20 DNA-directed RNA polymerase subunit beta |
| Rmet_3333, RpoC | 4709 ± 128 | 4801 ± 1105 | Q1LI21 DNA-directed RNA polymerase subunit beta' |
| Rmet_0857, RpoZ | 508 ± 287 | 569 ± 365 | B3R3N5, DNA-directed RNA polymerase subunit omega |
| Rmet_2606, RpoD | 1927 ± 96 | 1521 ± 394 | Q1LK43 RNA polymerase sigma factor |
| Rmet_4661, RpoD2 | 233 ± 70 | 1066 ± 814 | Q1LEA3 RNA polymerase sigma factor |
| Rmet_0303, RpoN | 330 ± 29 | 332 ± 23 | Q1LRN7 Sigma-54 (RpoN) |
| Rmet_0272, RpoH | 119 | NF | Q1LRR8 RNA polymerase sigma factor |
| Rmet_2115, RpoS | 252 ± 160 | 200 ± 80 | Q1LLI2 RNA polymerase sigma factor |
| Rmet_2425, RpoE | 273 ± 43 | 415 ± 198 | Q1LKM4 RNA polymerase sigma factor. ECF:RpoE cluster |
| Rmet_3702, FliA | NeF | NeF | Q1LH02 RNA polymerase sigma factor |
| Rmet_1120, RpoI | NeF | NeF | Q1LPC0 Sigma-24, ECF:FecI1 cluster |
| Rmet_4499, RpoJ | 53 | 151 ± 56 | Q1LER2 Sigma-24, ECF:FecI1 cluster |
| Rmet_4001, RpoK | NeF | NeF | Q1LG57 Sigma-24, ECF:FecI1 cluster |
| Rmet_0597, RpoO | NF | NF | Q1LQU3 Sigma-24, ECF:FecI2 cluster |
| Rmet_3280, RpoL | 279 ± 14 | NF | Q1LI74 Sigma-24, ECF:FecI2 cluster |
| Rmet_5400, RpoM | 10 | 150 | Q1LC67 Sigma-24, ECF:FecI2 cluster |
| Rmet_1648, RpoP | NeF | NeF | Q1LMU5 Sigma-24. ECF:RpoE cluster |
| Rmet_4686, RpoQ | NF | NF | Q1LE78 Sigma-24, “ECF” cluster |
| Rmet_0910, RpoR | 149 ± 3 | 150 ± 49 | Q1LPY0 Sigma-24, “ECF” cluster |
| Rmet_3844, “SigJ” | NeF | NeF | Q1LGL4 RNA polymerase, Sigma-24 subunit, ECF subfamily |
The number of F1F0 complexes in E. coli is 3200 per cell22 in the composition α3β3γδε of the F1 part and ab2c10 of the F0 part. In C. metallidurans, 3000 F1 complexes were identified with a ratio of 2.0 ± 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3.0 ± 0.8
3.0 ± 0.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.9 ± 0.2
0.9 ± 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.7 ± 0.3
0.7 ± 0.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 normalized to subunit ε (Table 1) as a mean value of all experiments performed. Soluble proteins could be accurately counted and the ratio of the subunits was correctly obtained. Of the F0 complex, ab2 was found in the correct ratio but in 5-fold lower numbers compared to the F1 subunits, ab2cn = 0.2 ± 0.1
1 normalized to subunit ε (Table 1) as a mean value of all experiments performed. Soluble proteins could be accurately counted and the ratio of the subunits was correctly obtained. Of the F0 complex, ab2 was found in the correct ratio but in 5-fold lower numbers compared to the F1 subunits, ab2cn = 0.2 ± 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.4 ± 0.2
0.4 ± 0.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.1 ± 0.1 (Table 1). We conclude that membrane proteins were difficult to count, especially when they were small and multimeric such as the c ring.
0.1 ± 0.1 (Table 1). We conclude that membrane proteins were difficult to count, especially when they were small and multimeric such as the c ring.
Judged from the number of RpoB and RpoC subunits, C. metallidurans should contain 5000 RNA polymerase core proteins per cell (RNAP, Table 2). RpoA, present in two copies in the RNAP, was found in a ratio of 1.53![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The values for E. coli were from 1500 in slow-growing cells up to 11
1. The values for E. coli were from 1500 in slow-growing cells up to 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 molecules in cells with a doubling time of 2.5 h−1,23,24 with the growth rate determining the number of molecules and their deployment.25 A number of 5000 RNAP molecules per cell in the 1.5-fold larger C. metallidurans cell growing slower (μ = 0.23 h−1, Fig. S2, ESI†) than E. coli (doubling time 0.5 h−1) again was a fair approximation of reality. Although the ΔzupT mutant strain contains a high number of misfolded RpoC subunits in inclusion bodies,5 these additional copies were not found here and probably removed during the low touring centrifugation of the cell-free extract or not solubilized from the ultracentrifugation sediment (Table 2). The number of RpoZ, responsible for the quality control of the RNAP assembly,26 was only 540 copies. This protein thus did not remain associated with the mature RNAP complexes after assembly. The number of RpoD subunits for the main housekeeping sigma factor was 1720 in both strains, twice as high as in E. coli.27,28 Of the remaining 16 sigma factors, 5 were not found in this experimental series, among those 5 the flagellum sigma factor FliA and RpoI were responsible for siderophore biosynthesis in C. metallidurans.29,30
400 molecules in cells with a doubling time of 2.5 h−1,23,24 with the growth rate determining the number of molecules and their deployment.25 A number of 5000 RNAP molecules per cell in the 1.5-fold larger C. metallidurans cell growing slower (μ = 0.23 h−1, Fig. S2, ESI†) than E. coli (doubling time 0.5 h−1) again was a fair approximation of reality. Although the ΔzupT mutant strain contains a high number of misfolded RpoC subunits in inclusion bodies,5 these additional copies were not found here and probably removed during the low touring centrifugation of the cell-free extract or not solubilized from the ultracentrifugation sediment (Table 2). The number of RpoZ, responsible for the quality control of the RNAP assembly,26 was only 540 copies. This protein thus did not remain associated with the mature RNAP complexes after assembly. The number of RpoD subunits for the main housekeeping sigma factor was 1720 in both strains, twice as high as in E. coli.27,28 Of the remaining 16 sigma factors, 5 were not found in this experimental series, among those 5 the flagellum sigma factor FliA and RpoI were responsible for siderophore biosynthesis in C. metallidurans.29,30
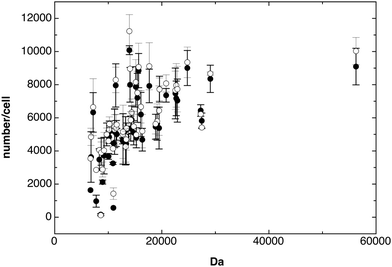
A number of 51 ribosomal proteins were reproducibly identified in cells of strain AE104 and the ΔzupT mutant with numbers between 140 per cell and 9560 per cell with not many differences between the strains (Fig. 1). The mean value of all ribosomal proteins was 5701 ± 774 per cell, however, some of the smaller ribosomal proteins gave low copy numbers (Fig. 1). When only the 25 largest ribosomal proteins (>14 kDa) were counted, the result was 7182 ± 905 ribosomes per cell. E. coli is slightly smaller but grows faster than C. metallidurans and would contain 6800 ribosomes per cell at a growth rate of 0.6 h−1.19
This all indicated that the individual protein numbers obtained were fair approximations of the in vivo numbers and their stoichiometry.
