Open Access Article
Open Access ArticleIdentification of major zinc-binding proteins from a marine cyanobacterium: insight into metal uptake in oligotrophic environments†
James Paul
Barnett
a,
David John
Scanlan
b and
Claudia Andrea
Blindauer
*a
aDepartment of Chemistry, University of Warwick, Coventry, CV4 7AL, UK. E-mail: c.blindauer@warwick.ac.uk; Fax: +44 (0)24 76 524112; Tel: +44 (0)24 76 528264
bSchool of Life Sciences, University of Warwick, Coventry, CV4 7AL, UK
First published on 21st May 2014
Abstract
Marine cyanobacteria make a significant contribution to primary production whilst occupying some of the most nutrient poor regions of the world's oceans. The low bioavailability of trace metals can limit the growth of phytoplankton in ocean waters, but only scarce data are available on the requirements of marine microbes for zinc. Recent genome mining studies suggest that marine cyanobacteria have both uptake systems for zinc and proteins that utilize zinc as a cofactor. In this study, the oligotrophic strain Synechococcus sp. WH8102 was grown at different zinc concentrations. Using metalloproteomics approaches, we demonstrate that even though this organism's growth was not affected by extremely low zinc levels, cells accumulated significant quantities of zinc, which was shown to be protein-associated by 2D liquid chromatography and ICP-MS. This indicates that the mechanisms for zinc uptake in Synechococcus sp. WH8102 are extremely efficient. Significantly, expression of SYNW2224, a putative porin, was up-regulated during growth in zinc-depleted conditions. Furthermore, along with 30 other proteins, SYNW2224 was captured by immobilised zinc affinity chromatography, indicating the presence of surface-exposed site(s) with metal-binding capacity. It is proposed that this porin plays a role in high-affinity zinc uptake in this and other cyanobacteria.
Introduction
Zinc is an important micronutrient required by almost all living organisms, functioning as a catalytic cofactor in enzymes and as a structural component of numerous proteins.1 Recent bioinformatic studies suggest that up to 15% of all proteins in a given genome contain Zn2+.2,3 Although prokaryotes have lower requirements for zinc than eukaryotes, zinc is thought to be essential for bacteria, too, and bacterial proteomes are predicted to include 5–6% zinc-binding proteins.2 However, these bioinformatic predictions are ultimately based on limited experimental biochemical and bio-inorganic data, and it has been concluded that “microbial metalloproteomes are largely uncharacterised”.4 The genomes of many organisms encode large numbers of uncharacterised conserved hypothetical proteins, some of which may contain novel metal binding sites.5,6 Even in cases where homologous proteins are known and permit structural modelling, it is often not possible to reliably predict the metal specificity of a particular protein. A further complication is introduced by “cambialistic” proteins,7 which may utilise different metal cofactors in different species or circumstances. These problems may be particularly pronounced in the case of organisms that live in extreme environments. One such extreme environment is the open ocean, one of the most nutrient-poor habitats on earth.Whilst there is ample evidence that the bioavailability of iron can limit phytoplankton growth8 and primary production in some open ocean regions,9 the impact of zinc is less clear.10 Only picomolar concentrations of free Zn2+ occur in surface waters of the world's oceans,11–13 with the vast majority of Zn2+ found complexed to organic ligands of unknown origin and identity.12,14,15 These free Zn2+ concentrations may be sufficiently low to directly limit the growth of some phytoplankton including diatoms, coccolithophores, and green algae.13,16–19 Based on the observation of zinc/carbon co-limitation in certain eukaryotic marine phytoplankton,20 a “zinc hypothesis” has been proposed, in analogy to Martin's “iron hypothesis”.21 This theory entails that on a global scale, high zinc levels in the oceans lead to increased CO2 sequestration – in turn, scarcity of zinc increases atmospheric CO2 levels.
Marine cyanobacteria are the predominant photosynthetic organisms in large parts of the oligotrophic regions of the world's oceans.22 Unfortunately, the specific requirements of marine cyanobacteria for zinc are scarcely studied,23–25 whereas for some freshwater cyanobacteria effects of zinc toxicity and deprivation have been examined in some detail.26,27 The genomes of over 30 marine cyanobacterial strains have been sequenced, and our genome mining work has discovered not only genes for several enzymes predicted to require a zinc cofactor, including alkaline phosphatase and one or more carbonic anhydrases, but also for proteins with likely roles in zinc uptake and trafficking, including putative periplasmic zinc binding proteins (ZnuA).28–30 All strains inspected also harboured a gene for the zinc uptake regulator Zur, with recognition sites for this zinc sensor protein predicted to occur in the upstream regions of several relevant genes. Whilst these bioinformatic studies strongly point towards dedicated networks for zinc uptake and utilisation, an absolute requirement of marine cyanobacteria for zinc has not been demonstrated conclusively.
In the present study we have used the oligotrophic open-ocean strain Synechococcus sp. WH810231 as a model cyanobacterium to study its growth and cellular zinc quota under zinc-depletion conditions, and to probe the presence of major Zn2+-binding proteins using solution-state metalloproteomics approaches. Metalloproteomics, a sub-discipline of metallomics, is dedicated to the provision of experimental evidence for metal–protein interactions.32–36 Combinations of inorganic and molecular mass-spectrometry are particularly powerful approaches, whilst separation techniques have posed the major experimental challenges.5
Previously, we examined the applicability of different liquid-chromatography-based approaches to probe the Fe-, Ni-, and Co-related proteome of Synechococcus sp. WH8102.5,37 In the present study, immobilised zinc affinity chromatography (Zn2+-IMAC) has enabled the capture and detection of several proteins with potential functions in zinc metabolism. Besides the detection of a predicted periplasmic zinc-binding protein (ZnuA) along with several other periplasmic binding proteins, the most significant finding concerns a predicted porin, a novel candidate for mediating zinc uptake across the outer membrane of marine cyanobacteria.
Materials and methods
Suppliers of chemicals and reagents
All chemicals used were of the highest grade available and were purchased from either Fisher Scientific (UK) or Sigma-Aldrich (UK) unless otherwise indicated.Bacterial strains and growth conditions
Axenic cultures of Synechococcus sp. WH8102 were grown in an artificial seawater medium based on Aquil,38,39 prepared using ultrapure (MilliQ 18 MΩ cm−1) water (Table S1, ESI†). The base medium (macronutrients) was Chelex-treated and then autoclaved. Micronutrient components were filter-sterilised before adding to the base medium. Media bottles and culture flasks were washed with 5% trace metal clean HCl prepared in house by sub-boiling point distillation and rinsed with MilliQ water before use.The theoretical free Zn2+ concentration in the growth medium at pH 8.0 was calculated from the total EDTA and Zn2+ concentrations using the “Species” module within the IUPAC stability constants database (Data version 4.56, L.D. Pettit, Academic Software, UK), taking also into account the concentrations of other metal ions. Stability constants and pKa values were also extracted from this database.
Stocks of Synechococcus sp. WH8102 were maintained through sub-culturing in the different Zn media that were tested, over a long period of time. Cells were initially transferred from standard medium to zinc-depleted medium by collecting cells by centrifugation, gently washing and resuspending into the zinc-depleted medium. Before the growth measurements were carried out, cells were acclimated over several serial transfers across at least 12 weeks into fresh medium with either no zinc or 80 nM added zinc. Cultures were grown at 25 °C with continuous illumination at 10 μE m−2 s−1 and shaken at 150 rpm. This light level corresponds to levels found at the bottom of the surface mixed layer, a region where clade III Synechococcus genotypes (of which Synechococcus sp. WH8102 is a member) proliferate. Such a light level also assists in stable culture maintenance, without inducing photodamage. Growth was monitored by measuring the optical density at 750 nm or by flow cytometry using a FACScan flow cytometer (Becton Dickinson, NJ, USA). Cultures were checked at regular intervals for contamination with other microorganisms by plating onto Aquil-Agar plates containing 500 mg l−1 yeast extract. Cells were harvested by centrifugation at 6000 × g. All growth and subsequent separation experiments were carried out in duplicate.
Preparation of whole cell lysates
Cell pellets were re-suspended in 1–10 ml of 10 mM HEPES pH 7.2, 0.5% (w/v) octyl β-D-glucopyranoside, with a dissolved Complete™ EDTA-free protease inhibitor cocktail tablet (Roche, UK). Cells were broken by sonication and debris was removed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. The BCA method40 was used to determine the total protein content of cell lysates.
000 × g. The BCA method40 was used to determine the total protein content of cell lysates.
Preparation of soluble and insoluble cell fractions
Cell pellets were re-suspended in 8 ml of 20 mM HEPES pH 7.2 containing a dissolved Complete™ EDTA-free protease inhibitor cocktail tablet (Roche, UK), and broken by sonication. Unbroken cells were removed by centrifugation at 6000 × g. The soluble and insoluble fractions were separated by ultracentrifugation at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 30 minutes using a Beckman TLA100.3 rotor. The supernatant (soluble fraction) was collected, and the pellet (insoluble fraction) was re-suspended in 1 ml of 20 mM HEPES pH 7.2, 0.5% (w/v) octyl β-D-glucopyranoside to solubilise membrane proteins. The prepared fractions were further clarified by filtration using 0.2 μm pore-sized filters.
000 × g for 30 minutes using a Beckman TLA100.3 rotor. The supernatant (soluble fraction) was collected, and the pellet (insoluble fraction) was re-suspended in 1 ml of 20 mM HEPES pH 7.2, 0.5% (w/v) octyl β-D-glucopyranoside to solubilise membrane proteins. The prepared fractions were further clarified by filtration using 0.2 μm pore-sized filters.
Preparation of a carboxysome enriched fraction
Carboxysomes were prepared according to the method described by Gonzales et al.,41 with some minor modifications. Briefly, cell pellets were resuspended in 3 ml of 20 mM HEPES pH 7.2 containing a Complete™ EDTA-free protease inhibitor cocktail tablet. Cells were disrupted by sonication and cell debris was removed by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g for 10 minutes. The collected supernatant was centrifuged at 40
000 × g for 10 minutes. The collected supernatant was centrifuged at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g at 4 °C for 30 minutes using a Beckman TLA100.3 rotor. The dark green pellet was resuspended in 3 ml of 20 mM HEPES pH 7.2, 20 mM MgSO4, 2% (v/v) Triton X-100 and incubated on ice for 45 minutes with occasional agitation. This step solubilises membrane lipids, whilst promoting carboxysome aggregation. The sample was centrifuged again at 40
000 × g at 4 °C for 30 minutes using a Beckman TLA100.3 rotor. The dark green pellet was resuspended in 3 ml of 20 mM HEPES pH 7.2, 20 mM MgSO4, 2% (v/v) Triton X-100 and incubated on ice for 45 minutes with occasional agitation. This step solubilises membrane lipids, whilst promoting carboxysome aggregation. The sample was centrifuged again at 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g to generate a yellow-brown carboxysome enriched pellet that was resuspended in 20 mM HEPES pH 7.2.
