Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Metallothionein-like peptides involved in sequestration of Zn in the Zn-accumulating ectomycorrhizal fungus Russula atropurpurea†
Tereza
Leonhardt
,
Jan
Sácký
,
Pavel
Šimek
,
Jiří
Šantrůček
and
Pavel
Kotrba
*
Institute of Chemical Technology, Prague, Department of Biochemistry and Microbiology, Technická 3, 166 28 Prague, Czech Republic. E-mail: pavel.kotrba@vscht.cz; Fax: +420 220445167; Tel: +420 220445134
First published on 26th June 2014
Abstract
Homeostatic mechanisms preventing the toxicity of free Zn ions in cells involve, among others, cytosolic Zn-binding ligands, particularly the cysteine-rich metallothioneins (MTs). Here we examined the Zn-binding peptides of Russula atropurpurea, an ectomycorrhizal fungus known for its ability to accumulate high amounts of Zn in its sporocarps. The Zn complexes and their peptide ligands were characterized using chromatography, electrophoresis after fluorescent labeling of cysteine residues, and tandem mass spectrometry. Functional complementation assays in Saccharomyces cerevisiae were used to obtain and characterize cDNA sequences. Zn-speciation analysis showed that nearly 80% of the Zn extracted from the sporocarps was associated with cysteine-containing peptides in a 5 kDa complex. Screening of an R. atropurpurea cDNA library for sequences encoding peptides capable of sequestering divalent heavy metals was conducted in the Cd-hypersensitive ycf1Δ yeast. This allowed identification of two cDNAs, RaZBP1 and RaZBP2, which protected the metal-sensitive yeast mutants against Cd and Zn, but not Co, Mn or Cu, toxicity. The corresponding RaZBP1 and RaZBP2 peptides consisting of 53 amino acid (AA) residues and sharing 77% identity showed only a limited sequence similarity to known MTs, particularly due to the absence of multiple Cys-AA-Cys motifs. Both RaZBPs were detected in a native Zn-complex of R. atropurpurea and the recombinant RaZBP1 was found associated with Zn and Cd in yeasts. Altogether, the results point to an important role of RaZBPs in the handling of a substantial portion of the Zn pool in R. atropurpurea.
Introduction
Ectomycorrhizal (EM) fungi, in their mutualistic associations with plant roots, benefit forest trees in a number of ways of which the most important is enhancing soil nutrient mobilization and uptake. There is evidence that EM fungi may increase the bioavailability of trace metals to the host plants by promoting the mobilization of metal ions in the soil and from minerals or, conversely, execute a metal barrier function through mechanisms such as extracellular precipitation or chelation, biosorption, exclusion, and cellular uptake of excess essential as well as non-essential metal species.1–3 The natural capacity of EM fungi to accumulate a wide range of heavy metals has been reported since the 1970s. In an extensive study, scoring the Zn contents of 383 species of basidiomycetous fungi, Vetter et al.4 have identified Russula atropurpurea, the EM fungus common in the northern temperate ecosystems, as a species with metal concentrations in the sporocarp tissue of up to 1067 mg Zn kg−1 dry weight (dwt). The sporocarp Zn concentrations ranging from 745 to 1062 mg kg−1 dwt, significantly exceeding the common levels found in other 86 EM species (median of 98.6 mg Zn kg−1 dwt), have been reported for R. atropurpurea also more recently.5Zinc is an essential catalytic and structural component of many proteins. However, an uncontrolled access of proteins to Zn under a transient or permanent (in Zn accumulators) metal overload would result in an aberrant binding of Zn ions to cysteinyl thiols or other functional groups, rendering the proteins dysfunctional. The mechanisms that evolved in eukaryotes to tightly control the intracellular concentration of free Zn principally involve compartmentalization, chelation, and efflux.6,7 Among fungi, the molecular mechanisms underlying the cellular Zn homeostasis and detoxification have been detailed in yeasts, while the knowledge about the biology of Zn in mycorrhizal fungi is still limited. In Saccharomyces cerevisiae, the detoxification of excess Zn relies largely upon the vacuolar Zrc1 and Cot1 transporters of the cation diffusion facilitator (CDF) family, and the sequestration in and remobilization from the vacuolar compartment play a dominant role in the buffering of the cytosolic Zn levels.8–10 Since glutathione (GSH) readily binds Zn,11 it has been proposed that the cytoplasmic Zn–GSH complex serves these transporters as the source of a labile Zn2+ ion for compartmentalization.6,9 There is evidence that the vacuolar Zn is bound with polyphosphate granules in EM Suillus bovinus and arbuscular mycorrhizal Rhizophagus intraradices (formerly Glomus intraradices).12,13 Interestingly, the Zn tolerant ecotypes of S. bovinus achieve efficient detoxification by an energy-dependent efflux of the metal out of the cell.2,14 Independently of Zrc1 and Cot1, S. cerevisiae can store Zn in small punctuated vesicles of unknown identity that transiently appear when Zn-limited cells are challenged with high Zn concentrations.15 Similar Zn-containing vesicles, so-called zincosomes, are implicated in zinc storage and detoxification in various mammalian cell types.6 In contrast to S. cerevisiae, the endoplasmic reticulum (ER) seems to be the major site of zinc deposition in Schizosaccharomyces pombe.16,17 Recently, Blaudez and Chalot18 characterized the HcZnT1 gene coding for a putative, ER-located, CDF Zn transporter of the EM Hebeloma cylindrosporum; they also demonstrated with this species that Zn in the EM fungi can be targeted into non-vacuolar zincosome-like vesicles.
S. cerevisiae and S. pombe further possess metallothioneins (MTs) Crs5 and Zym1, respectively, which were implicated in the handling of the cytosolic Zn pools under normal conditions, though they were critical only for the detoxification of excess Zn in cells subjected to metal overload.16,19 Noteworthily, Zym1, being transcriptionally activated upon both Zn and Cd exposure, also confers an increased tolerance to Cd. MTs are cytosolic, cysteine-rich peptides of distinct sizes,20 which differentiated up to variable levels of divalent (Zn2+ and Cd2+) or monovalent (Cu+) metal ion-binding specificity, often reflecting their particular roles in metal homeostasis and tolerance of eukaryotes and some prokaryotes.21–26
The Cd-, Cu- or Ag-responsive MTs have been characterized in different mycorrhizal species,27 including the metal-tolerant EM Paxillus involutus,28H. cylindrosporum,29Amanita strobiliformis30 and Hebeloma mesophaeum.31 We have also documented that H. mesophaeum can under experimental, Zn replete conditions deposit cellular Zn in both the zincosome-like vesicles and Zn–MT complexes and that the inventory of MT genes in this species involves Zn-inducible HmMT1.31 It was thus tempting to explore the involvement of MTs in the biology of Zn in the sporocarps of R. atropurpurea that are constantly subjected to Zn overload under natural conditions. Surprisingly, as shown in this paper, the majority of the Zn extracted from the sporocarps was sequestered by two isomorphic peptides, RaZBP1 and RaZBP2, which are (by sequence) only distantly related to MTs. The yeast complementation approach used in this study to search for cDNA coding for Zn ligands of R. atropurpurea thus allowed us to identify functional metal-binding peptides that would likely escape the homology-based search of genomic or transcriptomic data.