Comparison of the proteomes of the ΔzupT mutant and the parent strain AE104
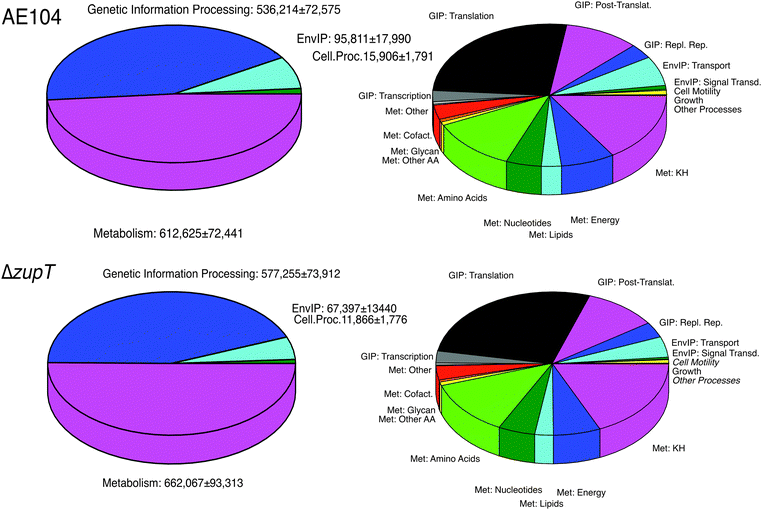
The mutant and parent proteomes were compared: (i) using the KEGG orthology (KO) system; (ii) the changes in the most abundant proteins; and (iii) the ratios of the protein numbers between the mutant and the parent cell. The KEGG orthology system sorts the proteins according to their function into functional categories at three different levels.31 Of the identified and quantified proteins, 65.7% could be assigned to the KO system. Of those, 48.6% of the proteins found in strain AE104 were involved in the KO level 1 category metabolism, 42.5% in genetic information processing (GIP), 7.6% in environmental information processing (EIP), and 1.3% in other cellular processes (Fig. 2 and Table S2, ESI†). The four most important metabolic level 2 categories were those for carbohydrates (32.8% metabolism), amino acids (24.8%), energy (15.3%) and nucleotides (10%). Genetic information processing was dominated by translation (61.7% GIP, 26.3% of all processes) and protein folding, sorting, and degradation (24.3% GIP), and that of environmental information processing by membrane transport (88.4% EIP). This visualized the different importance of the proteome fractions for the life process in C. metallidurans (Fig. 2).The ΔzupT mutant strain and its parent differed only in the lag phase of growth (Fig. S2, ESI†) and consequently no difference between the proteomes of both cells was evident at the top KO level (Fig. 2 and Table S2, ESI†), indicating the presence of some zinc-buffering system in mutant and parent cells that mollified the global effect of the zinc starvation. Despite this, the ΔzupT mutant cells synthesized 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 651 proteins involved in hydrogenase formation and Calvin cycle enzymes that were not found in the parent strain AE104 (Table S3, ESI†).
651 proteins involved in hydrogenase formation and Calvin cycle enzymes that were not found in the parent strain AE104 (Table S3, ESI†).
In contrast, (i) the glycerophospholipid and glycerolipid metabolism; (ii) chemotaxis, synthesis of the flagellum, components for twitching motility and of pili; (iii) two-component regulatory systems, and (iv) transport proteins were at lower abundance in the ΔzupT mutant (Table 3). The periplasmic CuZn superoxide dismutase SodC (Rmet_2757) was also found in lower abundance in the ΔzupT mutant cell (164 ± 43 proteins in AE104, 98 ± 12 in ΔzupT), explaining the lower tolerance of the ΔzupT mutant to reactive oxygen species.5
| Name | WT, cell−1 | Mut., cell−1 | Description |
|---|---|---|---|
| a The number of proteins per AE104 cell (WT) or ΔzupT cell (Mut.) is shown. The numbers in italic font indicate down-regulation and in bold font up-regulation. “NF”, not found in one strain. “NeF”, never found in all strains. | |||
| Secondary uptake systems | |||
| Rmet_2621, ZupT | NeF | NeF | Q1LK28 zinc/iron permease |
| Rmet_1973, PitA | NeF | NeF | Q1LLX4 phosphate transporter |
| Rmet_3052, CorA1 | 117 ± 8 | 50 ± 8 | Q1LIV2 magnesium and cobalt transport protein CorA |
| Rmet_0036, CorA2 | 38 ± 19 | NF | Q1LSF4 Mg2+ transporter protein, CorA-like protein |
| Rmet_3287, CorA3 | 37 ± 5 | NF | Q1LI67 Mg2+ transporter protein, CorA-like protein |
| Rmet_0549, ZntB | NeF | NeF | Q1LQZ1 Mg2+ transporter protein, CorA-like protein |
| Primary uptake systems | |||
| Rmet_2211, MgtA | 51 | 44 | Q1LL86 ATPase, P-type |
| Rmet_5396, MgtB | NF | 104 | Q1LC71 ATPase, P-type |
| Efflux systems/CDF | |||
| Rmet_0198, DmeF | NeF | NeF | Q1LRZ2 cation diffusion facilitator family transporter |
| Rmet_3406, FieF | 42 ± 5 | NF | Q1LHU8 cation diffusion facilitator family transporter |
| Efflux systems/P-type | |||
| Rmet_4594, ZntA | 396 ± 288 | NF | Q1LEH0 zinc translocating PIB2-type ATPase |
| Rmet_2303, CadA | 82 | NF | A7HYL0 cadmium translocating PIB2-type ATPase |
| Rmet_3524, CupA | 75 ± 12 | NF | Q1LHI0 copper translocating PIB1-type ATPase |
| Rmet_2379, CtpA1 | NF | NF | Q8GQ88 copper translocating PIB1-type ATPase |
| Rmet_2046, RdxI | NF | 516 | Q1LLQ1 copper translocating PIB1-type ATPase |
| Efflux systems/RND | |||
| Rmet_5319, ZniA | 76 ± 42 | NF | Q1LCE8 heavy metal efflux pump HME3a group |
| Rmet_4123, HmyA | 182 ± 112 | 55 | Q1LFT6 heavy metal efflux pump HME3c group |
| Efflux systems/ABC | |||
| Rmet_0391, AtmA | 38 ± 3 | NF | Q1LRE9 ABC transporter-related protein |
With 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per cell, the elongation factor Tu was the most abundant protein, followed by the GroEL chaperonin (11
000 copies per cell, the elongation factor Tu was the most abundant protein, followed by the GroEL chaperonin (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), ribosomal proteins, alkyl hydroperoxide reductase and citrate synthase (Data base, ESI†). The second and fourth most abundant proteins, however, were surprisingly two periplasmic binding proteins for citrate and related compounds (Rmet_0521, 13
000), ribosomal proteins, alkyl hydroperoxide reductase and citrate synthase (Data base, ESI†). The second and fourth most abundant proteins, however, were surprisingly two periplasmic binding proteins for citrate and related compounds (Rmet_0521, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per cell) and branched-chain amino acids (Rmet_2480, 11
000 per cell) and branched-chain amino acids (Rmet_2480, 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), respectively. These numbers were not different between mutant and parent strains.