000 × g to generate a yellow-brown carboxysome enriched pellet that was resuspended in 20 mM HEPES pH 7.2.
Inductively coupled plasma mass spectrometry (ICP-MS)
3% (v/v) trace metal grade nitric acid purified in house by sub-boiling point distillation was used as the sample matrix. For quantitation, calibration was performed in the range 0–500 ppb using external Zn and P ICP-MS standards. Er (Agilent Technologies, USA) was used as an internal standard. Measurements were taken using an Agilent 7500 series ICP mass spectrometer (Agilent Technologies, USA), equipped with a cross flow nebulizer, quartz spray chamber, and an Octopole Reaction System (ORS®) cell. All samples were measured in triplicate in helium gas-mode to remove matrix interferences. Cell lysates were prepared as described below from the combined cell pellets of 3 replicate cultures, and were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in matrix for analysis.
20 in matrix for analysis.
Two-dimensional liquid chromatography (2D-LC)
Two-dimensional liquid chromatography (2D-LC) was performed essentially as described previously.37 5 mg total protein from the soluble, insoluble, or carboxysome enriched cell fraction was applied to a Biosep S2000 HPLC gel filtration column (Phenomenex, UK) equilibrated in 10 mM HEPES pH 7.2 (containing 0.01% (w/v) octyl β-D-glucopyranoside for the insoluble fraction). Protein was eluted isocratically in the same buffer using a flow rate of 1 ml min−1 and collected in 1.0 ml fractions. The column was calibrated using 1 mg ml−1 solutions of bovine serum albumin – 66 kDa, Carbonic anhydrase – 29 kDa, lysozyme – 14 kDa, and substance P – 1 kDa standards in column buffer. Dextran blue dye (GE Healthcare, UK) was used to determine the void volume (V0), which was 5.1 ml. Protein containing fractions (5–13 ml) from 4 separate column runs (except for the carboxysome enriched cell fraction where just 1 column run was performed) were combined and further separated using strong anion exchange mini-spin columns (Thermo Scientific, UK). The spin columns were equilibrated using 2 mM HEPES pH 7.2 (plus 0.01% (w/v) octyl β-D-glucopyranoside for the insoluble fraction). Protein was eluted using a NaCl gradient of 0–2 M in 80 μl fractions. 50 μl of these samples were used for ICP-MS analysis and 30 μl were retained for analysis by SDS-PAGE and peptide mass fingerprinting. All stock solutions were prepared using 18 MΩ water and treated with Chelex-100 resin (Bio-Rad, UK) to remove traces of contaminating metal ions. Acid-washed plastic-ware was used throughout to prevent contamination of samples with external sources of zinc or other metals. For the 2D-LC experiments for soluble and insoluble fractions, two biological replicates were carried out, with reproducible results.Immobilized metal affinity chromatography (IMAC)
1 ml IMAC columns (GE Healthcare, UK) were prepared according to the manufacturer's instructions and either charged with Zn2+ or left un-charged. Columns were equilibrated with 10 mM HEPES pH 7.2, 0.5 M NaCl, 0.01% (w/v) octyl β-D-glucopyranoside. 20 mg of protein from crude cell lysates were loaded onto each column and the flow-through (FT) fraction collected. Unbound protein was washed through the column using 4 × 1 ml of buffer containing 2 mM imidazole (W1–W4), before bound proteins were eluted using 2 × 1 ml of buffer with 20 mM imidazole (E1–E2) and 2 × 1 ml of buffer containing 200 mM imidazole (E3–E4).SDS-polyacrylamide gel electrophoresis
SDS-PAGE was performed using mini-Protean® TGX™ precast 4–15% gels (Bio-Rad, UK) using standard protocols.42 Samples were mixed with an equal volume of gel loading buffer and heated to 80 °C for 5 minutes. 50 μl of sample was loaded per lane except where otherwise stated. Gels were stained using Coomassie brilliant blue R-250 (National Diagnostics, USA).Peptide mass fingerprinting
Protein bands were excised from SDS-PAGE gels using a scalpel blade and subjected to in-gel tryptic digestion using a commercially produced kit (Pierce, Thermo Scientific, UK). Peptide masses were determined by MALDI-TOF MS analysis. 2 μl of sample matrix (10 mg ml−1 α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, 0.1% trifluoroacetic acid) was mixed with 2 μl of sample and spotted onto a steel MALDI-target plate. Peptide masses were determined using a Bruker Ultraflex II MALDI-TOF/TOF mass spectrometer (Bruker Daltonics, Coventry, UK) with a 337 nm laser and operated in reflectron mode. Mass calibration was performed using PEG2000 and verified using bradykinin and substance P peptide standards. Internal mass accuracy was confirmed by the presence of the autolytic trypsin peaks at 845.2 and 2211.1 Da. Mass spectra were acquired over the range of 800–3500 Da. Mass lists were generated using Bruker Flex-analysis software with default parameters, and searched against either the NCBI or SwissProt databases using Mascot (Matrix Science, UK). The following search criteria were selected: Fixed modification of carbamidomethyl on cysteine, variable modification of oxidation of methionine, maximum of 1 missed cleavage, <50 ppm mass accuracy, “other bacteria” was selected for taxonomy. Only searches giving significant MOWSE scores were recorded.Structural models
Comparative modelling was employed to provide insight into the likely structures of the ZnuA, FutA, CynA, and cyanobacterial porin (CBP) proteins. Suitable templates were identified using the remote homology recognition server Phyre2,43 which also produces multiple and pairwise sequence alignments (see ESI†), and in favourable cases sound protein models. All initial models were generated by Phyre2. The model for ZnuA was based on pdb 2OV3 (ZnuA from Synechocystis sp. PCC6803; mutant devoid of His-rich loop),44 that for CynA on pdb 3UN6 (Ligand Binding Component of ABC-type Import System from Staphylococcus aureus),45 and that for FutA was based on pdb 2PT1 (FutA from Synechocystis sp. PCC6803).46 The ZnuA and CynA models were further developed using the molecular modelling program MOE2011.10. Hydrogen atoms, zinc ions, and a cyanate ion in the case of CynA were introduced, and the metal sites were then energy-minimised, using an in-house customised version of the AMBER99 force-field, in which Zn-specific parameters were incorporated. After energy minimisation of the position of the immediate zinc ligands, the adjacent environment was optimised, and finally, the entire molecule was subjected to energy minimisation. All minimisations were terminated based on the steepness of the RMS gradient (<0.5). Models were submitted to the WHATIF web interface (http://swift.cmbi.ru.nl/servers/html/index.html), ensuring that a physically reasonable model had been produced.The 3D model for the CBP SYNW2224 was generated using the “Intensive mode” on the Phyre2 server, and is based on three overlapping templates: residues 46–98 are based on pdb 3PYW (SLH domain from Bacillus anthracis),47 residues 92–141 on pdb 3SWF (a helix from a rod cyclic nucleotide-gated ion channel),48 and residues 134–501 on pdb 4GF4 (Pseudomonas putida OprB, a porin for carbohydrate uptake).49 Structural images were generated in MOLMOL v.2k.1.50
Results
Synechococcus sp. WH8102 grows well in zinc-depleted media
To investigate if depletion of zinc may affect the growth of Synechococcus sp. WH8102, cultures were studied in an artificial seawater medium based on Aquil38,39 that was supplemented with either 0 (zinc-depleted medium) or 80 nM (designated here as “replete” medium, although it should be noted that the resulting free zinc concentration is still very low) zinc. The presence of 100 μM EDTA leads to a theoretical free Zn2+ concentration of 16 pM in the replete medium, with 98.98% of the added Zn2+ complexed. Free Zn2+ concentrations in open ocean surface waters vary from 1 to 71 pM.11–13,16 Hence, the free Zn2+ concentration in our replete medium is not dissimilar to those that Synechococcus sp. WH8102 might encounter in its natural habitat. The free ion concentrations are considered the most relevant factor governing uptake.51,52The growth data in Fig. 1(a) show that the cultures grown with 0 or 80 nM zinc exhibited no significant difference in growth rate or final cell yield. This suggests that Synechococcus sp. WH8102 either has no absolute requirement for zinc, or that it is able to scavenge trace amounts of zinc from the depleted growth medium, which are unavoidably introduced as a contaminant with other media components.
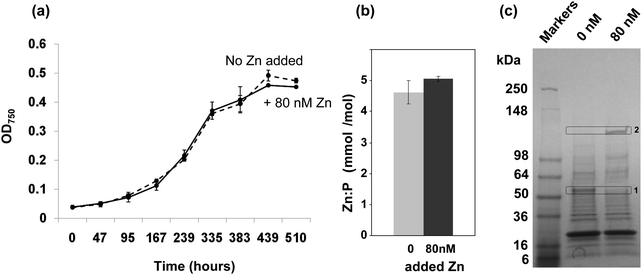 | ||
Fig. 1 Effect of zinc on the growth of Synechococcus sp. WH8102 and subcellular distribution of zinc. (a) Growth of Synechococcus sp. WH8102 in Aquil medium containing either 0 nM (dashed line), or 80 nM (solid line) added zinc. (b) Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) phosphorus ratios in whole cells. (c) CBB-stained SDS-PAGE gel comparing the protein profiles of crude cell lysates prepared from Synechococcus sp. WH8102 cells grown in zinc depleted or replete media. 32 μg of protein was loaded per lane. Proteins showing a marked difference in expression are boxed. Box 1 corresponds to a putative CBP (SYNW2224; identified with 14 matched peptides, 33% sequence coverage and a MOWSE score of 78). Box 2 yielded the large subunit of RuBisCO as possible hit (6 matched peptides, 14% sequence coverage, MOWSE score 68). The large deviation between the observed and expected molecular weight could be due to persistence of the homo-dimer or a larger complex.115 phosphorus ratios in whole cells. (c) CBB-stained SDS-PAGE gel comparing the protein profiles of crude cell lysates prepared from Synechococcus sp. WH8102 cells grown in zinc depleted or replete media. 32 μg of protein was loaded per lane. Proteins showing a marked difference in expression are boxed. Box 1 corresponds to a putative CBP (SYNW2224; identified with 14 matched peptides, 33% sequence coverage and a MOWSE score of 78). Box 2 yielded the large subunit of RuBisCO as possible hit (6 matched peptides, 14% sequence coverage, MOWSE score 68). The large deviation between the observed and expected molecular weight could be due to persistence of the homo-dimer or a larger complex.115 | ||
Efficient zinc uptake at extremely low zinc concentrations
In a subsequent experiment, following 19 days growth in either zinc depleted or replete medium, cells were harvested and the zinc and phosphorus content of crude cell lysates was measured by Inductively-Coupled-Plasma-Mass-Spectrometry (ICP-MS). The phosphorus levels were used as a proxy for biomass. The cells grown under zinc-depleted conditions had accumulated significant quantities of zinc, and no significant difference in the Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P ratio was recorded between the two conditions tested (Fig. 1(b)).