Materials and methods
Organisms and general culture conditions and procedures
The young sporocarps of R. atropurpurea (Krombh.) Britzelm., non Peck containing 640 to 650 mg Zn kg−1 and >1 mg Cd kg−1 dwt were obtained from Dr Borovička (harvested under a Quercus tree in Prague-Kobylisy, Czech Republic; a representative deposited in the herbarium of the Mycological Department, National Museum, Prague, under the number PRM 858109). The collected specimens were cleared of substrate debris, washed with distilled water and cut vertically into equivalent quarters (stipes and caps in a natural proportion). The parts to be used for the Zn speciation analysis were stored at −80 °C. The parts to be used for the molecular analyses were fixed by freeze-drying, homogenized with a mortar and pestle, and stored at −80 °C.The yeast strains used for the heterologous expression of the R. atropurpurea cDNAs were ycf1Δ strain DTY168 (MATα his6 leu2-3,-112 ura3-52 ycf1::hisG),32zrc1Δcot1Δ strain CM137 (MATα can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-52 zrc1::His3 cot1::KanR),8cup1Δ strain DTY113 (MATα trp1-1 leu2-3,-112 gal1 ura3-50 cup1Δ61),33 and BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) and its pmr1Δ derivative (pmr1::kanMX4) obtained from Euroscarf (http://web.uni-frankfurt.de/fb15/mikro/euroscarf/index.html). Transformed yeasts were grown at 30 °C on URA+ selective SD agar plates (SD medium: 0.7% [w/v] Difco yeast nitrogen base, 2% [w/v] glucose, 0.005% [w/v] adenine hemisulfate, and 0.003% [w/v] of each of L-histidine, L-tryptophan, L-methionine and L-leucine). For the metal tolerance plate assays, the mid-log cultures of S. cerevisiae transformants were adjusted to an optical density at 590 nm (OD590) of 0.05, and 5 μl of serial dilutions were spotted on SD medium plates without metal addition or supplemented with 100 μM CdCl2, 0.1 to 2 mM CoCl2, 5 to 50 μM CuSO4, 0.1 to 2 mM MnSO4 or 250 μM ZnCl2. The growth of S. cerevisiae BY4741 transformants in the presence of 2.5 mM Zn or 200 μM Cd in a liquid SD medium was initiated by addition of the metal to cultures that reached the OD590 of 0.5. The cultures were further propagated for 12 h (final OD590 of 3.7 to 4.2), and the cells were harvested by centrifugation at 4000 × g and 25 °C for 3 min. To determine the concentration of the metal accumulated in the yeasts from 50 ml culture aliquots, the surface-bound metal was removed by two washes with 5 ml of 5 mM EDTA and the cells were digested with 65% nitric acid for 16 h. The metal content of the supernatant resulting from a 20 min centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g was analyzed by atomic absorption spectrometry (AAS; model Spectr AA300, Varian, Inc.).
000 × g was analyzed by atomic absorption spectrometry (AAS; model Spectr AA300, Varian, Inc.).
To isolate total RNA and chromosomal DNA from 50 mg of freeze-dried tissue, an RNeasy Plant Mini Kit (Qiagen) with the RLT buffer and a NucleoSpin Plant II Kit (Macherey-Nagel) were used according to the manufacturer's instructions, respectively. The RNase free DNase set (Qiagen) was used to digest the residual DNA during RNA purification. The integrity of the isolated RNA was checked by formaldehyde agarose gel electrophoresis and concentration was determined by measuring the absorbance at 260 nm. The DNA manipulations in E. coli DH5α, gene expression in E. coli BL21(DE3) and routine DNA, RNA and protein work were performed according to the standard protocols. Recombinant DNAs were subjected to custom DNA sequencing on both strands with vector-specific primers.
Speciation analysis of intracellular metal
The sporocarps were ground in liquid N2 with a mortar and pestle, and the disintegrated tissue was extracted with 0.7 ml of 50 mM HEPES (pH 7.0) per 1 g of tissue fresh weight (fwt). Tissue debris was removed by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 4 °C for 10 min. To fractionate the extracted Zn species, 2 ml of the extract (6 to 8 mg of the total protein as determined using a BCA Protein Assay Kit [Thermo Scientific] with BSA and lysozyme as standards) was loaded onto a Superdex Peptide 10/300 GL column (GE Healthcare). The SEC separation was performed with a BioLogic DuoFlow fast protein liquid chromatography (FPLC) system (BioRad) and 50 mM HEPES, 25 mM KNO3 (pH 7.0) as a mobile phase at a flow rate of 0.5 ml min−1. Ribonuclease A (GE Healthcare), ubiquitin (Sigma-Aldrich), a synthetic 2.1 kDa peptide and glutathione (GSH; Merck) were used as molecular mass standards. The metal contents in the aliquots of the 0.5 ml fractions from SEC were analyzed by AAS. To assess the portion of Zn that escaped the extraction, the tissue debris was extracted with 65% nitric acid for 16 h, and the concentration of released Zn was determined by AAS.
000 × g and 4 °C for 10 min. To fractionate the extracted Zn species, 2 ml of the extract (6 to 8 mg of the total protein as determined using a BCA Protein Assay Kit [Thermo Scientific] with BSA and lysozyme as standards) was loaded onto a Superdex Peptide 10/300 GL column (GE Healthcare). The SEC separation was performed with a BioLogic DuoFlow fast protein liquid chromatography (FPLC) system (BioRad) and 50 mM HEPES, 25 mM KNO3 (pH 7.0) as a mobile phase at a flow rate of 0.5 ml min−1. Ribonuclease A (GE Healthcare), ubiquitin (Sigma-Aldrich), a synthetic 2.1 kDa peptide and glutathione (GSH; Merck) were used as molecular mass standards. The metal contents in the aliquots of the 0.5 ml fractions from SEC were analyzed by AAS. To assess the portion of Zn that escaped the extraction, the tissue debris was extracted with 65% nitric acid for 16 h, and the concentration of released Zn was determined by AAS.