000), respectively. These numbers were not different between mutant and parent strains.
Requirement of zinc for efficient identification of nickel ions by HypA
41![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 900 proteins of the NAD-reducing hydrogenases, maturation enzymes and the Calvin cycle proteins were produced in the ΔzupT strain but only 1282 in the parent, half of these being the NtrC-type response regulator HoxA (Table S3, ESI†), which is truncated in C. metallidurans (data not shown). We can conclude that three of the four processes generally controlling synthesis of hydrogenases in bacteria32 did not contribute to these differences in hydrogenase production between AE104 and ΔzupT: (i) hydrogenases in C. metallidurans and related bacteria such as Ralstonia eutropha are oxygen-tolerant33 and produced under oxic conditions,34,35 excluding molecular oxygen as an inhibitor of hydrogenase synthesis; (ii) molecular hydrogen cannot control hydrogenase synthesis because the sensory hydrogenase, histidine kinase and full-length HoxA required for this process36 are absent in C. metallidurans; and (iii) the proteome analysis suggests that the general metabolism is not different between AE104 and ΔzupT (Fig. 2 and Table S2, ESI†), making differences in metabolic control unlikely.34 The remaining fourth control process is nickel-dependent via the GTPase and nickel chaperone HypB (Fig. S3, ESI†). This protein is necessary for allocation of the metal to the active center of hydrogenases. Nickel control is not relayed by the sensory hydrogenase in R. eutropha,36,37 which is missing in C. metallidurans anyhow. Instead, HypB interacts with HypA.38,39 X-ray absorption spectroscopy demonstrated that the Zn site in HypA changes the structure, from a S3(O/N)-donor ligand environment to a S4-donor ligand environment, while Ni(II) is a six-coordinate complex composed of O/N-donors including two histidines.40 HypA controls the flow of nickel traffic in the cell, and a ligand exchange of zinc from Zn(Cys)4 to Zn(Cys)2(His)2 is part of this process.41 Zinc in HypA seems to be crucial to discriminate nickel and to control further allocation of Ni(II) to nickel-dependent proteins. Thus, in the ΔzupT mutant with its low zinc content5 and high cobalt content,10 zinc-dependent identification of nickel ions by the kinetics and thermodynamics of complex formation in comparison to ligand transfer to Zn(II) is hampered, leading to misregulation of hydrogenase synthesis, maybe by surplus cobalt ions. These data complement the biochemical evidence for the function of HypA41 with bottom-up system insights and they indicate that zinc is also involved in allocation control for other transition metal cations.
900 proteins of the NAD-reducing hydrogenases, maturation enzymes and the Calvin cycle proteins were produced in the ΔzupT strain but only 1282 in the parent, half of these being the NtrC-type response regulator HoxA (Table S3, ESI†), which is truncated in C. metallidurans (data not shown). We can conclude that three of the four processes generally controlling synthesis of hydrogenases in bacteria32 did not contribute to these differences in hydrogenase production between AE104 and ΔzupT: (i) hydrogenases in C. metallidurans and related bacteria such as Ralstonia eutropha are oxygen-tolerant33 and produced under oxic conditions,34,35 excluding molecular oxygen as an inhibitor of hydrogenase synthesis; (ii) molecular hydrogen cannot control hydrogenase synthesis because the sensory hydrogenase, histidine kinase and full-length HoxA required for this process36 are absent in C. metallidurans; and (iii) the proteome analysis suggests that the general metabolism is not different between AE104 and ΔzupT (Fig. 2 and Table S2, ESI†), making differences in metabolic control unlikely.34 The remaining fourth control process is nickel-dependent via the GTPase and nickel chaperone HypB (Fig. S3, ESI†). This protein is necessary for allocation of the metal to the active center of hydrogenases. Nickel control is not relayed by the sensory hydrogenase in R. eutropha,36,37 which is missing in C. metallidurans anyhow. Instead, HypB interacts with HypA.38,39 X-ray absorption spectroscopy demonstrated that the Zn site in HypA changes the structure, from a S3(O/N)-donor ligand environment to a S4-donor ligand environment, while Ni(II) is a six-coordinate complex composed of O/N-donors including two histidines.40 HypA controls the flow of nickel traffic in the cell, and a ligand exchange of zinc from Zn(Cys)4 to Zn(Cys)2(His)2 is part of this process.41 Zinc in HypA seems to be crucial to discriminate nickel and to control further allocation of Ni(II) to nickel-dependent proteins. Thus, in the ΔzupT mutant with its low zinc content5 and high cobalt content,10 zinc-dependent identification of nickel ions by the kinetics and thermodynamics of complex formation in comparison to ligand transfer to Zn(II) is hampered, leading to misregulation of hydrogenase synthesis, maybe by surplus cobalt ions. These data complement the biochemical evidence for the function of HypA41 with bottom-up system insights and they indicate that zinc is also involved in allocation control for other transition metal cations.