P ratio was recorded between the two conditions tested (Fig. 1(b)).
The fact that significant amounts of zinc were captured by Synechococcus sp. WH8102 even at extremely low concentrations points to the existence of highly efficient uptake mechanisms. Significantly, analysis of 1-dimensional SDS-PAGE gels of the complete proteomes from cells grown under the two regimes revealed that the amount of an outer-membrane protein (SYNW2224) was markedly increased under zinc-depleted conditions suggesting a potential role in zinc acquisition for this protein (Fig. 1(c)). A second protein band was visibly diminished under zinc-depletion conditions; this was tentatively identified as the large subunit of RuBisCO, although it is not clear why the observed molecular weight was considerably larger than expected, or why differences in zinc levels should affect the abundance of this protein.
Fractionation of the zinc proteome by 2-dimensional liquid chromatography
In an attempt to isolate intact Zn-bound proteins from Synechococcus sp. WH8102, the soluble and insoluble proteomes, as well as carboxysomal fractions, were further fractionated using native 2-dimensional liquid chromatography (2D-LC). This approach was selected as it allows proteins to be separated rapidly and under mild conditions that promote the likelihood for keeping proteins folded and intact including moderately strongly bound metal cofactors. It should therefore in principle be suitable for the separation of Zn-containing proteins that form relatively stable complexes.53 However, a drawback is the resolution of separation achievable by 2D-LC, which is considerably lower than that obtained using traditional denaturing 2D-gel electrophoresis methods.5In the present study, proteins were initially separated using size exclusion chromatography (SEC), followed by anion exchange chromatography (AEX). The “insoluble” fraction was solubilised and separated in the presence of the mild detergent octyl glucoside, which has been used extensively for the isolation of native membrane proteins.54 In each case, 5 mg total protein was applied to a BioSep S2000 HPLC gel filtration column and protein was eluted in 13 × 1 ml fractions. The elution fractions from four separate SEC runs were combined and applied to mini-spin anion exchange columns. These columns allowed the rapid and simultaneous separation and concentration of the gel-filtration fractions. Bound proteins were eluted from the anion exchange columns in 80 μl fractions in a step-wise fashion using a NaCl gradient of 0–2 M. Each of the collected fractions was then analysed by ICP-MS for zinc content (Fig. 2). In the soluble fraction, zinc eluted from the gel filtration column as a single broad peak between ca. 14 kDa and 70 kDa. In the insoluble fraction, two distinct peaks were observed, one at ca. 30–70 kDa and a second at around 1 kDa, which contains peptides, other small molecules, and possibly also free metal ions. The proteins present in fractions with the highest zinc concentrations were analysed by SDS-PAGE (Fig. 2). For the soluble fraction, the anion exchange samples obtained from separation of the 9 ml gel filtration fraction were analysed. Despite the concentration step, very few proteins were observed on the gel, but one protein migrating to ca. 18 kDa on the gel could be observed even before staining by its light pink colour. This protein was identified by peptide mass fingerprinting and found to be a subunit of c-phycoerythrin, a component of the light harvesting phycobilisome complex. After the gel was stained with Coomassie, only one further protein (at ca. 10 kDa) could be visualised. This protein was identified as ribosome recycling factor, and neither of these two proteins is predicted to bind metal ions. However, like other biliproteins, c-phycoerythrin contains linear tetrapyrrole chromophores (phycobilins) with considerable metal-binding ability. It appears likely that cellular zinc has partially been redistributed to the chromophores on these proteins. In support of this hypothesis, several biliproteins were also captured on Zn2+-IMAC columns (see below). The redistribution of zinc in our samples might explain why no predicted zinc binding proteins including alkaline phosphatase or carbonic anhydrase were detected in the soluble fractions following 2D-LC. Similar results were obtained from the analysis of the insoluble cell fraction, with c-phycoerythrin again the most abundant protein present in the zinc enriched cell fractions (Fig. 2). The only other protein detected in the peak zinc fractions was PstS, a periplasmic phosphate binding protein. Whilst this protein is not predicted to bind zinc or any other metals in vivo, it was captured using a Zn2+-IMAC column (see below), suggesting that it has an affinity for zinc in vitro. Full details of the proteins identified are given in Table S2 (ESI†). In the carboxysome fraction, zinc was only detected in lower molecular weight sub-fractions (20 kDa and below), but the concentrations of proteins were too low for identification by peptide mass fingerprinting.
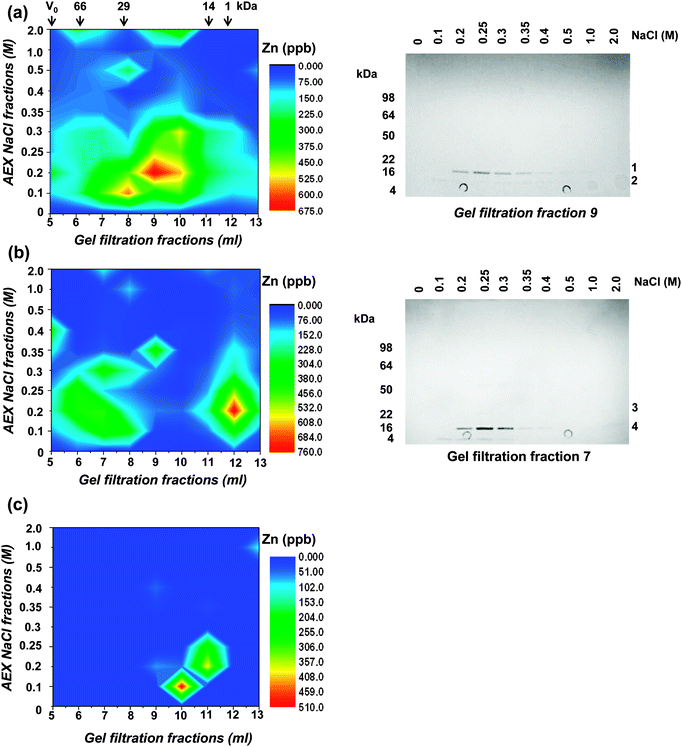 | ||
| Fig. 2 Two-dimensional liquid chromatography separation of the zinc proteome of Synechococcus sp. WH8102. Proteins present in the (a) soluble, (b) insoluble, and (c) carboxysome cell fractions were separated by gel filtration and anion exchange chromatography. The zinc contents of collected fractions were measured by ICP-MS and the data plotted. The elution of molecular weight markers (BSA – 66 kDa, carbonic anhydrase – 29 kDa, lysozyme – 14 kDa, and substance P – 1 kDa) from the gel filtration column is indicated with arrows. Right hand panels are CBB-stained SDS-PAGE gels showing proteins present in peak zinc containing fractions following 2D-LC separation. The motilities of molecular weight markers are indicated on the left of the gels and the proteins identified (Table S2, ESI†) are numbered on the right. Bands 1 and 4 are MpeA, a component of c-phycoerythrin, band 2 is ribosome recycling factor, and band 3 (very faint) is PstS. None of these proteins are predicted to bind metal ions. | ||
In conclusion, despite the rapid and mild separation conditions used for the chromatography and the relatively high stability of Zn2+ complexes according to the Irving–Williams series,53 no major zinc-binding proteins in any of the sub-cellular fractions could be identified by the 2D-LC approach. Major drawbacks were the low resolution of the separation steps and insufficient sensitivity in protein detection, both of which are exacerbated by the presence of highly abundant biliproteins. These pose a significant challenge for native metalloproteomics in this organism and likely other related cyanobacteria. In terms of classical mass-spectrometry-based proteomics, biliproteins cause dynamic range problems, since they can account for as much as 60% of total cellular protein; hence, they are inherently likely to impede the detection of low abundance proteins. In terms of native metalloproteomics, their demonstrated metal-binding ability (also see below) causes additional problems, as this may lead to metal redistribution in cell lysates – a problem unlikely to be solvable by depletion strategies.
Enrichment of zinc-binding proteins by immobilized metal ion affinity chromatography
Metal-trafficking proteins often bind their cargo in a kinetically labile fashion and close to the protein surface, to enable facile transfer to and from other molecules. This is an ideal prerequisite to capture such proteins by immobilized metal ion affinity chromatography (IMAC). Similar approaches have been used in previous studies to isolate proteins that have an affinity for the immobilised metal ion in vitro, e.g. proteins with copper affinity from human liver cells,55Arabidopsis thaliana,56 and microalgae,57 and metal-binding proteins from plant mitochondria.58Eluates from a Zn2+-IMAC column were analysed by 1D SDS-PAGE (Fig. 3), and proteins were identified by peptide mass fingerprinting. A total of 30 different proteins with zinc-binding ability were identified (Table 1), including several enzymes, biliproteins, carboxysomal shell proteins, periplasmic binding proteins, and two outer membrane proteins. Based on biochemical data from homologous proteins, some of these proteins are predicted to bind metal ions; others, including biliproteins and carboxysomal shell proteins, are not. Identified proteins are described and discussed below.