The cell-free extracts from Zn- and Cd-exposed S. cerevisiae were prepared from 1 g (fwt) of the metal-exposed cells washed with 10 ml of fresh SD medium. The yeasts were resuspended in 50 mM HEPES (pH 7.0) at a density of 0.25 g (fwt) ml−1, and combined with 3 ml of glass beads (0.5 mm i.d.) for cell disruption in a FastPrep-24 device (M. P. Biomedicals). This was conducted in four cycles of disintegration at maximum speed at room temperature for 1 min and 5 min cooling on ice. The glass beads and cell debris were separated by centrifugation at 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g and 4 °C for 30 min. The supernatant (2 ml) was resolved by SEC as already described.
000 × g and 4 °C for 30 min. The supernatant (2 ml) was resolved by SEC as already described.
Labeling and electrophoresis of cysteinyl-containing peptides
To analyze the sulfhydryl-containing ligands of R. atropurpurea, the pooled metal-containing fractions were brought to a final volume of 60 ± 5 μl by ultrafiltration with the Microcon YM-3. Pooled aliquots (200 μl) of eligible fractions obtained from SEC of yeast extracts were freeze-dried and dissolved in 50 μl of distilled water. The ligands were labeled fluorescent in a reaction with sulfhydryl-specific 7-fluorobenzofurazan-4-sulfonic acid (SBD-F; Sigma-Aldrich) and resolved by sodium dodecylsulfate-polyacrylamide gel electrophoresis (SDS-PAGE) as described previously.30 The electrophoresis was conducted in a discontinuous 10% (upper) and 16% (lower) acrylamide gel in a Tris-Tricine buffer system with 6 M urea, and the fluorescence signal (>605 nm) was scanned with an LAS1000 (Fuji) after excitation at 312 nm. A rabbit liver MT1a (Enzo Life Sci.) was used as the standard.cDNA expression library construction and screening
To constitutively express a cDNA library in S. cerevisiae, the centromeric yeast vector p416GPD was employed, which contains a yeast glyceraldehyde-3-phosphate dehydrogenase promoter, cytochrome c oxidase terminator and URA3 gene.34 An Oligotex mRNA Mini Kit (Qiagen) was used according to the manufacturer's instructions to obtain 5 μg of R. atropurpurea mRNA from 300 μg of total sporocarp RNA. Double stranded cDNAs flanked by 5′ EcoRI and 3′ XhoI sites were produced following the cDNA synthesis protocol of a ZAP-cDNA® Gigapack® III Gold Cloning Kit (Stratagene). This library was inserted into an approximately twofold stoichiometric amount of EcoRI-/XhoI-digested p416GPD and transformed into E. coli for amplification. Approximately 2 × 105 transformant colonies (0.5 to 1 mm i.d.) were washed from the plates with Luria-Bertani media, pooled, and stored in 20% glycerol (v/v) at −80 °C. The plasmid cDNA library was isolated from an aliquot (approximately 8 × 1011 cells) of these primary clones by using the Qiagen Plasmid Midi Kit (Qiagen) and 1 μg of the isolated plasmids was used to transform the Δycf1 S. cerevisiae strain DTY168. The URA3+ yeasts were selected on SD medium, and approximately 6 × 104 transformants were washed from the plates with the same media. To select for Cd-tolerant transformants from this pool, aliquots were replated on SD media amended with 75 μM Cd2+. The plasmids harboring the cDNAs that conferred Cd-tolerance were extracted using a QiaPrep Spin Miniprep Kit (Qiagen), amplified in E. coli, and retransformed into the yap1Δ cells to confirm their function.Amplification of 5′ cDNA ends, isolation of a genomic clone and sequence analysis
The entire 5′ ends of the individual RaZBP cDNAs were obtained from the total RNA by using an ExactSTART Eukaryotic mRNA 5′- & 3′-RACE Kit (Epicentre) according to the manufacturer's instructions. The transcript-specific reverse primers were 5′-CTGACTCGAGGGGATTACTAACCGGAATGCTG-3′ for RaZBP1 and 5′-CTGACTCGAGGGGATTACTAATCGGAATGCTG-3′ for RaZBP2. The chromosomal clone of the RaZBP1 gene with an adjacent 0.7 kb DNA sequence upstream of the start codon was isolated from PvuII-digested, adapter-flanked genomic DNA using a GenomeWalker Universal Kit (Clontech) according to the manufacturer's instructions. The PCR amplification procedure used an Advantage 2 Proofreading DNA Polymerase Mix (Clontech) with adaptor-specific primers and primer sets designed to match the 3′ untranslated sequences of RaZBP1. The gene-specific primers used in primary and secondary nested PCR were 5′-AACGCATITCATTCGAAAACACAATCC-3′ and 5′-CCTTCCICTCITTGCTAACCGACTCAG-3′, respectively. The resulting 5′ cDNAs and the chromosomal clones were inserted into the pGEM-T Easy vector (Promega). The amino acid sequences deduced from cDNAs were repeatedly BLASTed against the UniProt databases and the translated nucleotide database at GenBank (last search on 20. 2. 2014). The abundance of potential transcription factor-binding sites was investigated by using TESS software35 and Patch software with TRANSFAC® Public 6.0 database.36 The nucleotide sequences were deposited in GenBank under the accession numbers KF477286 (RaZBP1 gene), KF477287 (RaZBP1 cDNA) and KF477288 (RaZBP2 cDNA).Mass spectroscopy (MS) analysis of Zn complex ligands