Zinc proteome
The number of potential zinc-binding or -containing proteins in the C. metallidurans cell was predicted from the orthologs of zinc-binding proteins in E. coli4,42 or the NCBI database (Table S4, ESI†), assuming that the orthologs of zinc-containing proteins in E. coli are also zinc-binding proteins in C. metallidurans. The strain AE104 contained 109![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 11
000 ± 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 potential zinc-binding proteins and the ΔzupT strain 123
000 potential zinc-binding proteins and the ΔzupT strain 123![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ± 14
000 ± 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Using the theoretical value of 4.5 million proteins per cell instead of the experimental value of 1.86 million proteins per cell would more than double this number, assuming more than one zinc-binding site per protein even more. The number of zinc binding sites in zinc-binding proteins of C. metallidurans is thus larger than 115
000. Using the theoretical value of 4.5 million proteins per cell instead of the experimental value of 1.86 million proteins per cell would more than double this number, assuming more than one zinc-binding site per protein even more. The number of zinc binding sites in zinc-binding proteins of C. metallidurans is thus larger than 115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, possibly even between 200
000, possibly even between 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 300
000 and 300![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.
000.
Half of the predicted zinc-binding proteins in the strain AE104 belonged to the KO group Genetic Information Processing and here mainly zinc-binding ribosomal proteins (30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) and the RNA polymerase subunits (17
000) and the RNA polymerase subunits (17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) are included. A smaller number (37.5%) were metabolic proteins, mainly involved in amino acid metabolism (19
000) are included. A smaller number (37.5%) were metabolic proteins, mainly involved in amino acid metabolism (19![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) especially of methionine (11
000) especially of methionine (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), energy metabolism (7500) including carbonic anhydrases, and carbohydrate metabolism (5800). 12% of the proteins were not assigned to a KO group in the database, including the translational elongation factor Tsf (4300) and the putative metal-binding proteins CobW2 and CobW3, zinc-containing alcohol dehydrogenases and proteases (Table S4, ESI†). Since (i) the number of zinc-binding proteins was between 110
000), energy metabolism (7500) including carbonic anhydrases, and carbohydrate metabolism (5800). 12% of the proteins were not assigned to a KO group in the database, including the translational elongation factor Tsf (4300) and the putative metal-binding proteins CobW2 and CobW3, zinc-containing alcohol dehydrogenases and proteases (Table S4, ESI†). Since (i) the number of zinc-binding proteins was between 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and 120
000 and 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, (ii) some of these proteins contain more than one zinc-binding site per monomer, and (iii) the assumed 1.86 million proteins per cell used for normalization was the lower limit of protein per cell, the number of zinc-binding sites in C. metallidurans AE104 and its ΔzupT mutant is higher than the number of 70
000, (ii) some of these proteins contain more than one zinc-binding site per monomer, and (iii) the assumed 1.86 million proteins per cell used for normalization was the lower limit of protein per cell, the number of zinc-binding sites in C. metallidurans AE104 and its ΔzupT mutant is higher than the number of 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell.5
000 zinc atoms per cell.5
In contrast, cells of strain AE104 contained 1302 ± 169 putative Co(II)-binding proteins per cell, and the mutant strain 1035 ± 413 (Table S4, ESI†). The number of cobalt atoms per cell was 3760 in cells of AE104 cultivated without added cobalt and 6290 in ΔzupT cells under the same conditions. C. metallidurans AE104 cells contained more cobalt atoms per cell than the known cobalt binding sites. Nevertheless, the strain could accumulate about 124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Co(II) when 50 μM cobalt chloride was added,5 100-fold more than that available for cobalt-binding proteins. The strain AE104 contained 1000 nickel binding sites per cell but the ΔzupT mutant contained 11
000 Co(II) when 50 μM cobalt chloride was added,5 100-fold more than that available for cobalt-binding proteins. The strain AE104 contained 1000 nickel binding sites per cell but the ΔzupT mutant contained 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 due to the increased number of hydrogenases (Data base, ESI†). The number of nickel atoms in strain AE104 was 5300 per cell.5,10 For the ΔzupT mutant the two determinations with different washing procedures resulted in 2500 ± 330 nickel atoms5 and 12
500 due to the increased number of hydrogenases (Data base, ESI†). The number of nickel atoms in strain AE104 was 5300 per cell.5,10 For the ΔzupT mutant the two determinations with different washing procedures resulted in 2500 ± 330 nickel atoms5 and 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 ± 5000 nickel atoms,10 which are in agreement with the number of nickel-binding sites found. There were more zinc binding sites in C. metallidurans cells than the measured zinc ions but more cobalt and nickel ions are present than predicted by the specific binding sites for these metals when the cells were cultivated in medium without added metals.
500 ± 5000 nickel atoms,10 which are in agreement with the number of nickel-binding sites found. There were more zinc binding sites in C. metallidurans cells than the measured zinc ions but more cobalt and nickel ions are present than predicted by the specific binding sites for these metals when the cells were cultivated in medium without added metals.
Thus, C. metallidurans cells may contain a zinc repository formed by the totality of all zinc binding sites of its cytoplasmic proteins. According to this hypothesis, the repository is able to accommodate at least 110![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc ions but probably many more. The E. coli ribosome bound 8 equivalents of zinc per ribosome that was required for synthesis of zinc-containing proteins,43 and between 2000 and 70
000 zinc ions but probably many more. The E. coli ribosome bound 8 equivalents of zinc per ribosome that was required for synthesis of zinc-containing proteins,43 and between 2000 and 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ribosomes, depending on the growth conditions.44 These ribosomes could bind 16
000 ribosomes, depending on the growth conditions.44 These ribosomes could bind 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 560
000 to 560![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc ions. In rapidly growing E. coli cells, the number of zinc binding sites solely provided by the ribosomes would exceed the number of 114
000 zinc ions. In rapidly growing E. coli cells, the number of zinc binding sites solely provided by the ribosomes would exceed the number of 114![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms measured.10 In C. metallidurans, 8 zinc atoms per ribosome would lead to 56
000 zinc atoms measured.10 In C. metallidurans, 8 zinc atoms per ribosome would lead to 56![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc ions instead of 30
000 zinc ions instead of 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bound to the 7000 ribosomes in each cell. This alone would raise the number of binding sites to 140
000 bound to the 7000 ribosomes in each cell. This alone would raise the number of binding sites to 140![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. In contrast, the strain AE104 contains 70
000. In contrast, the strain AE104 contains 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms at the middle of its exponential phase of growth when only nM concentrations of zinc were added, which fills up the repository only by half. The ΔzupT mutant contains only 20
000 zinc atoms at the middle of its exponential phase of growth when only nM concentrations of zinc were added, which fills up the repository only by half. The ΔzupT mutant contains only 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell under these conditions, less than 1/7 of the repository is filled. Since ribosomes need the available zinc for efficient synthesis of zinc-requiring proteins,43 this explains the inefficient folding of the RpoC subunit of the RNA polymerase5 and the gratuitous synthesis of the nickel-containing hydrogenases by HypB and the zinc enzyme HypA.