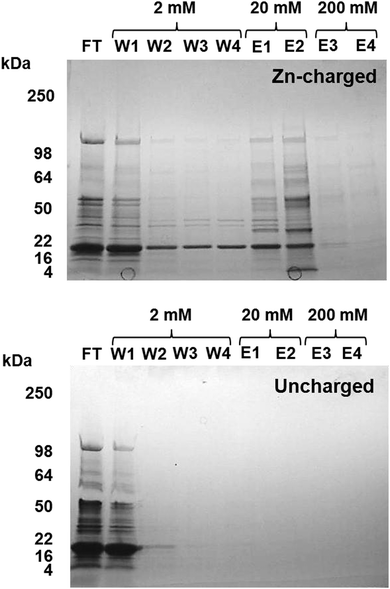 | ||
| Fig. 3 Isolation of zinc binding proteins by Zn2+-IMAC. CBB-stained gel of Zn-IMAC column fractions. 20 mg of crude cell lysate was applied to a 1 ml Zn-IMAC column and the flow-through (FT) collected. Unbound protein was washed through the column using buffer containing 2 mM imidazole (W1–W4). Strongly bound protein was eluted using buffer containing 20–200 mM imidazole (E1–E4). Proteins present in the elution fractions were identified by peptide mass fingerprinting (Table 1). | ||
| Mass (kDa) | Gene (cluster)a | Protein name | No. peptides matched | % Sequence coverage | MOWSE score |
|---|---|---|---|---|---|
| a This is the protein cluster number in the Cyanorak database which is publically accessible at http://www.sb-roscoff.fr/Phyto/cyanorak/. | |||||
| Enzymes | |||||
| 9.4 | synw2310 445 | Glutaredoxin | 7 | 67 | 70 |
| 25.9 | rbrA 1682 | Rubrerythrin | 15 | 55 | 153 |
| 34.3 | hemC 643 | Porphobilinogen deaminase | 12 | 43 | 80 |
| 38.5 | cbbA 976 | Fructose bisphosphate aldolase | 7 | 25 | 71 |
| 47.1 | cysD 1284 | O-Acetyl homoserine sulfhydrylase | 12 | 42 | 67 |
| 51.3 | lpdA 102 | Dihydrolipoyl dehydrogenase | 9 | 30 | 78 |
| 52.0 | atpB 1107 | ATP synthase subunit beta | 7 | 21 | 90 |
| 52.8 | ccbL 681 | Ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit | 11 | 49 | 87 |
| 52.5 | glnA 103 | Glutamine synthetase, glutamate ammonia ligase | 9 | 23 | 100 |
| 58.9 | pgm 1145 | Phosphoglucomutase | 13 | 39 | 76 |
| 63 | synw2391 5416 | Alkaline phosphatase | 9 | 24 | 77 |
| 60.3 | pgi 827 | Glucose-6-phosphate isomerase | 12 | 29 | 95 |
| 72.2 | tktA 291 | Transketolase | 25 | 42 | 165 |
| 95 | glgP 118 | Phosphorylase | 11 | 18 | 76 |
| 148.4 | rpoC2 1059 | DNA directed RNA polymerase subunit beta' | 32 | 31 | 190 |
| Components of phycobilisomes | |||||
| 18 | mpeB 8005 | C-phycoerythrin class 2 subunit beta | 9 | 54 | 75 |
| 17.7 | mpeA 7994 | C-phycoerythrin class 2 subunit alpha | 12 | 71 | 112 |
| 32 | mpeC 8012 | C-phycoerythrin class II gamma chain, linker polypeptide | 21 | 74 | 218 |
| 59.4 | mpeD 8022 | Phycobilisome linker polypeptide | 24 | 43 | 159 |
| Components of carboxysomes | |||||
| 10.6 | ccmK1 8056 | CcmK1 | 16 | 99 | 169 |
| 18.3 | ccmK 237 | CcmK2 | 9 | 62 | 124 |
| Transcription and translation factors | |||||
| 27.7 | rpaB 8013 | Two component response regulator | 11 | 48 | 75 |
| 43.6 | tuf 494 | Elongation factor Tu | 22 | 64 | 140 |
| 75.1 | fusA 495 | Elongation factor G | 28 | 46 | 160 |
| Periplasmic binding proteins | |||||
| 33.2 | znuA 2462 (synw2481) | Zn ABC transporter, substrate binding protein | 6 | 25 | 79 |
| 33.8 | pstS 23 (synw1018) | ABC transporter, substrate binding protein, phosphate | 11 | 33 | 142 |
| 37.7 | futA 68 | Fe ABC transporter, substrate binding protein | 17 | 60 | 154 |
| 60.5 | synw2487 2165 | Cyanate ABC transporter, substrate binding protein | 8 | 35 | 85 |
| Porins | |||||
| 53.8 | synw2224 8 | Porin | 18 | 43 | 135 |
| 51.2 | synw2227 8 | Porin | 7 | 24 | 79 |
The enzymes transketolase (TktA) and dihydrolipoyl dehydrogenase (LpdA) from E. coli were previously found to bind 65Zn after separation of the proteome by denaturing 2D gel electrophoresis.61 More recently, in vivo zinc binding was demonstrated for both enzymes by in vivo labelling with 65Zn and subsequent separation by native 2D gel electrophoresis.62 TktA from E. coli (pdb 2R8O)63 contains at least six His residues in its substrate-binding pocket, which are fully conserved in the homologue from Synechococcus sp. WH8102. These His residues are involved in binding the diphosphate moiety, the sugar hydroxyls, and the phosphate of the D-fructose-6 phosphate thiamine diphosphate adduct. Thus, although they might contribute to the observed affinity for zinc, an in vivo zinc-binding role is unlikely. LpdA (pdb 4JDR for the enzyme from E. coli)64 abounds with surface-exposed metal-binding residues. The CHED server for automatic metal site recognition, which takes into account the main metal-binding residues (Cys, His, Glu, Asp; http://ligin1.weizmann.ac.il/%E2%88%BClpgerzon/mbs4/mbs.cgi)65 detected no less than seven possible metal sites, even though the published structure is devoid of any metal ions. With the exception of one site involving two Asp and one Glu residue, none of these sites are conserved in the enzyme from Synechococcus sp. WH8102. Whether the latter site is of significance regarding enzymatic activity is unknown.
In addition, several further enzymes were also captured on the IMAC column. Using structural models, CHED analysis, and manual inspection, possible reasons for this are explored below. Glutaredoxin (modelled on pdb 3QMX, from Synechocystis66) contains three Cys residues, and one of them is flanked by a His and an Asp residue, both surface-accessible, hence in principle suitable for IMAC capture. Rubrerythrin from Desulfovibrio vulgaris (pdb 1DVB67) contains binding sites for three iron ions, one Cys4 site, and a binuclear site, which are conserved in RbrA from Synechococcus sp. WH8102. As a relatively weakly-binding metal ion, Fe2+ is prone to be lost during protein separation. Thus, in principle, sites in Fe-proteins may become available for binding to immobilised metal. The most surface-accessible residues are two Cys residues, whereas the His and Glu residues in the binuclear site are buried in the folded protein, and are not clustered in the primary sequence, so relatively unlikely to be responsible for IMAC binding.
SYNW2391 is annotated as a putative alkaline phosphatase, is structurally related to 7-bladed β-propeller oxidoreductases, and contains an abundance of potential metal sites (CHED detected seven sites in total). Similarly, at least five metal-binding sites can be identified in glutamine synthetase (GlnA). The protein is 55% identical to GlnA from Salmonella typhimurium (pdb 1FPY68). One large, surface-exposed site involving several Glu and His residues is the ATP-binding site, which also requires two M2+ ions for binding and ATP hydrolysis. Essentially similar considerations hold for the beta' subunit of RNA polymerase RpoC2 which also harbours a Mg-requiring ATP-binding site. In addition, a structural ZnCys4 site is present, but this is deeply buried (RpoC2 was modelled on pdb 4G7O, RNA polymerase from Thermus thermophilus69). Similarly, the large subunit of RuBisCO harbours three potential metal sites, one of them coinciding with the binding site for ribulose-1,5-bisphosphate, which also requires a Mg2+ ion.70 It is hence conceivable that similar ternary complexes can also be formed with NTA-immobilised Zn2+ (or indeed other immobilised metal ions). O-Acetyl-homoserine sulfhydrylase also displayed three potential metal sites, one of them containing a Cys residue, but not related to enzymatic activity. Phosphoglucomutase, glucose-6-phosphate isomerase, and the oligosaccharide phosphorylase GlgP are all part of sugar metabolism, and are enzymes that work in sequence. GlgP is required for the breakdown of oligosaccharides, resulting in glucose-1-phosphate. This is converted to glucose-6-phosphate by phosphoglucomutase, and this is converted to fructose-6-phosphate by the isomerase. Several oligosaccharide phosphorylases are reported to be activated, stimulated, or inhibited by various metal ions (http://www.brenda-enzymes.info/php/ result_flat.php4?ecno=2.4.1.1), suggesting the presence of metal binding sites, and indeed, six potential sites were detected by CHED. Most phosphoglucomutases require Mg2+ for activity, but show limited activity with various other metal ions. The Synechococcus sp. WH8102 phosphoglucomutase shares 51% identity with that from the ciliate Paramecium tetraaurelia (pdb 1KFI71), which has been crystallised with Zn2+ bound. One of the sites identified by CHED coincides with this Zn-binding site, but it is deeply buried and hence unlikely to explain the IMAC interaction. Although archaeal glucose-6-phosphate isomerases have been isolated with zinc and iron bound,72,73 they are structurally not related to the corresponding bacterial and eukaryotic enzymes, for which no metal requirements or inhibition are reported. CHED identified nevertheless four potential binding sites, one of them comprising five amino acid side-chains.
The two-component response regulator RpaB is a two-domain protein, and two potential metal-binding sites in its N-terminal domain were detected by CHED. It shares 42% identity with the RegX3 regulator from Mycobacterium tuberculosis (pdb 2OQR74), and the crystal structure of the latter has been stabilised by La3+ ions, although metals are not thought to play a role in the activity of this protein. The metal-binding residues in RpaB and RegX3 are not identical, but the location of the exposed N-terminal sites is roughly similar. Elongation factor Tu displays an abundance of clustered, surface-exposed His and Glu residues forming at least three sites, with no clear functional significance, and not related to the GTP-binding site.75 Two potential metal binding sites can be predicted for elongation factor G, again without relationship to known protein function.76 CHED analysis of structural models of subunit beta of ATP synthase and porphobilinogen deaminase did not reveal any pertinent metal binding sites.