To isolate the 5 kDa Zn-complex on a preparative scale, the extract originating from 10 g (fwt) of the R. atropurpurea tissue was brought to a final volume of 2 ml by ultrafiltration with a Microcon YM-3 membrane (Millipore) prior to SEC conducted as described above. For the separation of the complex ligands, the eligible fractions from SEC were pooled and concentrated with the Microcon YM-3 to a volume of 500 μl, and tris(carboxyethyl)phosphine (TCEP; Sigma-Aldrich) was added at 140 mM to perform a reduction reaction for 1 h at room temperature. The ligands were released by acidification with trifluoroacetic acid (TFA; Sigma-Aldrich) added to a final concentration of 1% (pH 1.05) and resolved on the Superdex Peptide 10/300 GL column with 1% TFA in water as a mobile phase at 0.5 ml min−1. The fractions containing a peptide ligand were pooled and freeze-dried, and the lyophilized material was resolved in 80 μl of 50 mM HEPES (pH 7.0). The Zn-associated peptides were reduced with 5 mM dithiothreitol for 30 min at 50 °C and alkylated by 25 mM iodoacetamide for 30 min at room temperature. Mass spectra were recorded on an electrospray ionization quadrupole time-of-flight (ESI-Q-TOF) Maxis Impact (Bruker Daltonics) linked to an Ultra HPLC (UHPLC) UltiMate 3000 RSLCnano (Dionex). For the UHPLC, the alkylated peptides were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 with 3% acetonitrile in 0.1% formic acid water solution (v/v) and washed on a 20 mm Acclaim PepMap100 C18 trap column (100 μm ID; Dionex) with the same solution for 5 min at 5 μl min−1. The peptides were then separated on an analytical 150 mm Acclaim PepMapRSLC C18 nanoViper column (75 μm ID; Dionex). The acetonitrile proportion in water (both with 0.1% formic acid) during elution at 300 nl min−1 was 3–40% linear gradient from 0 to 25 min, 90% from 25 to 35 min, and 3% from 35 to 50 min. The peptides were eluted directly to the ESI source (Captive spray) and the MS measurement was carried out in a positive ion mode with the capillary voltage set at 1500 V, flow of drying gas at 3 l min−1 and temperature 150 °C. Precursors were selected in the range of 400–2200 m/z and tandem MS (MS/MS) spectra recorded in the range of 50–2200 m/z. Up to five precursors from each spectrum were selected for fragmentation using N2 as a collision gas (exclusion time set to 30 s) and the raw MS data were analyzed using Data Analysis version 4.1 (Bruker Daltonics). The in-house Mascot server version 2.4.1 (Matrix Science, UK) was used to identify peptides from MS/MS data through search in a custom made database created by combining Swiss-Prot database (version 20130129) with the RaZBP sequences predicted from the coding cDNAs. Carbamidomethylation of cysteine and oxidation of methionine residues were set as fixed and variable modifications, respectively. Tolerations of 5 ppm and 0.05 Da were used in the MS and the MS/MS mode, respectively. Peptides with the Mascot scores higher than 43 (Table S1, ESI†) were considered to be statistically significant (p ≤ 0.05). The identified sequences were further inspected manually using BioTools 3.2 (Bruker Daltonics). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium37via the PRIDE partner repository with the dataset identifier PXD001073 and DOI: http://10.6019/PXD001073.
20 with 3% acetonitrile in 0.1% formic acid water solution (v/v) and washed on a 20 mm Acclaim PepMap100 C18 trap column (100 μm ID; Dionex) with the same solution for 5 min at 5 μl min−1. The peptides were then separated on an analytical 150 mm Acclaim PepMapRSLC C18 nanoViper column (75 μm ID; Dionex). The acetonitrile proportion in water (both with 0.1% formic acid) during elution at 300 nl min−1 was 3–40% linear gradient from 0 to 25 min, 90% from 25 to 35 min, and 3% from 35 to 50 min. The peptides were eluted directly to the ESI source (Captive spray) and the MS measurement was carried out in a positive ion mode with the capillary voltage set at 1500 V, flow of drying gas at 3 l min−1 and temperature 150 °C. Precursors were selected in the range of 400–2200 m/z and tandem MS (MS/MS) spectra recorded in the range of 50–2200 m/z. Up to five precursors from each spectrum were selected for fragmentation using N2 as a collision gas (exclusion time set to 30 s) and the raw MS data were analyzed using Data Analysis version 4.1 (Bruker Daltonics). The in-house Mascot server version 2.4.1 (Matrix Science, UK) was used to identify peptides from MS/MS data through search in a custom made database created by combining Swiss-Prot database (version 20130129) with the RaZBP sequences predicted from the coding cDNAs. Carbamidomethylation of cysteine and oxidation of methionine residues were set as fixed and variable modifications, respectively. Tolerations of 5 ppm and 0.05 Da were used in the MS and the MS/MS mode, respectively. Peptides with the Mascot scores higher than 43 (Table S1, ESI†) were considered to be statistically significant (p ≤ 0.05). The identified sequences were further inspected manually using BioTools 3.2 (Bruker Daltonics). The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium37via the PRIDE partner repository with the dataset identifier PXD001073 and DOI: http://10.6019/PXD001073.
Vectors for the expression of RaZBPs in S. cerevisiae
The coding sequences of RaZBPs were amplified from their individual cDNAs using a Pfu DNA polymerase (Promega) and gene-specific primers. To construct the plasmids for yeast complementation assays, the primer sets introducing BamHI and XhoI sites at the 5′ and 3′ amplicon ends, respectively, were 5′-CATGGATCC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CCCGCTCAAGAGACTATC-3′ and 5′-CTGACTCGAGGGGA
CCCGCTCAAGAGACTATC-3′ and 5′-CTGACTCGAGGGGA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CTAACCGGAATGCTG-3′ for RaZBP1 and 5′-CTTGGATCC
CTAACCGGAATGCTG-3′ for RaZBP1 and 5′-CTTGGATCC![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[G with combining low line]](https://www.rsc.org/images/entities/char_0047_0332.gif) CCCGCTCAAGAGACTATC-3′ and 5′-CTGACTCGAGGGGA
CCCGCTCAAGAGACTATC-3′ and 5′-CTGACTCGAGGGGA![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif)
![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CTAATCGGAATGCTG-3′ for RaZBP2 (start and stop codons underlined and endonuclease target sequences italicized on the primer sequences). The resulting amplicons were inserted into a BamHI-/XhoI-treated plasmid p426GPD (the same as p416GPD but containing 2μ origin of replication34).
CTAATCGGAATGCTG-3′ for RaZBP2 (start and stop codons underlined and endonuclease target sequences italicized on the primer sequences). The resulting amplicons were inserted into a BamHI-/XhoI-treated plasmid p426GPD (the same as p416GPD but containing 2μ origin of replication34).