000 zinc atoms per cell under these conditions, less than 1/7 of the repository is filled. Since ribosomes need the available zinc for efficient synthesis of zinc-requiring proteins,43 this explains the inefficient folding of the RpoC subunit of the RNA polymerase5 and the gratuitous synthesis of the nickel-containing hydrogenases by HypB and the zinc enzyme HypA.
When 100 μM Zn(II) were added, both strains contain 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell, and the repository starts to fill up. In a titration experiment, the number of zinc ions per cell increased with the external zinc chloride concentration from 50 to 100 μM but the cellular zinc content was saturated at 125
000 zinc atoms per cell, and the repository starts to fill up. In a titration experiment, the number of zinc ions per cell increased with the external zinc chloride concentration from 50 to 100 μM but the cellular zinc content was saturated at 125![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 zinc atoms per cell in strain AE104 (Fig. 3) up to 150 μM. Only mutants with deletions in zinc efflux systems contain more zinc atoms per cell, 250
000 zinc atoms per cell in strain AE104 (Fig. 3) up to 150 μM. Only mutants with deletions in zinc efflux systems contain more zinc atoms per cell, 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000,5 indicating a saturated or overflowing repository, and these cells are not able to tolerate more zinc. This indicates that the repository also serves as a counterbalance of the transport systems. Finally, 50 μM Co(II) also leads to 124
000,5 indicating a saturated or overflowing repository, and these cells are not able to tolerate more zinc. This indicates that the repository also serves as a counterbalance of the transport systems. Finally, 50 μM Co(II) also leads to 124![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cobalt atoms per cell,5 and similar numbers have also been observed for copper ions and even gold complexes (Wiesemann and Nies, unpublished), indicating that the zinc repository might also initially accommodate other transition metal cations until they can be suitably allocated or exported again. Moreover, misregulation of hydrogenase synthesis also indicates that allocation pipelines for other divalent transition metal cations such as Ni(II) start at the zinc repository, and that comparison of those ions with Zn(II) could be an essential part of the identification process.
000 cobalt atoms per cell,5 and similar numbers have also been observed for copper ions and even gold complexes (Wiesemann and Nies, unpublished), indicating that the zinc repository might also initially accommodate other transition metal cations until they can be suitably allocated or exported again. Moreover, misregulation of hydrogenase synthesis also indicates that allocation pipelines for other divalent transition metal cations such as Ni(II) start at the zinc repository, and that comparison of those ions with Zn(II) could be an essential part of the identification process.
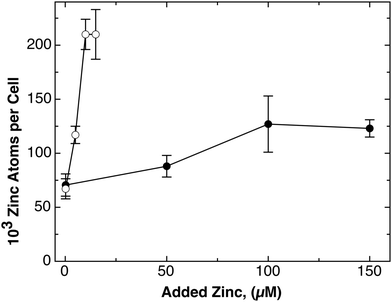 | ||
| Fig. 3 Titration of the cellular zinc content with the external zinc concentration. The cellular zinc content of strain AE104 (closed circles) and its Δe4 deletion strain (open circles) that has the four most important zinc efflux systems deleted (ΔzntA ΔcadA ΔdmeF ΔfieF) were cultivated in Tris-buffered mineral salt medium containing various concentrations of added zinc chloride, and the cellular zinc content was determined by ICP-MS as published.5 | ||
The hypothesis of the zinc repository predicts a high number of unsaturated metal-binding proteins in C. metallidurans. These binding sites might be required for rapid binding of transition metal cations in a tetrahedral complex, using only the 4sp orbitals and no 3d orbitals of the respective metal for binding to keep this process high-rate albeit of low specificity. These metals could be just sequestered or even act as metal cofactors. Indeed, enzymes with a low metallation quotient in vivo have been described.45 Alternatively, some enzymes may be promiscuous with regard to their metal cofactor4 or use in vivo Fe(II), which is present as 750![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Fe atoms per cell in strain AE104 (ref. 5) but renders the respective enzyme strongly sensitive to H2O2.46 It is, however, unclear at this stage if iron also needs the zinc repository for initial binding and sorting. C. metallidurans uses nearly no manganese, does not contain a Mn-dependent superoxide dismutase and no MntH uptake system to limit the import of cadmium as one pre-requisite of its cadmium resistance.10
000 Fe atoms per cell in strain AE104 (ref. 5) but renders the respective enzyme strongly sensitive to H2O2.46 It is, however, unclear at this stage if iron also needs the zinc repository for initial binding and sorting. C. metallidurans uses nearly no manganese, does not contain a Mn-dependent superoxide dismutase and no MntH uptake system to limit the import of cadmium as one pre-requisite of its cadmium resistance.10
Regulatory zinc binding sites responsible for flux control of transport proteins2 or for transcriptional control such as ZntR1 but also primary substrate binding sites of efflux systems can be pictured as part of the zinc repository, which also explains the zinc content of bacterial cells despite the femtomolar sensitivity of zinc regulatory proteins.1 Incoming zinc is immediately accommodated by empty binding sites of the repository, traverses down the affinity gradient from loosely to more strongly bound as shown for copper,47 thereby filling up the levels of tightly bound zinc first, e.g. in the interior of the proteins such as RpoC {PDB: 4IGC48}. With an increase in the filling levels, regulatory sites and the substrate binding sites of efflux systems are being filled, leading to a decreased import and an increased export of zinc, which keeps the number of zinc atoms in the cytoplasm at the same level as observed (Fig. 3). Additionally, low weight thiols such as glutathione in E. coli or bacillithiol in Bacillus subtilis may also play a role as a zinc pool, almost certainly at least under conditions of metal excess.49–51 Transport systems and the zinc repository are thus the key players of zinc homeostasis in C. metallidurans, and may be also in other organisms.