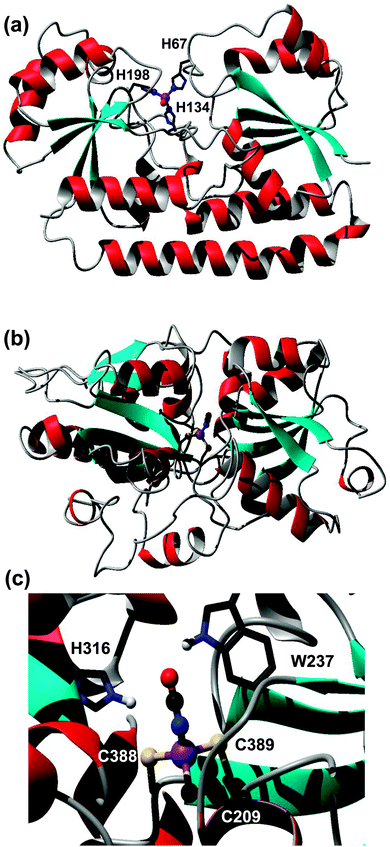 | ||
| Fig. 4 Modelled metal-binding sites in periplasmic binding proteins captured on a Zn-IMAC column. (a) Putative ZnuA (SYNW2481) from Synechococcus sp. WH8102. The zinc ion is coordinated to three His residues (H67, H139, H198) and a fourth non-proteinaceous ligand, modelled as water. (b) Putative cyanate transporter CynA (SYNW2487). The model (residues 197–520) is based on pdb 3UN6,45 an uncharacterised protein from Staphylococcus aureus. Zinc is coordinated to three Cys residues and a cyanate ion. The fold of the modelled CynA protein is very similar to that of periplasmic nitrate and hydrogen carbonate binding proteins, but none of the metal-binding residues are conserved in either nitrate or hydrogen carbonate transporters from Synechocystis sp. PCC6803. (c) Detailed view of the metal- and cyanate-binding site, with His316 and Trp237 as potential H-bond donors. | ||
However, several other periplasmic binding proteins were also captured on the Zn-IMAC column, including putative iron (FutA/IdiA; SYNW1797), cyanate (CynA; SYNW2487), and phosphate (PstS; SYNW1018) transporters. The interaction of the iron-binding protein FutA with the immobilised Zn2+ is unsurprising, because a certain degree of affinity for a non-cognate metal ion can be expected based on simple coordination chemistry principles. In contrast, the capture of the predicted cyanate (CynA) and phosphate (PstS) binding proteins was surprising. Therefore, homology models for both latter proteins were generated to inspect potential metal-binding sites. The model for the phosphate binding protein PstS did not reveal any clear metal sites (not shown). In contrast, one of the templates identified for the cyanate transporter CynA contained zinc ions in the binding cleft, one of them coordinated to two Cys and one His residue plus one water molecule. The template (pdb 3UN6)45 refers to an uncharacterised protein from Staphylococcus aureus. Like CynA, this protein is most closely related to the COG0715 cluster which contains periplasmic components of ABC-type nitrate–sulfonate–bicarbonate transport systems. The two Cys residues are conserved in CynA, and the His residue is replaced by another Cys residue (Fig. 4(b) and (c)). It is likely that at least some of these residues are responsible for the observed interaction with the immobilised Zn2+, and considering the nature of the ligands, in vivo Zn2+ and/or Cd2+ binding is most likely. It is plausible that the metal ion facilitates the binding of the cyanate anion to be transported. Considering that Zn2+ (or any other metal ion) is needed only in catalytic quantities inside cells, whilst cyanate provides the macronutrient nitrogen, co-transport of the metal appears less likely. It is noteworthy that the orthologous bicarbonate transporter CmpA from Synechocystis sp. PCC6803 utilises a Ca2+ ion to enhance bicarbonate binding.81
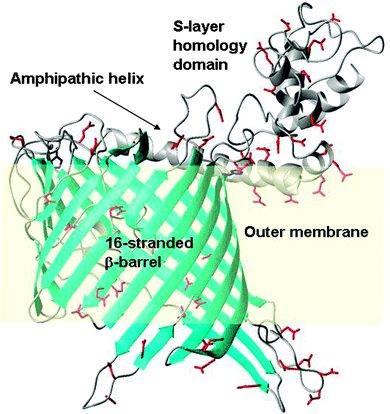
To enable the inspection of SYNW2224 for potential metal-binding sites, we generated a coarse model with the aid of the Phyre2 server (Fig. 5). The model shows a 16-stranded β-barrel, an N-terminal S-layer homology domain, connected by a long amphipathic helix. The S-layer domain, the helix, as well as several loops (both extra- and intracellular) abound in carboxylate groups, which might provide metal ion interaction sites, although it should be noted that the number of positively and negatively charged side-chains in SYNW2224 is overall similar. Many other marine CBPs (Table S3, ESI†) have a pronounced over-abundance of negatively charged side-chains. The model also suggests that the ‘pore’ is probably constricted by several loops, consistent with the hypothesis that these porins transport simple inorganic ions.
Discussion
The ability of Synechococcus sp. WH8102 cultures to grow at extremely low zinc concentrations and maintain cellular zinc homeostasis, as judged by total zinc quotas, suggests that Synechococcus sp. WH8102 has extremely efficient zinc uptake mechanisms capable of scavenging zinc even from depleted media. Only slight, if any, growth limitation at extremely low free zinc concentrations has previously been observed for Synechococcus bacillaris23 and Prochlorococcus MED4,24 especially when cobalt concentrations were also low, pointing to potential co-limitation by these two metal ions, and to possible substitution of zinc by cobalt but not vice versa.24,85The mechanisms for metal ion uptake in marine cyanobacteria are only partially understood. Genes for DNA-binding metal sensor proteins of the Fur family are present in all available genomes,29 as are those for ABC transporters for Fe, Mn, and Zn.28,30 Little is known regarding transport across the outer membrane, but considering that metal ions such as Fe3+ and Zn2+ must be bio-accumulated by a factor of at least 105, it follows that uptake mechanisms, including transport across the outer membrane, must be highly efficient.
One principal possibility is that zinc is transported into the cell as a complex with a chelating ligand. This option typically involves TonB-dependent receptors,86 which actively (i.e. under consumption of ATP) transport metal complexes with organic ligands across the outer membrane. TonB-dependent receptors are widely involved in bacterial iron uptake, but have also been implicated in bacterial zinc uptake, including four examples in Pseudomonas protegens PF-5 in zinc-depleted soil,87 two examples in the opportunistic pathogen Acinetobacter baumannii,88 and a protein named “ZnuD” produced by pathogenic Neisseria meningitidis under zinc limitation.89 In the freshwater cyanobacterium Anabaena sp. PCC 7120, zinc starvation caused the upregulation of a Zur-regulated TonB-like receptor.27 However, even though tonB-like genes have been discovered in Prochlorococcus,90 most marine Synechococcus strains lack the respective proteins.30,31,91,92 This absence of TonB-dependent receptors means that no specific outer membrane transporters for any M2+ or M3+ metal ion, including Fe3+, are known for marine Synechococcus.
Our discovery that the putative porins SYNW2224 and SYNW2227 have metal-binding ability offers a new hypothesis regarding the uptake of scarce trace metal ions by Synechococcus from oligotrophic waters, namely that at least some cyanobacterial porins of the CBP family play a central role in this process. The SYNW2224 model generated (Fig. 5) does not reveal any clearly defined binding sites, but many surface-accessible negatively charged carboxylate residues. On the basis of the nature and distribution of these potential metal-binding residues, the specificity of CBPs is expected to be limited, since all M2+ or M3+ species have significant affinities for clusters of carboxylate groups. Indeed, SYNW2224 was also captured on a Co2+-IMAC column in our previous study.37 The S-layer homology domain portion, and the amphipathic helix of SYNW2224, are also rich in negative charges, and hence may contribute to attracting and scavenging metal ions. In fact, a role for S-layers in biosorption of metals has been shown for bacilli,93 and the metal uptake process in cyanobacteria has recently been shown to involve a surface-adsorption step.94
Evidence that porins may function in metal uptake is available at the transcriptional level for other bacteria; for example, the expression of the porin OmpT in Vibrio cholerae is dependent on the level of iron in the environment, and is positively regulated by the ferric uptake regulator, Fur.95 The outer-membrane protein MnoP in Bradyrhizobium japonicum is expressed under conditions of manganese limitation, and is required for high-affinity manganese uptake.96 In Mycobacterium tuberculosis the porin MspA has been shown to be required for copper uptake across the outer cell membrane, with mspA deletion mutants showing severe growth defects when grown in a trace copper medium.97 The expression of several porins was also zinc-dependent in Pseudomonas protegens,87 and that of the OprD porin in Pseudomonas aeruginosa is down-regulated by excess zinc.98
Significantly, in freshwater Synechococcus sp. PCC7942, the gene for the CBP somB, but not its neighbouring CBP gene somA, has been found to be up-regulated under iron starvation.99somA and somB do not form an operon, but each have their own transcription start sites.100 Importantly, the upstream region of the somB gene in the PCC7942 strain is predicted by RegPrecise101 to contain a recognition site for the zinc-uptake regulator protein Zur. Hence, its expression is likely also zinc-regulated. RegPrecise also predicts zur boxes for CBP homologues in Thermosynechococcus elongatus (tlr1246), Microcystis aeruginosa (MAE_10010), and Synechocystis sp. PCC6803 (sll1550), and therefore proposed the name OmpZ (for zinc-regulated outer-membrane protein) for these homologues. Most cyanobacterial strains, including marine strains, have multiple CBP genes (Table S3, ESI†), with Synechococcus sp. WH8102 having at least four (synw2128, synw2223, synw2224, synw2227). The most divergent cyanobacterial strain, Gloeobacter violaceus, also harbours six CBP genes; four of them with and two without an SLH domain, indicating that duplication and divergence of these porins occurred even before the evolution of chloroplasts. Prochlorococcus sp. CCMP1375 (SS120), a strain with one of the smallest genomes, contains only two CBP genes, but there is no simple relationship between genome size and the number of CBP genes. Inspection of the genomic neighbourhoods of these genes (Table S3, ESI†) reveals that they are frequently localised in the vicinity of genes for the periplasmic binding proteins for Zn, Fe, or phosphate, or associated with genes suggesting a relationship with the metabolism of other metal ions, including Mn, Co and Ni. Whilst analyses of genomic neighbourhoods, or regulation by a particular nutrient cannot directly infer function, we suggest that our demonstrated zinc-binding ability of a CBP, and the high abundance of SYNW2224 under zinc-depleted conditions, adds two strong pieces of experimental evidence towards at least some of these porins playing an important role in the transport of essential metal ions such as zinc.