Results and discussion
Intracellular Zn in the sporocarp of R. atropurpurea
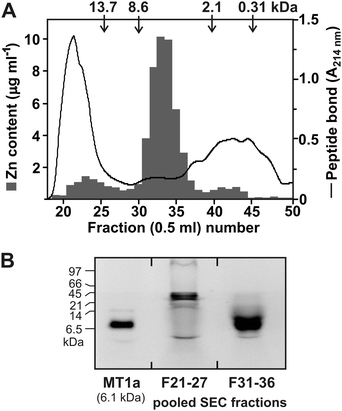
To obtain information regarding soluble, intracellular Zn species, the tissue of the R. atropurpurea PRM 858109 sporocarp was disintegrated under the mild conditions of neutral pH, which allowed 55% of the total Zn to be extracted. The size exclusion chromatography (SEC) revealed that 79% of the extracted Zn was contained in a peak of molecular mass of approximately 5 kDa (Fig. 1A). A minor portion (6%) of Zn eluted with fractions 40 to 45 (peak maximum close to 1 kDa) and may correspond to cytosolic Zn transients and compartmentalized Zn2+ bound, e.g., with glutathione and polycarboxylic or amino acids.3,6,31 Nearly 15% of Zn was excluded from the column with proteins of molecular mass of ≥20 kDa. However, these data have to be interpreted with caution, since our approach does not allow discriminating between proteins metalated in vivo and those that might eventually associate with Zn2+ released from compartments upon extraction. It is also remarkable that nearly half of the sporocarp Zn remained repeatedly associated with the cell debris. Although incomplete disintegration could not be excluded, inspection of the cell debris by microscopy revealed thorough disruption of the tissue. The reduced recovery of Zn may also be attributable to biosorption of the metal ion liberated from the compartments on the cell wall and other biopolymers in the cell debris and/or, the existence of insoluble Zn species such as the vacuolar Zn-(poly)phosphate granules described in other mycorrhizal fungi.12,13 The same results were obtained with an extract from the R. atropurpurea sporocarp (821 mg Zn kg−1 dwt), originating from a different locality, in which the proportions of the soluble Zn sequestered in ≥20 kDa, 5 kDa, and 1 kDa fractions were approximately 18%, 74% and 11%, respectively (data not shown).The size of the major, 5 kDa Zn complex that contained more than 40% of the total Zn of the PRM 858109 sporocarp suggested binding with peptidaceous ligands, such as MTs; therefore, the corresponding fractions were concentrated, labeled with the sulfhydryl-specific SBD fluorochrome and resolved using SDS-PAGE. As shown in Fig. 1B, the double band observed after the separation in the 16% acrylamide gel indicated that the compounds associated with Zn in the 5 kDa complex might be peptides of a molecular mass close to 6.1 kDa of the rabbit MT1a.
Isolation and sequence analysis of RaZBPs
In an attempt to identify R. atropurpurea cDNAs capable of conferring increased tolerance to divalent heavy metal ions, a yeast complementation screen using the ycf1Δ mutant was employed. Disruption of the ycf1 gene, which encodes an ABC-type transporter involved in the vacuolar sequestration of Cd, renders S. cerevisiae highly sensitive towards Cd.32 The reason for the choice of a Cd-sensitive strain in our search for an expected Zn-binding MT was that, unlike Zn-sensitive strains, ycf1Δ cells respond sharply to the metal toxicity and this was expected to enhance the selection. We also considered the fact that both Cd and Zn bind, albeit with some exceptions, to known MTs in an isostructural manner.38 The yeast cells were transformed with the p416GPD-based, sporocarp cDNA expression library and the transformants were plated on a uracil-deficient SD agar medium without metal supplement to fix the cDNA library in the cells. Approximately 108 of the individual primary transformants were selected to be screened for their capability to grow on the same agar media in the presence of 75 μM Cd2+. Among the positive transformants, sequencing of cDNAs that were still able to complement Δycf1 after retransformation revealed two individual clones, which contained 159-nucleotide sequences coding for two predicted 53-amino-acid (AA) peptides. As documented below, these peptides can bind with Zn and the cDNAs were thus designated RaZBP1 and RaZBP2 (Fig. 2).Sequencing of the 5′ ends of the respective cDNAs isolated via 5′-RACE from R. atropurpurea indicated that both RaZBPs obtained from the library harbored full-length coding sequences. The deduced peptide sequences share more than 77% identity and appear to conserve the positions of the cysteinyl (Cys), histidyl (His) and some glutamyl (Glu) and aspartyl (Asp) residues. These residues are known to form stable Zn-binding centers of many metalloproteins in which they are present within the sequence motifs characterizing specific coordination groups.39 However, neither BLAST searches against various databases nor the comparison with the Cys/His/Glu/Asp coordination groups of Zn, Cu, Co, Fe or Mn metalloproteins indicated a reasonable similarity of the RaZBP peptides to proteins or domains that have been assigned a specific function. A significant similarity (expect value of 10−13) at the amino acid, but not nucleotide, sequence level was found with the C-terminal Cys- and His-containing sequences of hypothetical peptides deduced from the GenBank EST sequences FR708292 and FR713564l, expressed by uncultured eukaryotes from a forest soil.40 The former of the two nearly identical peptides is shown in Fig. 2.
The alignment of RaZBP1 and RaZBP2 with MTs suggested certain similarity with some fungal MTs, particularly when the distribution of the metal-binding residues was considered (Fig. 2). In spite of a high diversity of MT sequences, the feature particularly characteristic of MTs is the presence of multiple Cys-X-Cys motifs.20 With a single exception of the C-Q-C motif, these are absent from both RaZBP peptides. A second such motif could be inferred considering His a Cys mimic. Given the facts that similar Zn binding constants pertain at physiological pH for both Zn-Cys4 and mixed Zn-Cys3His or Cys2His2 coordination environments,41 and that His contributes to the high-affinity coordination of Zn in SmtA of the cyanobacterium Synechococcus PCC 7942,21,42 wheat MT Ec-138,43 and nematode CeMT1,23 this assumption seems to be valid. RaZBPs then appear most closely related to HcMT2 of the EM H. cylindrosporum and HmMT3 of Hebeloma mesophaeum (Fig. 2), sharing the sequence pattern Cys-X-Cys-X9-Cys-X4-Cys-X-Cys/His (X stands for any residue other than Cys or His).
Genome walking experiments allowed us to isolate a 1 kb genomic clone harboring the RaZBP1 gene and a sequence of 0.7 kb upstream of the start codon. The mRNA-to-genomic sequence alignment revealed that RaZBP1 contained two exons, with the intron flanked by conserved |GT-AG| junctions. While the TATA box consensus was absent from the RaZBP1 core promoter, a nonconsensus 5′-TATTTAAA-3′ element was observed at the position −37 relative to the potential transcription start site. It should be noted that in S. cerevisiae, the same element is a target of the TATA-binding protein and facilitates a high rate of transcription.44 Two heptanucleotide sequences matching a conserved core motif of MRE (metal responsive element; 5′-TGCRCNC-3′ with R = A/G and N = any nucleotide) were observed at positions −279 and −473. The cis-acting MREs of animals are recognized by MRE-binding transcription factor 1 (MTF-1) and, like in plants, they are essential and sufficient for the transcriptional activation of heavy metal-responsive genes under metal replete conditions.45,46 Taken together, these data provided support to the notion that RaZBPs might be authentic metal binding peptides of R. atropurpurea.