Experimental
Bacterial strains and growth conditions
Strains used for the experiments were Cupriavidus metallidurans CH34(pMOL28, pMOL30) wild type strain, its plasmid-free derivative strain AE104,34 the AE104 ΔzupT deletion mutant,10 and the Δe4 (ΔzntA ΔcadA ΔdmeF ΔfieF) quadruple deletion mutant.11 The bacteria were cultivated in Tris-buffered mineral salt medium34 containing 2 g sodium gluconate L−1 (TMM) aerobically with shaking at 30 °C. TMM contained 34 nM zinc chloride as part of an added trace element solution. Solid Tris-buffered media contained 20 g agar L−1. The metal content of the cells was determined by ICP-MS as published.5Quantitative proteome analysis
For the quantitative proteome analysis, the strains were cultivated in the mid-exponential phase, harvested, and the crude extract was prepared by ultra-sonication as published.52 After low-touring centrifugation (30 min 4500g), ultracentrifugation (1 h 540![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) was used to separate soluble and membrane proteins. The protein concentration was determined as described.53 100 μg of proteins of both fractions were precipitated in 80% acetone at −80 °C overnight and pelleted (4 °C, 21
000g) was used to separate soluble and membrane proteins. The protein concentration was determined as described.53 100 μg of proteins of both fractions were precipitated in 80% acetone at −80 °C overnight and pelleted (4 °C, 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500g, 10 min). Both fractions were solubilized in 233.5 μL of 25 mM ammonium bicarbonate containing 0.1% (w/v) RapiGest SF (Waters, Eschborn, Germany) for 10 min at 80 °C, and reduced by adding 6.2 μL of 10 mM dithiothreitol. Incubation was continued for 10 min at 60 °C. Cysteine residues were alkylated using 6.2 μL of 30 mM iodoacetamide for 30 min in the dark. After another ultracentrifugation (20 min 135
500g, 10 min). Both fractions were solubilized in 233.5 μL of 25 mM ammonium bicarbonate containing 0.1% (w/v) RapiGest SF (Waters, Eschborn, Germany) for 10 min at 80 °C, and reduced by adding 6.2 μL of 10 mM dithiothreitol. Incubation was continued for 10 min at 60 °C. Cysteine residues were alkylated using 6.2 μL of 30 mM iodoacetamide for 30 min in the dark. After another ultracentrifugation (20 min 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g) 4 μL of trypsin (0.25 g L−1, final concentration: 1
000 × g) 4 μL of trypsin (0.25 g L−1, final concentration: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 (w/w) corresponding to 1 μg of protease per vial) was added. Samples (final volume 250 μL) were digested at 37 °C for 16 h. To stop the digestion at pH values below 2.0, 2 μL of 37% HCl was added. To avoid loading insoluble materials onto the C18 column, the peptide solutions were thoroughly centrifuged prior to sample loading (4 °C, 21
100 (w/w) corresponding to 1 μg of protease per vial) was added. Samples (final volume 250 μL) were digested at 37 °C for 16 h. To stop the digestion at pH values below 2.0, 2 μL of 37% HCl was added. To avoid loading insoluble materials onto the C18 column, the peptide solutions were thoroughly centrifuged prior to sample loading (4 °C, 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 × g, 30 min).
500 × g, 30 min).
Nano-LC-HD-MSE data were acquired on a SYNAPT G2-S (Waters, Eschborn, Germany) (same protein amount per sample, three biological repeats for AE104 and ΔzupT, one for CH34, at least three technical repeats per biological repeat). Glycogen phosphorylase B (P00489) was used as an internal standard at a concentration of 10 fmol (Waters, Eschborn, Germany) as well as common contaminants (http://ftp://ftp.thegpm.org/fasta/cRAP/crap.fasta). 1 μL of each in-solution digest (containing ∼400 ng of protein) were separated on an ACQUITY nanoUPLC System (Waters, Eschborn, Germany). Peptides were trapped for 5 min at 5 μL min−1 at 1% B (0.1% formic acid in ACN) and 99% A (0.1% trifluoroacetic acid in water) on a 200 mm × 180 μm fused silica trap column packed with 5 μm Symmetry C18 (Waters, Eschborn, Germany), and separated at 300 nL min−1 in a linear gradient of 7–35% B (0.1% formic acid in ACN, A: 0.1% formic acid in water) within 140 min on a 250 mm × 75 μm fused silica separation column packed with 1.8 μm HSS T3 C18 (Waters, Eschborn, Germany).
Data independent acquisition (HD-MSE) with ion mobility separation
The eluting peptides were ionized (nanoESI source using a Pre-cut PicoTip Emitter; 360 μm OD × 20 μm ID, 10 μm tip; 2.5′′ long (Waters, Eschborn, Germany) at 2.1 kV, 80 °C, cone voltage: 40 V, N2 was used for the cone (10 L h−1), nanoflow (0.4 bar) and purging (450 L h−1 gases). Intact peptide mass spectra as well as fragmentation spectra were acquired on a SYNAPT G2-S mass spectrometer (Waters, Eschborn, Germany, resolution mode, positive ionization) for 140 min (50–2000 Da, 1 s scan time). Ion accumulation (ion mobility separation) was performed in the trap cell (release time: 500 μs) followed by mobility separation delay after trap release (“cooling”) in the helium cell (located between trap and ion mobility separation (IMS) cell): 1000 μs He2: 4.7 mbar). Ion separation (based on ion size, shape and charge) occurred in the IMS cell (N2: 2.87 mbar, wave height: 38 V, wave velocity: ramped from 1200 m s−1 to 400 m s−1). Other parameters were maintained at the predetermined, instrument-specific settings. CID was achieved by Ar2 (collision gas, function 2 high energy with ramp transfer collision energy of 25–55 V). Lock mass (Glu-1-Fibrinopeptide B, 250 fmol μL−1, 0.3 μL min−1, m/z = 785.8426, z = 2) mass correction was applied to the spectra during data processing.Data analysis