CBPs have previously been found to be up-regulated under nitrogen or sulfur starvation conditions in Synechococcus sp. PCC7942,102 and under phosphorus starvation in Synechococcus sp. WH810225,103,104 and Prochlorococcus spp. MED4 and MIT9313.105 This has led to suggestions that these proteins might increase the permeability of the outer membrane for enhanced nitrogen and sulfur uptake,102 or that they might transport phosphate.103 Cation- and anion-selective porins from the same organism, e.g. OmpF and PhoE from E. coli, share a high degree of similarity. Without direct biophysical studies, it is not possible to assign or predict particular selectivities for CBPs, but it may be expected that separate membrane channels for cationic and anionic nutrients should exist.106 If SYNW2224 proves to function in metal transport, then its increased expression under P-limitation25,103–105 could be an indirect consequence of the increased requirement for metal-dependent alkaline phosphatases, which are also up-regulated under these conditions.103 The increased expression of these enzymes would likely also increase cellular demand for zinc and/or calcium.107,108 Upregulation of the definitively zinc-related metallothionein SmtA in response to P-limitation in Synechococcus sp. WH8102 has recently been observed,25 indicating that Zn and P metabolic processes are linked in this strain. A more general link between Zn and P metabolism is also reflected in global biogeochemical cycling.109
Regarding the transported species, for eukaryotic phytoplankton zinc uptake, the free ion concentration has been thought to determine bioavailability,51 but a recent study demonstrated that weak organic ligands in the presence of a much stronger ligand such as EDTA increased the rate of zinc uptake by zinc-limited cultures of Emiliania huxleyi and Thalassiosira weissflogii, via a mechanism that likely involves the formation of ternary complexes between the weak metal–ligand complexes and a cell surface uptake molecule.110 Such a mechanism is also conceivable for zinc uptake by CBPs. SYNW2224 and SYNW2227 clearly have the ability to interact with partially complexed Zn2+, as presented by a nitrilotriacetic acid-based IMAC column. The conductance data characteristic of small solutes found for porins closely similar to SYNW2224 and SYNW222784 suggests that free Zn2+ (and likely other metal ions), or complexes with water, chloride, and other small anions, may be the entities transported, hence transport through the outer membrane could involve a decomplexation step.
The periplasm could then act as the first sorting point for different ions. Four periplasmic binding proteins were captured by Zn-IMAC, only two of them predicted to bind metal ions. Besides the periplasmic ZnuA component of a predicted zinc ABC transporter (SYNW2481), the Fe3+-binding protein FutA (SYNW1797) was also identified. The genome of Synechococcus sp. WH8102 contains an additional putative znuABC gene cluster (synw0969, 0970, and 0971) which according to RegPrecise101 harbours a predicted Zur recognition sequence in its promoter region. The periplasmic component of this cluster is synw0971; the respective protein was not detected in our metalloproteomics study.
The phosphate-binding protein PstS (SYNW1018) and the cyanate-binding protein CynA (SYNW2487) were also captured by Zn-IMAC. Whilst the observed metal affinity of PstS remains enigmatic, a very obvious metal-binding site ideally suited to bind Zn2+ (or related ions) was detected in CynA. Considering the scarcity of Zn2+ (and related ions such as Co2+ and Cd2+) in the natural habitat of Synechococcus sp. WH8102, it appears counter-intuitive that the bacterium should “waste” a potentially catalytically active metal ion for a merely supporting role in transport of an unusual nitrogen source. It is noteworthy that Synechococcus sp. WH8102 is one of the few strains that contains an active uptake system for cyanate,111,112 and is also able to utilise cyanate as sole nitrogen source.113 Several Prochlorococcus strains also have this capability. Transcripts for cynA (likely most closely related to HLII types) were abundant in stratified surface waters of the Gulf of Aqaba.112 These waters are characterised by nitrogen depletion, but also by high Prochlorococcus abundance. It has been suggested that the ability to utilise an additional nitrogen source may give a competitive advantage in extremely N-depleted surface layers of stratified ocean waters. It is intriguing that this may involve the participation of zinc or a closely related metal.
Conclusions
Our understanding of the mechanisms of zinc uptake into Gram-negative bacteria is incomplete, even though these are of particular interest for environments in which the concentration of essential zinc is extremely low, for example in oligotrophic oceans, but also during the acute phase response of a mammalian host to bacterial infection.114This study has demonstrated the increased expression of a putative cyanobacterial porin (CBP) under conditions of zinc depletion, and its zinc-binding ability – a property not previously demonstrated for any bacterial porin. Together with bioinformatic evidence for likely regulation of CBP homologues by the zinc uptake regulator Zur, this suggests a role for at least some of these proteins in Zn2+ uptake across the outer membrane of cyanobacteria, although the transport of other inorganic cations is also likely. CBPs have previously been implicated in the uptake of carbon, nitrogen, and phosphorus. This work has shown that their expression may not only be regulated by lack of macronutrients, but also of micronutrients.
Furthermore, the isolation of a putative periplasmic zinc-binding protein, ZnuA, by Zn2+-IMAC has provided the first experimental evidence that this protein is expressed by Synechococcus sp. WH8102, and has the ability to bind zinc in vitro. Together with the finding that Synechococcus sp. WH8102 accumulates appreciable quantities of zinc even under extreme zinc depletion, and further bioinformatic information, this augments our understanding of zinc homeostasis in this and other marine cyanobacterial strains (Fig. 6). In addition, combining Zn-IMAC with comparative modelling has led to the discovery of a novel metal-binding site in the periplasmic cyanate-binding protein CynA that enables Synechococcus sp. WH8102 to exploit cyanate as nitrogen source.
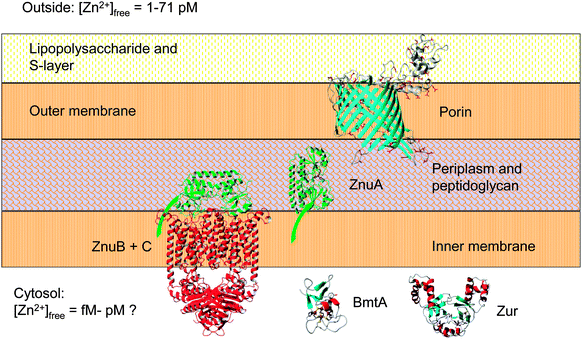 | ||
| Fig. 6 Major components of the zinc uptake and retention system in Synechococcus sp. WH8102. The zinc uptake regulator Zur controls the transcription of the cytosolic bacterial metallothionein BmtA, as well as that of at least one ZnuABC zinc uptake transporter. Expression levels of the putative CBP (SYNW2224) are also zinc-dependent (this work). Note that Synechococcus sp. WH8102 likely utilises more than one porin and more than one ZnuABC transporter. In cyanobacteria, the S-layer is on the outside of the outer membrane;84,116 this would suggest that CBPs are oriented as shown. The extracellular S-layer homology domain of SYNW2224 may function in scavenging and adsorbing trace metal ions; this may subsequently enable passage through the porin into the periplasm. Eventually, mediated by the periplasmic binding protein ZnuA (illustrated with its membrane anchor), zinc is thought to pass through the inner membrane via one of its two ZnuABC systems. The bacterial metallothionein, predicted to be Zur-regulated,29 may provide an intracellular zinc reservoir. | ||
Although no intact zinc-bound enzymes or other proteins were identified from any of the fractions analysed by native liquid chromatography, the alternative approach, IMAC, has enabled the enrichment of at least one protein involved in zinc uptake, as well as several candidates that warrant further detailed biophysical studies, including the porins SYNW2224 and 2227, the carboxysomal proteins CcmK1 and CcmK2, and the periplasmic cyanate binding protein CynA. It should be emphasised again that binding to a particular IMAC column does not allow any firm conclusions regarding any in vivo binding partners. It is possible that in several instances, the observed interaction with Zn2+ corresponds to adventitious binding. Careful inspection of each candidate, as a minimum via homology modelling and metal site analysis, should be carried out to eliminate false positives. Even then, caution must be applied regarding any statements pertaining to metal specificity, and further meta-data (such as genomic neighbourhood, synteny, and presence of transcription factor recognition sites) must be considered. Taking all these caveats into consideration, the experimental metalloproteomics in conjunction with bioinformatic approaches employed in the present work have uncovered expected and unexpected players in the zinc-binding network of a representative of an environmentally important class of marine phytoplankton.
Acknowledgements
This work was supported by the Leverhulme Trust (F/00 215/AY) and NERC (NE/F004249/1). Part of the equipment used in the research was obtained through Birmingham Science City with support from Advantage West Midlands and the European Regional Development Fund.References
- J. E. Coleman, Annu. Rev. Biochem., 1992, 61, 897–946 CrossRef CAS PubMed
.
- C. Andreini, L. Banci, I. Bertini and A. Rosato, J. Proteome Res., 2006, 5, 3173–3178 CrossRef CAS PubMed
.
- M. Brylinski and J. Skolnick, Proteins, 2011, 79, 735–751 CrossRef CAS PubMed
.
- A. Cvetkovic, A. L. Menon, M. P. Thorgersen, J. W. Scott, F. L. Poole, F. E. Jenney, W. A. Lancaster, J. L. Praissman, S. Shanmukh, B. J. Vaccaro, S. A. Trauger, E. Kalisiak, J. V. Apon, G. Siuzdak, S. M. Yannone, J. A. Tainer and M. W. W. Adams, Nature, 2010, 466, 779–782 CrossRef CAS PubMed
.
- J. P. Barnett, D. J. Scanlan and C. A. Blindauer, Anal. Bioanal. Chem., 2012, 402, 3311–3322 CrossRef CAS PubMed
.
- S. M. Yannone, S. Hartung, A. L. Menon, M. W. Adams and J. A. Tainer, Curr. Opin. Biotechnol., 2012, 23, 89–95 CrossRef CAS PubMed
.
- C. L. Dupont, A. Butcher, R. E. Valas, P. E. Bourne and G. Caetano-Anolles, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10567–10572 CrossRef CAS PubMed
.
- E. L. Mann and S. W. Chisholm, Limnol. Oceanogr., 2000, 45, 1067–1076 CrossRef CAS
.