Identification of RaZBP1 and RaZBP2 in the major Zn-complex of R. atropurpurea
To investigate whether RaZBPs were present in the major Zn complex of R. atropurpurea, a hydrogen ion (1% TFA, pH 1.1)/Zn2+ competition for metal binding site(s) was used to release the metal from the ligands, which were further isolated using SEC and proved metal-free through AAS measurements. The isolated peptides were alkylated by carbamidomethylation to protect thiols from oxidation and analyzed by UHPLC coupled with ESI-Q-TOF tandem mass spectrometry (MS/MS) as described in Materials and methods. As shown in Fig. 3, the fragmentation spectra of the two molecular ions selected as precursors at m/z corresponding to 4090.62 and 4033.60 of non-alkylated [M + H]+ ions displayed a series of b and y ions, which identified these compounds as the fragments of RaZBP1 (AA 12–53) and RaZBP2 (AA 13–53), respectively. The MS/MS analysis of peptide fragments selected at lower m/z values revealed the presence of several shorter forms of both peptides (Table S1, ESI†). Although we were unable to detect a precursor corresponding to the entire sequences of the RaZBP peptides, these data confirmed that the 5 kDa Zn complex contained both RaZBPs. It is also worth noting that the UHPLC-ESI-Q-TOF analysis of the Zn complex peptides did not detect the presence of either MTs predicted from the transcriptome sequence of the same sporocarp (GenBank accession no. KF278561 and KF278562). | ||
| Fig. 3 Sequence analysis of the peptides of the 5 kDa Zn complex according to Fig. 1 by tandem mass (MS/MS) spectrometry. Shown are ESI-Q-TOF MS/MS spectra identifying the partial sequences of carbamidomethylated RaZBP1 (m/z of alkylated [M + H]+ of 4375.72; UHPLC retention time of 18.6 min) and RaZBP2 (m/z of alkylated [M + H]+ of 4318.71; UHPLC retention time of 19.4 min). The signals of singly protonated molecular b and y ions are labeled and the collision-induced fragmentation pattern of the corresponding peptides is indicated. Truncated, ≥37-AA RaZBP sequences identified by MS/MS analysis (spectra not shown) are also annotated (for a complete list of identified peptide fragments, see Table S1, ESI†). | ||
Functional expression of RaZBPs in S. cerevisiae
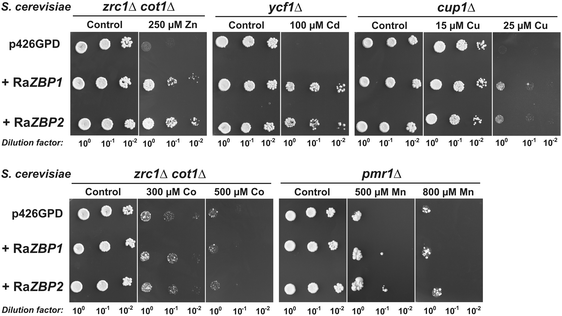
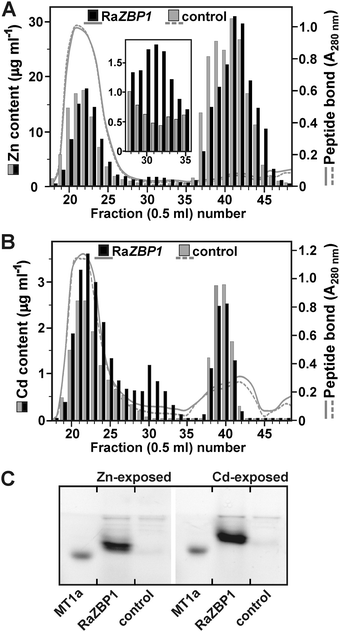
In order to gain information regarding the heavy-metal binding capability of RaZBP peptides, corresponding coding sequences were inserted into the p426GPD vector and constitutively expressed in metal-sensitive S. cerevisiae strains grown on media with or without metal supplement. The well-characterized yeast mutants were Cd-sensitive ycf1Δ strain; Zn-sensitive zrc1Δcot1Δ double mutant (cot1 coding for a vacuolar CDF transporter also confers increased Co tolerance);8,47cup1Δ61 strain carrying a deletion of its MT gene cup1, which renders the cells particularly sensitive to Cu;33 and pmrΔ strain that is unable to grow under Mn replete conditions due to the lack of Ca2+, Mn2+-ATPase for the efflux of the cytosolic metal into Golgi.48 As documented in Fig. 4, RaZBPs fully complemented the Zn-hypersensitive phenotype of zrc1Δcot1Δ on 250 μM Zn2+. While both RaZBP cDNAs protected the ycf1Δ host from the Cd2+ toxicity, they were unable to restore full growth of cup1Δ cells in the presence of 25 μM Cu2+, an external Cu2+ concentration only slightly higher than 15 μM tolerated by cup1Δ transformed with an empty expression vector. These data suggested that, unlike with divalent Zn and Cd ions, the RaZBP peptides failed to form stable, functional Cu+-binding center(s) in vivo (note that Cu+ is the predominating intracellular Cu species22,41,49). It is worth noting that while some MTs do not exhibit clear metal-binding preferences, there are MTs that show strong preferences for binding with divalent Zn or Cd ions and others that favor monovalent Cu.22–25,50,51 Consequently, the Cd- and Zn-specific HpCdMT of the Roman snail Helix pomatia protects yeasts from the Cd, but not Cu, toxicity and vice versa for its Cu-specific isoform HpCuMT.25 The idea of the Zn/Cd-specific binding sites of RaZBPs is further reinforced by the observation that the transformation with RaZBPs did not result in an increased Co2+ tolerance in the zrc1Δcot1Δ strain nor better growth of the pmr1Δ cells on high Mn2+ (Fig. 4).Since the yeast complementation assays indicated that RaZBPs can sequester the excess intracellular Zn and Cd, the ability of RaZBP1 to bind with these metals was assessed in S. cerevisiae BY4741 cultured in the presence of subtoxic concentrations of 2.5 mM Zn2+ or 200 μM Cd2+. This strain lacks a functional gene for Zn-binding MT Crs5,19 but carries Zrc1 plus Cot1 and Ycf1 functions for the vacuolar sequestration of Zn and Cd, respectively. The SEC of the extracts from the yeasts expressing RaZBP1, but not from the control cells harboring p426GPD, revealed the presence of Zn-containing (Fig. 5A) and Cd-containing (Fig. 5B) complexes eluting with fractions corresponding to those of the major Zn complex of R. atropurpurea (Fig. 1A). The electrophoretic separation of SBD-F-labeled complexes from the RaZBP1-expressing yeasts further identified the peptide double band (Fig. 5C), corresponding to that seen with the peptides associated with Zn in the R. atropurpurea sporocarp extract (Fig. 1B). Considering the facts that such peptides were absent from the controls and that two peptide bands of the same intensity were also observed with the recombinant RaZBP1 produced in E. coli (Fig. S1, ESI†), we assumed that both bands originated from the recombinant RaZBP1. The reason why RaZBPs may resolve into a double band remains unknown. We can only speculate that it would be a consequence of a different extent of their derivatization with SBD-F.