Data analysis was carried out using ProteinLynx Global Server (PLGS 3.0, Apex3D algorithm v. 2.128.5.0, 64 bit, Waters, Eschborn, Germany) with automated determination of the chromatographic peak width as well as MS TOF resolution with the following values: lock mass: 785.8426 Da e−1, lock mass window: 0.25 Da, low/high energy threshold: 200/20 counts, elution start time: 5 min, intensity threshold: 750 counts. Databank search query (PLGS workflow) was carried out as follows: peptide and fragment tolerance was set to automatic, two fragment ions match per peptide, at least five fragment ions for protein identification, and minimum 2 peptides per protein. A maximum protein mass was set to 250 kDa. The primary digest reagent was trypsin with one missed cleavage allowed. According to the digest protocol fixed (carbamidomethyl on Cys) as well as variable (oxidation on Met) modifications were set. The false discovery rate (FDR) was set to 4% at the protein level. Rabbit glycogen phosphorylase B (P00489, 10 fmol per injection) was defined as an internal calibration standard. The modified NCBI database (http://www.ncbi.nlm.nih.gov) to identify proteins contained 5804 proteins encoded by the open reading frames Rmet_0001 to Rmet_5941 of the two chromosomes of C. metallidurans, glycogen phosphorylase B (P00489, added as an internal standard at a concentration of 10 fmol, Waters, Eschborn, Germany) as well as the common contaminants (http://ftp://ftp.thegpm.org/fasta/cRAP/crap.fasta). ProteinLynx Global SERVER generated (“PRIDE plugin”) xml data have been deposited at the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the data set identifier PXD001356.Statistical analysis, reference protein mixtures and data normalization
The accumulated amounts of each Hi3-quantified protein in each of the three technical repeats, out of the cytoplasmic as well as membrane fraction, were used to calculate the mean values and deviations of the protein amounts per cell fraction, biological repeats and bacterial strains. The mean protein amounts and deviations of the cytoplasmic and the membrane fraction were added to give the cellular protein amount and deviation per biological repeat and bacterial strain. These values were not normalized to 1 million proteins per bacterial cell but instead to 1.88 million proteins per cell. This number was calculated from the number of bacterial cells before harvest and the protein concentration in the crude extract,53 which gave an average protein content of 115 fg per cell. This number was divided by the average protein mass of 37.2 kDa of the synthesized proteome, ∑[(fmol(proteini))/∑fmol(allproteins)·mass(proteini)], yielding 3.088 amol protein per cell, which was multiplied by the Avogadro number. The resulting 1.86 million proteins per cell was lower than the number of 2.6 million proteins per E. coli cell, although E. coli has a 1.5-fold lower cell volume and should have a lower dry mass.18–21The resulting cellular numbers and deviations for each protein per biological repeat and bacterial strain were used to calculate the mean protein numbers per bacterial cell and strain. The deviation was the mean value of the deviations of the biological repeats. Finally, the protein numbers of the strains were compared. A protein number was judged as up- or down-regulated, if the quantities were at least 2-fold lower or higher in the mutant-derived extract, respectively. These values were labeled in italic font (down-regulated) or bold font (up-regulated). Additionally, a “D” value was calculated as a measure of the significance of the difference. The “D” value gives the difference in the protein numbers, mutant minus parent, divided by the sum of the deviation of these mean values. If D > 1, the deviation bars of the mean values do not overlap, leading to a significant result (>95%) if n > 3 as published.54 If a protein was up- and down-regulated, respectively, and the difference between the protein numbers was significant (D ≥ 1), the value was judged as significantly different (bold font). If not, the value was counted as not significant (italic font). All values are provided in the supplementary data base (ESI†).
Conclusions
All cells, from bacteria to human, need zinc ions as essential trace elements. Although essential, zinc ions and other transition metal ions are also toxic when present at high concentrations. This paper describes how a bacterial cell is able to adjust the correct number of zinc ions per cell, how zinc ions are discriminated and finally allocated to zinc-requiring proteins. The newly identified key element here is the zinc depository, a huge number of empty zinc binding sites in cellular proteins, which accepts the incoming zinc and other metal ions, stores, discriminates and allocates them, or interacts with export systems to release surplus ions again.Acknowledgements
Funding for this work was provided by the Deutsche Forschungsgemeinschaft (Ni262/10 and INST 271/266-1 FUGG for the ICP-MS). We thank Grit Schleuder and Karola Otto for skillful technical assistance. SB gratefully acknowledges DFG support for the acquisition of a Synapt G2-S mass spectrometer (INST 271/283-1 FUGG).Notes and references
- C. E. Outten and T. V. O'Halloran, Science, 2001, 292, 2488–2492 CrossRef CAS PubMed.
- D. H. Nies, Science, 2007, 317, 1695–1696 CrossRef CAS PubMed.
- W. Maret, BioMetals, 2011, 24, 411–418 CrossRef CAS PubMed.
- D. H. Nies and G. Grass, in EcoSal—Escherichia coli and Salmonella: cellular and molecular biology, ed. A. Böck, R. Curtiss, J. B. Kaper, F. C. Neidhardt, T. Nyström, K. E. Rudd and C. L. Squires, ASM Press, Washington, D.C., 2009 DOI:10.1128/ecosal.5.4.4.3.
- M. Herzberg, L. Bauer and D. H. Nies, Metallomics, 2014, 6, 421–436 RSC.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, J. Proteome Res., 2006, 5, 3173–3178 CrossRef CAS PubMed.
- L. Banci and I. Bertini, Met. Ions Life Sci., 2013, 12, 1–13 CrossRef PubMed.
- P. J. Janssen, R. Van Houdt, H. Moors, P. Monsieurs, N. Morin, A. Michaux, M. A. Benotmane, N. Leys, T. Vallaeys, A. Lapidus, S. Monchy, C. Medigue, S. Taghavi, S. McCorkle, J. Dunn, D. van der Lelie and M. Mergeay, PLoS One, 2010, 5, e10433 Search PubMed.
- F. Reith, S. L. Rogers, D. C. McPhail and D. Webb, Science, 2006, 313, 233–236 CrossRef CAS PubMed.
- A. Kirsten, M. Herzberg, A. Voigt, J. Seravalli, G. Grass, J. Scherer and D. H. Nies, J. Bacteriol., 2011, 193, 4652–4663 CrossRef CAS PubMed.
- J. Scherer and D. H. Nies, Mol. Microbiol., 2009, 73, 601–621 CrossRef CAS PubMed.
- D. H. Nies, in Molecular microbiology of heavy metals, ed. D. H. Nies and S. Silver, Springer-Verlag, Berlin, 2007, vol. 6, pp. 118–142 Search PubMed.
- D. H. Nies, A. Nies, L. Chu and S. Silver, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7351–7355 CrossRef CAS.
- W. Busch and M. H. J. Saier, Crit. Rev. Biochem. Mol. Biol., 2002, 37, 287–337 CrossRef CAS PubMed.
- M. H. J. Saier, C. V. Tran and R. D. Barabote, Nucleic Acids Res., 2006, 34, D181–D186 CrossRef CAS PubMed.
- T. von Rozycki, D. H. Nies and M. H. J. Saier, Comp. Funct. Genomics, 2005, 6, 17–56 CrossRef CAS PubMed.
- S. Helm, D. Dobritzsch, A. Rodiger, B. Agne and S. Baginsky, J. Proteomics, 2014, 98, 79–89 CrossRef CAS PubMed.