- P. W. Boyd, T. Jickells, C. S. Law, S. Blain, E. A. Boyle, K. O. Buesseler, K. H. Coale, J. J. Cullen, H. J. de Baar, M. Follows, M. Harvey, C. Lancelot, M. Levasseur, N. P. Owens, R. Pollard, R. B. Rivkin, J. Sarmiento, V. Schoemann, V. Smetacek, S. Takeda, A. Tsuda, S. Turner and A. J. Watson, Science, 2007, 315, 612–617 CrossRef CAS PubMed
.
- M. Sinoir, E. C. V. Butler, A. R. Bowie, M. Mongin, P. N. Nesterenko and C. S. Hassler, Mar. Freshwater Res., 2012, 63, 644–657 CAS
.
- K. W. Bruland, Earth Planet. Sci. Lett., 1980, 47, 176–198 CrossRef CAS
.
- J. R. Donat and K. W. Bruland, Mar. Chem., 1990, 28, 301–323 CrossRef CAS
.
- R. W. Jakuba, M. A. Saito, J. W. Moffett and Y. Xu, Global Biogeochem. Cycles, 2012, 26, GB2015 CrossRef
.
- K. W. Bruland, Limnol. Oceanogr., 1989, 34, 269–285 CrossRef CAS
.
- M. J. Ellwood and C. M. G. Van den Berg, Mar. Chem., 2000, 68, 295–306 CrossRef CAS
.
- L. E. Brand, W. G. Sunda and R. R. L. Guillard, Limnol. Oceanogr., 1983, 28, 1182–1198 CrossRef CAS
.
- W. G. Sunda and S. A. Huntsman, Limnol. Oceanogr., 1992, 37, 25–40 CrossRef CAS
.
- K. G. Schulz, I. Zondervan, L. J. A. Gerringa, K. R. Timmermans, M. J. W. Veldhuis and U. Riebesell, Nature, 2004, 430, 673–676 CrossRef CAS PubMed
.
- D. Malasarn, J. Kropat, S. I. Hsieh, G. Finazzi, D. Casero, J. A. Loo, M. Pellegrini, F. A. Wollman and S. S. Merchant, J. Biol. Chem., 2013, 288, 10672–10683 CrossRef CAS PubMed
.
- F. M. M. Morel, J. R. Reinfelder, S. B. Roberts, C. P. Chamberlain, J. G. Lee and D. Yee, Nature, 1994, 369, 740–742 CrossRef CAS
.
- J. H. Martin, Paleoceanography, 1990, 5, 1–13 CrossRef
.
- P. Flombaum, J. L. Gallegos, R. A. Gordillo, J. Rincón, L. L. Zabala, N. Jiao, D. M. Karl, W. K. W. Li, M. W. Lomas, D. Veneziano, C. S. Vera, J. A. Vrugt and A. C. Martiny, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 9824–9829 CrossRef CAS PubMed
.
- W. G. Sunda and S. A. Huntsman, Limnol. Oceanogr., 1995, 40, 1404–1417 CrossRef CAS
.
- M. A. Saito, J. W. Moffett, S. W. Chisholm and J. B. Waterbury, Limnol. Oceanogr., 2002, 47, 1629–1636 CrossRef CAS
.
- A. Cox and M. Saito, Front. Microbiol., 2013, 4, 387 Search PubMed
.
- J. S. Cavet, G. P. M. Borrelly and N. J. Robinson, FEMS Microbiol. Rev., 2003, 27, 165–181 CrossRef CAS
.
- M. Napolitano, M. A. Rubio, J. Santamaria-Gomez, E. Olmedo-Verd, N. J. Robinson and I. Luque, J. Bacteriol., 2012, 194, 2426–2436 CrossRef CAS PubMed
.
- C. A. Blindauer, Chem. Biodiversity, 2008, 5, 1990–2013 CAS
.
- J. P. Barnett, A. Millard, A. Z. Ksibe, D. J. Scanlan, R. Schmid and C. A. Blindauer, Front. Microbiol., 2012, 3, 142 Search PubMed
.
- D. J. Scanlan, M. Ostrowski, S. Mazard, A. Dufresne, L. Garczarek, W. R. Hess, A. F. Post, M. Hagemann, I. Paulsen and F. Partensky, Microbiol. Mol. Biol. Rev., 2009, 73, 249–299 CrossRef CAS PubMed
.
- B. Palenik, B. Brahamsha, F. W. Larimer, M. Land, L. Hauser, P. Chain, J. Lamerdin, W. Regala, E. E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. A. Webb and J. Waterbury, Nature, 2003, 424, 1037–1042 CrossRef CAS PubMed
.
- M. A. da Silva, A. Sussulini and M. A. Z. Arruda, Expert Rev. Proteomics, 2010, 7, 387–400 CrossRef CAS PubMed
.
- W. Shi and M. R. Chance, Curr. Opin. Chem. Biol., 2011, 15, 144–148 CrossRef PubMed
.
- D. Fu and L. Finney, Expert Rev. Proteomics, 2014, 11, 13–19 CrossRef CAS PubMed
.
- A. Sussulini and J. S. Becker, Metallomics, 2011, 3, 1271–1279 RSC
.
- S. Mounicou, J. Szpunar and R. Lobinski, Chem. Soc. Rev., 2009, 38, 1119–1138 RSC
.
- J. P. Barnett, D. J. Scanlan and C. A. Blindauer, Anal. Bioanal. Chem., 2012, 402, 3371–3377 CrossRef CAS PubMed
.
- F. M. M. Morel, J. G. Rueter, D. M. Anderson and R. R. L. Guillard, J. Phycol., 1979, 15, 135–141 CrossRef CAS PubMed
.
- N. M. Price, G. I. Harrison, J. G. Hering, R. J. Hudson, P. M. V. Nirel, B. Palenik and F. M. M. Morel, Biol. Oceanogr., 1989, 5–6, 443–461 Search PubMed
.
- P. K. Smith, R. I. Krohn, G. T. Hermanson, A. K. Mallia, F. H. Gartner, M. D. Provenzano, E. K. Fujimoto, N. M. Goeke, B. J. Olson and D. C. Klenk, Anal. Biochem., 1985, 150, 76–85 CrossRef CAS
.
- A. D. Gonzales, Y. K. Light, Z. Zhang, T. Iqbal, T. W. Lane and A. Martino, Can. J. Bot. Rev. Can. Bot., 2005, 83, 735–745 CrossRef CAS
.
- U. K. Laemmli, Nature, 1970, 227, 680–685 CrossRef CAS
.
- L. A. Kelley and M. J. E. Sternberg, Nat. Protoc., 2009, 4, 363–371 CrossRef CAS PubMed
.
- B. X. Wei, A. M. Randich, M. Bhattacharyya-Pakrasi, H. B. Pakrasi and T. J. Smith, Biochemistry, 2007, 46, 8734–8743 CrossRef CAS PubMed
.
-
G. Minasov, Z. Wawrzak, A. Halavaty, L. Shuvalova, I. Dubrovska, J. Winsor, O. Kiryukhina, F. Bagnoli, F. Falugi, M. Bottomley, G. Grandi and W. F. Anderson, Protein Data Bank, 2011 Search PubMed
.
- N. Koropatkin, A. M. Randich, M. Bhattacharyya-Pakrasi, H. B. Pakrasi and T. J. Smith, J. Biol. Chem., 2007, 282, 27468–27477 CrossRef CAS PubMed
.
- J. Kern, R. Wilton, R. G. Zhang, T. A. Binkowski, A. Joachimiak and O. Schneewind, J. Biol. Chem., 2011, 286, 26042–26049 CrossRef CAS PubMed
.
- N. G. Shuart, Y. Haitin, S. S. Camp, K. D. Black and W. N. Zagotta, Nat. Commun., 2011, 2, 457 CrossRef PubMed
.
- B. van den Berg, J. Biol. Chem., 2012, 287, 41044–41052 CrossRef CAS PubMed
.
- R. Koradi, M. Billeter and K. Wuthrich, J. Mol. Graphics, 1996, 14, 51–55 CrossRef CAS
.
- W. G. Sunda, Biol. Oceanogr., 1988, 6, 411–442 Search PubMed
.
- W. G. Sunda, Front. Microbiol., 2012, 3, 204 Search PubMed
.
- H. Irving and R. J. P. Williams, J. Chem. Soc., 1953, 3192–3210 RSC
.
- D. V. Tulumello and C. M. Deber, Biochim. Biophys. Acta, Biomembr., 2012, 1818, 1351–1358 CrossRef CAS PubMed
.
- S. D. Smith, Y. M. She, E. A. Roberts and B. Sarkar, J. Proteome Res., 2004, 3, 834–840 CrossRef CAS
.
- C. C. S. Kung, W. N. Huang, Y. C. Huang and K. C. Yeh, Proteomics, 2006, 6, 2746–2758 CrossRef CAS PubMed
.
- C. L. Smith, J. L. Stauber, M. R. Wilson and D. F. Jolley, Anal. Bioanal. Chem., 2014, 406, 305–315 CrossRef CAS PubMed
.
- A. H. Millar, Y. F. Tan, N. O'Toole and N. L. Taylor, Plant Physiol., 2010, 152, 747–761 CrossRef PubMed
.
- A. Galkin, Z. Li, L. Li, L. Kulakova, L. R. Pal, D. Dunaway-Mariano and O. Herzberg, Biochemistry, 2009, 48, 3186–3196 CrossRef CAS PubMed
.
- K. Nakahara, H. Yamamoto, C. Miyake and A. Yokota, Plant Cell Physiol., 2003, 44, 326–333 CrossRef CAS PubMed
.
- A. Katayama, A. Tsujii, A. Wada, T. Nishino and A. Ishihama, Eur. J. Biochem., 2002, 269, 2403–2413 CrossRef CAS
.
- A. M. Sevcenco, M. W. Pinkse, H. T. Wolterbeek, P. D. Verhaert, W. R. Hagen and P. L. Hagedoorn, Metallomics, 2011, 3, 1324–1330 RSC
.
- P. Asztalos, C. Parthier, R. Golbik, M. Kleinschmidt, G. Huebner, M. S. Weiss, R. Friedemann, G. Wille and K. Tittmann, Biochemistry, 2007, 46, 12037–12052 CrossRef CAS PubMed
.
- K. Chandrasekhar, J. J. Wang, P. Arjunan, M. Sax, Y. H. Park, N. S. Nemeria, S. Kumaran, J. Y. Song, F. Jordan and W. Furey, J. Biol. Chem., 2013, 288, 15402–15417 CrossRef CAS PubMed
.