Altogether, these data indicate that RaZBPs are functional Zn- and Cd-binding peptides. On the other hand, the observation that the majority of Zn and Cd was in the yeasts expressing RaZBP1 contained in the low molecular mass fractions (Fig. 5A and B) attributable to compartmentalized metal31 would suggest that the metal binding with the peptide was not under the used experimental conditions able to efficiently compete with the vacuolar compartmentalization. However, considering the proposed metal-chaperone functions of GSH or MTs in the transport across endomembranes in yeasts6,9 and animals,7 the possibility that the metal–RaZBP1 complexes served, at least in part, as a source of metal ions for transport cannot be rejected.
Conclusions
The natural capacity of R. atropurpurea to accumulate Zn in large quantities from unpolluted environments has been well documented.4,5 It prompted our efforts towards the identification of the molecules involved in the intracellular sequestration of the (over)accumulated metal. The results presented here provide evidence that the handling of a substantial portion of the sporocarp Zn under natural conditions involves the RaZBP1 and RaZBP2 peptides. The majority of the extractable sporocarp Zn was contained in a 5 kDa complex in which we confirmed the presence of both peptides at the protein level. The notion that RaZBPs may participate in the sequestration of surplus cellular Zn ions in the sporocarps is reinforced by the observation that RaZBPs conferred increased Zn tolerance to yeasts and that production of RaZBP1 in the metal exposed yeasts resulted in the formation of Zn complexes resembling those identified in R. atropurpurea. Our data further show that RaZBPs are efficient Cd-binding peptides. Considering this, RaZBPs appear to be functionally related to MTs. Moreover, the sequence analyses also suggested certain similarity of RaZBPs to MTs but not to other characterized peptides or protein metal binding motifs. Hence, it may be reasonable to expect that, like the Zn–MT complexes, the Zn-RaZBP resides in R. atropurpurea in the cytoplasm with Zn stably coordinated with Cys and His residues. The confirmation of the analogy between MTs and RaZBPs will rely on the analysis of the tertiary structure of metalated peptides and metal-binding stoichiometry and affinity measurements. These analyses will also provide further information regarding the observed Zn-binding preferences of the RaZBPs, the property that would be important to eventually avoid interference of the RaZBPs (produced to provide the fungus with a sink for accumulated Zn) with the homeostasis of other metals.Acknowledgements
We thank Dr Jan Borovička (Institute of Geology and Nuclear Physics Institute, Academy of Science of the Czech Republic) for the provision of characterized R. atropurpurea sporocarps and helpful discussions, Prof. David Eide (University of Wisconsin-Madison) for the gift of CM137 strain and Prof. Dennis J. Thiele (Duke University Medical Center) for the gift of strains DTY113 and DTY168. This work was funded by the research project P504-11-0484 from the Czech Science Foundation.References
- M. Bellion, M. Courbot, C. Jacob, D. Blaudez and M. Chalot, FEMS Microbiol. Lett., 2006, 254, 173–181 CrossRef CAS PubMed.
- J. V. Colpaert, J. H. L. Wevers, E. Krznaric and K. Adriaensen, Ann. For. Sci., 2011, 68, 17–24 CrossRef PubMed.
- G. M. Gadd, Y. J. Rhee, K. Stephenson and Z. Wei, Environ. Microbiol. Rep., 2012, 4, 270–296 CrossRef CAS PubMed.
- J. Vetter, I. Siller and Z. Horváth, Mycologia, 1997, 89, 481–483 CrossRef CAS.
- J. Borovička and Z. Řanda, Mycol. Prog., 2007, 6, 249–259 CrossRef.
- D. J. Eide, Biochim. Biophys. Acta, 2006, 1763, 711–722 CrossRef CAS PubMed.
- R. A. Colvin, W. A. Holmes, C. P. Fontaine and W. Maret, Metallomics, 2010, 2, 306–317 RSC.
- C. W. MacDiarmid, L. A. Gaither and D. J. Eide, EMBO J., 2000, 19, 2845–2855 CrossRef CAS PubMed.
- C. W. MacDiarmid, M. A. Milanick and D. J. Eide, J. Biol. Chem., 2002, 277, 39187–39194 CrossRef CAS PubMed.
- C. W. MacDiarmid, M. A. Milanick and D. J. Eide, J. Biol. Chem., 2003, 278, 15065–15072 CrossRef CAS PubMed.
- L. Feretti, L. Elviri, M. A. Pelinghelli, G. Prediery and M. Tegoni, J. Inorg. Biochem., 2007, 101, 1442–1456 CrossRef PubMed.
- H. Bücking and W. Heyser, Mycol. Res., 1999, 103, 31–39 CrossRef.
- P. A. Olsson, E. C. Hammer, J. Pallon, I. M. Van Aarle and H. Wallander, Fungal Biol., 2011, 115, 643–648 CrossRef CAS PubMed.
- J. Ruytinx, H. Nguyen, M. Van Hees, M. Op De Beeck, J. Vangronsveld, R. Carleer, J. V. Colpaert and K. Adriaensen, Metallomics, 2013, 5, 1225–1233 RSC.
- C. Devirgiliis, C. Murgia, G. Danscher and G. Perozzi, Biochem. Biophys. Res. Commun., 2004, 323, 58–64 CrossRef CAS PubMed.
- G. P. M. Borrelly, M. D. Harrison, A. K. Robinson, S. G. Cox, N. J. Robinson and S. K. Whitehall, J. Biol. Chem., 2002, 277, 30394–30400 CrossRef CAS PubMed.
- S. Clemens, T. Bloss, C. Vess, D. Neumann, D. H. Nies and U. Zur Nieden, J. Biol. Chem., 2002, 277, 18215–18221 CrossRef CAS PubMed.