- S. Sundararaj, A. Guo, B. Habibi-Nazhad, M. Rouani, P. Stothard, M. Ellison and D. S. Wishart, Nucleic Acids Res., 2004, 32(Database issue), D293–D295 CrossRef CAS PubMed.
- F. C. Neidhardt and H. E. Umbarger, in Escherichia coli and Salmonella: cellular and molecular biology, ed. F. C. Neidhardt, American Society of Microbiology, Washington, 1996 Search PubMed.
- J. Goris, P. De Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters and P. Vandamme, Int. J. Syst. Evol. Microbiol., 2001, 51, 1773–1782 CrossRef CAS PubMed.
- A. Legatzki, S. Franke, S. Lucke, T. Hoffmann, A. Anton, D. Neumann and D. H. Nies, Biodegradation, 2003, 14, 153–168 CrossRef CAS.
- K. von Meyenburg, B. B. Jørgensen and B. van Deurs, EMBO J., 1984, 3, 1791–1797 CAS.
- N. S. Shepherd, G. Churchward and H. Bremer, J. Bacteriol, 1980, 141, 1098–1108 CAS.
- N. S. Shepherd, P. Dennis and E. Bremer, J. Bacteriol., 2001, 183, 2527–2534 CrossRef CAS PubMed.
- S. Klumpp and T. Hwa, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20245–20250 CrossRef CAS PubMed.
- P. Sarkar, A. A. Sardesai, K. S. Murakami and D. Chatterji, J. Biol. Chem., 2013, 288, 25076–25087 CrossRef CAS PubMed.
- M. Jishage and A. Ishihama, J. Bacteriol., 1995, 177, 6832–6835 CAS.
- H. Maeda, M. Jishage and A. Ishihama, Nucleic Acids Res., 2000, 28, 3497–3503 CrossRef CAS PubMed.
- C. Große, S. Friedrich and D. H. Nies, J. Mol. Microbiol. Biotechnol., 2007, 12, 227–240 CrossRef PubMed.
- D. H. Nies, Arch. Microbiol., 2004, 181, 255–268 CrossRef CAS PubMed.
- M. Kanehisa, S. Goto, M. Hattori, K. F. Aoki-Kinoshita, M. Itoh, S. Kawashima, T. Katayama, M. Araki and M. Hirakawa, Nucleic Acids Res., 2006, 34, D354–D357 CrossRef CAS PubMed.
- P. M. Vignais and A. Colbeau, Curr. Issues Mol. Biol., 2004, 6, 159–188 CAS.
- J. Fritsch, P. Scheerer, S. Frielingsdorf, S. Kroschinsky, B. Friedrich, O. Lenz and C. M. T. Spahn, Nature, 2011, 479, 249–252 CrossRef CAS PubMed.
- M. Mergeay, D. Nies, H. G. Schlegel, J. Gerits, P. Charles and F. van Gijsegem, J. Bacteriol., 1985, 162, 328–334 CAS.
- T. Burgdorf, O. Lenz, T. Buhrke, E. van der Linden, A. K. Jones, S. P. J. Albracht and B. Friedrich, J. Mol. Microbiol. Biotechnol., 2005, 10, 181–196 CrossRef CAS PubMed.
- M. Bernhard, T. Buhrke, B. Bleijlevens, A. L. De Lacey, V. M. Fernandez, S. P. J. Albracht and B. Friedrich, J. Biol. Chem., 2001, 276, 15592–15597 CrossRef CAS PubMed.
- J. W. Olson and R. J. Maier, J. Bacteriol., 2000, 182, 1702–1705 CrossRef CAS.
- M. Hube, M. Blokesch and A. Bock, J. Bacteriol., 2002, 184, 3879–3885 CrossRef CAS.
- Y. Li and D. B. Zamble, Chem. Rev., 2009, 109, 4617–4643 CrossRef CAS PubMed.
- D. C. Kennedy, R. W. Herbst, J. S. Iwig, P. T. Chivers and M. J. Maroney, J. Am. Chem. Soc., 2007, 129, 16–17 CrossRef CAS PubMed.
- R. W. Herbst, I. Perovic, V. Martin-Diaconescu, K. O'Brien, P. T. Chivers, S. S. Pochapsky, T. C. Pochapsky and M. J. Maroney, J. Am. Chem. Soc., 2010, 132, 10338–10351 CrossRef CAS PubMed.
- A. Katayama, A. Tsujii, A. Wada, T. Nishino and A. Ishihama, Eur. J. Biochem., 2002, 269, 2403–2413 CrossRef CAS.
- M. P. Hensley, D. L. Tierney and M. W. Crowder, Biochemistry, 2011, 50, 9937–9939 CrossRef CAS PubMed.
- M. Kaczanowska and M. Ryden-Aulin, Microbiol. Mol. Biol. Rev., 2007, 71, 477–494 CrossRef CAS PubMed.
- A. Anjem, S. Varghese and J. A. Imlay, Mol. Microbiol., 2009, 72, 844–858 CrossRef CAS PubMed.
- A. Anjem and J. A. Imlay, J. Biol. Chem., 2012, 287, 15544–15556 CrossRef CAS PubMed.
- L. Banci, I. Bertini, S. Ciofi-Baffoni, T. Kozyreva, K. Zovo and P. Palumaa, Nature, 2010, 465, 645–648 CrossRef CAS PubMed.
- F. Y. H. Wu, W. J. Huang, R. B. Sinclair and L. Powers, J. Biol. Chem., 1992, 267, 25560–25567 CAS.
- K. Helbig, C. Bleuel, G. J. Krauss and D. H. Nies, J. Bacteriol., 2008, 190, 5431–5438 CrossRef CAS PubMed.
- C. Große, G. Schleuder, C. Schmole and D. H. Nies, Appl. Environ. Microbiol., 2014, 80 DOI:10.1128/AEM.02842-14.
- Z. Ma, P. Chandrangsu, T. C. Helmann, A. Romsang, A. Gaballa and J. D. Helmann, Mol. Microbiol., 2014 DOI:10.1111/mmi.12794.
- C. Rensing, U. Kües, U. Stahl, D. H. Nies and B. Friedrich, J. Bacteriol., 1992, 174, 1288–1292 CAS.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, J. Biol. Chem., 1951, 193, 265–275 CAS.
- N. Wiesemann, J. Mohr, C. Grosse, M. Herzberg, G. Hause, F. Reith and D. H. Nies, J. Bacteriol., 2013, 195, 2298–2308 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00171k |
| This journal is © The Royal Society of Chemistry 2014 |