- D. Blaudez and M. Chalot, Fungal Genet. Biol., 2011, 48, 496–503 CrossRef CAS PubMed.
- A. Pagani, L. Villarreal, M. Capdevila and S. Atrian, Mol. Microbiol., 2007, 63, 256–269 CrossRef CAS PubMed.
- M. Capdevila and S. Atrian, J. Biol. Inorg. Chem., 2011, 16, 977–989 CrossRef CAS PubMed.
- C. A. Blindauer, J. Biol. Inorg. Chem., 2011, 16, 1011–1024 CrossRef CAS PubMed.
- R. Bofill, M. Capdevila and S. Atrian, Metallomics, 2009, 1, 229–234 RSC.
- R. Bofill, R. Orihuela, M. Romagosa, J. Domènech, S. Atrian and M. Capdevila, FEBS J., 2009, 276, 7040–7056 CrossRef CAS PubMed.
- Ò. Palacios, S. Atrian and M. Capdevila, J. Biol. Inorg. Chem., 2011, 16, 991–1009 CrossRef PubMed.
- Ò. Palacios, A. Pagani, S. Pérez-Rafael, M. Egg, M. Höcker, A. Brandstätter, M. Capdevila, S. Atrian and E. Dallinger, BMC Biol., 2011, 9, 4, DOI:10.1186/1741-7007-9-4.
- J. Loebus, B. Leitenmaier, D. Meissner, B. Braha, G.-J. Krauss, G. Dobritzsch and E. Freisinger, J. Inorg. Biochem., 2013, 127, 253–260 CrossRef CAS PubMed.
- L. Lafranco, New Phytol., 2007, 174, 3–6 CrossRef PubMed.
- M. Bellion, M. Courbot, F. Guinet, C. Jacob, D. Blaudez and M. Chalot, New Phytol., 2007, 174, 151–158 CrossRef CAS PubMed.
- G. Ramesh, G. K. Podila, G. Gay, R. Marmeisse and M. S. Reddy, Appl. Environ. Microbiol., 2009, 75, 2266–2274 CrossRef CAS PubMed.
- M. Osobová, V. Urban, P. L. Jedelský, J. Borovička, M. Gryndler, T. Ruml and P. Kotrba, New Phytol., 2011, 190, 916–926 CrossRef PubMed.
- J. Sácký, T. Leonhardt, J. Borovička, M. Gryndler, A. Briksí and P. Kotrba, Fungal Genet. Biol., 2014, 67, 3–14 CrossRef PubMed.
- M. S. Szczypka, J. A. Wemmie, W. S. Moye-Rowley and D. J. Thiele, J. Biol. Chem., 1994, 269, 22853–22858 CAS.
- K. T. Tamai, E. B. Gralla, L. M. Ellerby, J. S. Valentine and D. J. Thiele, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 8013–8017 CrossRef CAS.
- D. Mumberg, R. Müller and M. Funk, Gene, 1995, 156, 119–122 CrossRef CAS.
- J. Schug, in Current protocols in bioinformatics, ed. A. D. Baxevanis, J. Wiley and Sons, New York, 2003, unit 2.6 Search PubMed.
- V. Matys, O. V. Kel-Margoulis, E. Fricke, I. Liebich, S. Land, A. Barre-Dirrie, I. Reuter, D. Chekmenev, M. Krull, K. Hornischer, N. Voss, P. Stegmaier, B. Lewicki-Potapov, H. Saxel, A. E. Kel and E. Wingender, Nucleic Acids Res., 2006, 34, D108–D110 CrossRef CAS PubMed.
- J. A. Vizcaíno, E. W. Deutsch, R. Wang, A. Csordas, F. Reisinger, D. Ríos, J. A. Dianes, Z. Sun, T. Farrah, N. Bandeira, P. A. Binz, I. Xenarios, M. Eisenacher, G. Mayer, L. Gatto, A. Campos, R. J. Chalkley, H. J. Kraus, J. P. Albar, S. Martinez-Bartolomé, R. Apweiler, G. S. Omenn, L. Martens, A. R. Jones and H. Hermjakob, Nat. Biotechnol., 2014, 30, 223–226 CrossRef PubMed.
- C. A. Blindauer, J. Inorg. Biochem., 2013, 121, 145–155 CrossRef CAS PubMed.
- M. M. Harding, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2004, 60, 849–859 Search PubMed.
- C. Damon, F. Lehembre, C. Oger-Desfeux, P. Luis, J. Ranger, L. Frassinet-Tachet and R. Marmeisse, PLoS One, 2012, 7, e28967 CAS.
- A. R. Reddi and B. R. Gibney, Biochemistry, 2007, 46, 3745–3758 CrossRef CAS PubMed.
- C. A. Blindauer, M. R. Razi, D. J. Campopiano and P. J. Sadler, J. Biol. Inorg. Chem., 2007, 12, 393–405 CrossRef CAS PubMed.
- E. A. Peroza, A. Al Kaabi, W. Mayer-Klaucke, G. Waéllemreuther and E. Freisinger, J. Inorg. Biochem., 2009, 103, 342–353 CrossRef CAS PubMed.
- G. Bjornsdottir and L. C. Myers, Nucleic Acids Res., 2008, 36, 2906–2916 CrossRef CAS PubMed.
- X. Qi, Y. Zhang and T. Chai, Plant Physiol., 2007, 143, 50–59 CrossRef CAS PubMed.
- V. Günther, U. Lindert and W. Schaffer, Biochim. Biophys. Acta, 2011, 1823, 1416–1425 CrossRef PubMed.
- D. S. Conklin, J. A. McMaster, M. R. Culbertson and C. Kung, Mol. Cell. Biol., 1992, 12, 3678–3688 CAS.
- P. J. Lapinskas, K. W. Cunningham, X. F. Liu, G. R. Fink and V. C. Culotta, Mol. Cell. Biol., 1995, 15, 1382–1388 CAS.
- S. Puig and D. J. Thiele, Curr. Opin. Chem. Biol., 2002, 6, 171–180 CrossRef CAS.
- M. Tomas, J. Domènech, M. Capdevila, R. Bofill and S. Atrian, FEBS Open Bio, 2013, 3, 89–100 CrossRef CAS PubMed.
- Ò. Palacios, A. Espart, J. Espín, C. Ding, D. J. Thiele, S. Atrian and M. Capdevila, Metallomics, 2014, 6, 279–291 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00141a |
| This journal is © The Royal Society of Chemistry 2014 |