- M. Babor, S. Gerzon, B. Raveh, V. Sobolev and M. Edelman, Proteins, 2008, 70, 208–217 CrossRef CAS PubMed
.
- S. G. Kim, J. S. Chung, R. B. Sutton, J. S. Lee, L. Lopez-Maury, S. Y. Lee, F. J. Florencio, T. Lin, M. Zabet-Moghaddam, M. J. Wood, K. Nayak, V. Madem, J. N. Tripathy, S. K. Kim and D. B. Knaff, Biochim. Biophys. Acta, 2012, 1824, 392–403 CrossRef CAS PubMed
.
- L. C. Sieker, M. Holmes, I. Le Trong, S. Turley, M. Y. Liu, J. LeGall and R. E. Stenkamp, J. Biol. Inorg. Chem., 2000, 5, 505–513 CrossRef CAS
.
- H. S. Gill and D. Eisenberg, Biochemistry, 2001, 40, 1903–1912 CrossRef CAS PubMed
.
- Y. Zhang, Y. Feng, S. Chatterjee, S. Tuske, M. X. Ho, E. Arnold and R. H. Ebright, Science, 2012, 338, 1076–1080 CrossRef CAS PubMed
.
- H. Sugawara, H. Yamamoto, N. Shibata, T. Inoue, S. Okada, C. Miyake, A. Yokota and Y. Kai, J. Biol. Chem., 1999, 274, 15655–15661 CrossRef CAS PubMed
.
- S. Müller, K. Diederichs, J. Breed, R. Kissmehl, K. Hauser, H. Plattner and W. Welte, J. Mol. Biol., 2002, 315, 141–153 CrossRef PubMed
.
- J. J. Jeong, S. Fushinobu, S. Ito, B. S. Jeon, H. Shoun and T. Wakagi, FEBS Lett., 2003, 535, 200–204 CrossRef CAS
.
- J. M. Berrisford, J. Akerboom, A. P. Turnbull, D. de Geus, S. E. Sedelnikova, I. Staton, C. W. McLeod, C. H. Verhees, J. van der Oost, D. W. Rice and P. J. Baker, J. Biol. Chem., 2003, 278, 33290–33297 CrossRef CAS PubMed
.
- J. King-Scott, E. Nowak, E. Mylonas, S. Panjikar, M. Roessle, D. I. Svergun and P. A. Tucker, J. Biol. Chem., 2007, 282, 37717–37729 CrossRef CAS PubMed
.
- D. Takeshita and K. Tomita, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15733–15738 CrossRef CAS PubMed
.
- Y. Chen, R. K. Koripella, S. Sanyal and M. Selmer, FEBS J., 2010, 277, 3789–3803 CrossRef CAS PubMed
.
-
Y. Fan, L. Volkart, M. Gu, S. Axen, W. B. Greenleaf, C. Kerfeld and A. Joachimiak, Protein Data Bank, 2010 Search PubMed
.
- Y. Tsai, M. R. Sawaya, G. C. Cannon, F. Cai, E. B. Williams, S. Heinhorst, C. A. Kerfeld and T. O. Yeates, PLoS Biol., 2007, 5, 1345–1354 CAS
.
- M. Takenoya, K. Nikolakakis and M. Sagermann, J. Bacteriol., 2010, 192, 6056–6063 CrossRef CAS PubMed
.
- V. V. Bartsevich and H. B. Pakrasi, EMBO J., 1995, 14, 1845–1853 CAS
.
- N. M. Koropatkin, D. W. Koppenaal, H. B. Pakrasi and T. J. Smith, J. Biol. Chem., 2007, 282, 2606–2614 CrossRef CAS PubMed
.
- H. Umeda, H. Aiba and T. Mizuno, Microbiology, 1996, 142, 2121–2128 CrossRef CAS PubMed
.
- A. Hansel and M. H. Tadros, Curr. Microbiol., 1998, 36, 321–326 CrossRef CAS
.
- E. Hoiczyk and A. Hansel, J. Bacteriol., 2000, 182, 1191–1199 CrossRef CAS
.
- M. A. Saito, T. J. Goepfert and J. T. Ritt, Limnol. Oceanogr., 2008, 53, 276–290 CrossRef CAS
.
- K. D. Krewulak and H. J. Vogel, Biochem. Cell Biol., 2011, 89, 87–97 CrossRef CAS PubMed
.
- C. K. Lim, K. A. Hassan, A. Penesyan, J. E. Loper and I. T. Paulsen, Environ. Microbiol., 2013, 15, 702–715 CrossRef CAS PubMed
.
- M. I. Hood, B. L. Mortensen, J. L. Moore, Y. F. Zhang, T. E. Kehl-Fie, N. Sugitani, W. J. Chazin, R. M. Caprioli and E. P. Skaar, PLoS Pathog., 2012, 8, e1003068 CAS
.
- M. Stork, M. P. Bos, I. Jongerius, N. de Kok, I. Schilders, V. E. Weynants, J. T. Poolman and J. Tommassen, PLoS Pathog., 2010, 6, e1000969 Search PubMed
.
- R. R. Malmstrom, S. Rodrigue, K. H. Huang, L. Kelly, S. E. Kern, A. Thompson, S. Roggensack, P. M. Berube, M. R. Henn and S. W. Chisholm, ISME J., 2013, 7, 184–198 CrossRef CAS PubMed
.
- E. A. Webb, J. W. Moffett and J. B. Waterbury, Appl. Environ. Microbiol., 2001, 67, 5444–5452 CrossRef CAS PubMed
.
- A. R. Rivers, R. W. Jakuba and E. A. Webb, Environ. Microbiol., 2009, 11, 382–396 CrossRef CAS PubMed
.
- M. C. Allievi, S. Florencia, P. A. Mariano, P. M. Mercedes, S. M. Ruzal and S. R. Carmen, J. Microbiol. Biotechnol., 2011, 21, 147–153 CrossRef CAS
.
- L. Hudek, S. Rai, A. Michalczyk, L. C. Rai, B. A. Neilan and M. L. Ackland, BioMetals, 2012, 25, 893–903 CrossRef CAS PubMed
.
- S. A. Craig, C. D. Carpenter, A. R. Mey, E. E. Wyckoff and S. M. Payne, J. Bacteriol., 2011, 193, 6505–6511 CrossRef CAS PubMed
.
- T. H. Hohle, W. L. Franck, G. Stacey and M. R. O'Brian, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 15390–15395 CrossRef CAS PubMed
.
- A. Speer, J. L. Rowland, M. Haeili, M. Niederweis and F. Wolschendorf, J. Bacteriol., 2013, 195, 5133–5140 CrossRef CAS PubMed
.
- M. C. Conejo, I. Garcia, L. Martinez-Martinez, L. Picabea and A. Pascual, Antimicrob. Agents Chemother., 2003, 47, 2313–2315 CrossRef CAS
.
- A. Nodop, D. Pietsch, R. Hocker, A. Becker, E. K. Pistorius, K. Forchhammer and K. P. Michel, Plant Physiol., 2008, 147, 747–763 CrossRef CAS PubMed
.
- A. Hansel, F. Pattus, U. J. Jurgens and M. H. Tadros, Biochim. Biophys. Acta, Gene Struct.
Expression, 1998, 1399, 31–39 CrossRef CAS
.
- P. S. Novichkov, T. S. Brettin, E. S. Novichkova, P. S. Dehal, A. P. Arkin, I. Dubchak and D. A. Rodionov, Nucleic Acids Res., 2012, 40, W604–W608 CrossRef CAS PubMed
.
- J. Sauer, U. Schreiber, R. Schmid, U. Volker and K. Forchhammer, Plant Physiol., 2001, 126, 233–243 CrossRef CAS PubMed
.
- S. G. Tetu, B. Brahamsha, D. A. Johnson, V. Tai, K. Phillippy, B. Palenik and I. T. Paulsen, ISME J., 2009, 3, 835–849 CrossRef CAS PubMed
.
- M. Ostrowski, S. Mazard, S. G. Tetu, K. Phillippy, A. Johnson, B. Palenik, I. T. Paulsen and D. J. Scanlan, ISME J., 2010, 4, 908–921 CrossRef CAS PubMed
.
- A. C. Martiny, M. L. Coleman and S. W. Chisholm, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12552–12557 CrossRef CAS PubMed
.
- D. Duy, J. Soll and K. Philippar, Biol. Chem., 2007, 388, 879–889 CrossRef CAS PubMed
.
- J. E. Coleman, Annu. Rev. Biophys. Biomol. Struct., 1992, 21, 441–483 CrossRef CAS PubMed
.
- S. Kathuria and A. C. Martiny, Environ. Microbiol., 2011, 13, 74–83 CrossRef CAS PubMed
.
- R. W. Jakuba, J. W. Moffett and S. T. Dyhrman, Global Biogeochem. Cycles, 2008, 22, GB4012 CrossRef
.
- L. Aristilde, Y. Xu and F. M. M. Morel, Environ. Sci. Technol., 2012, 46, 5438–5445 CrossRef CAS PubMed
.
- G. S. Espie, F. Jalali, T. Tong, N. J. Zacal and A. K. C. So, J. Bacteriol., 2007, 189, 1013–1024 CrossRef CAS PubMed
.
- N. A. Kamennaya, M. Chernihovsky and A. F. Post, Limnol. Oceanogr., 2008, 53, 2485–2494 CrossRef CAS
.
- Z. C. Su, F. L. Mao, P. Dam, H. W. Wu, V. Olman, I. T. Paulsen, B. Palenik and Y. Xu, Nucleic Acids Res., 2006, 34, 1050–1065 CrossRef CAS PubMed
.
- E. K. LeGrand and J. Alcock, Q. Rev. Biol., 2012, 87, 3–18 CrossRef
.
- F. R. Tabita, S. Satagopan, T. E. Hanson, N. E. Kreel and S. S. Scott, J. Exp. Bot., 2008, 59, 1515–1524 CrossRef CAS PubMed
.
- J. Šmarda, D. Šmajs, J. Komrska and V. Krzyžánek, Micron, 2002, 33, 257–277 CrossRef
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4mt00048j |
| ‡ Since WH8102 contains α carboxysomes, the term CsoS1 should be used (Badger and Price, 2003) – ccm (for carbon-concentrating mechanism) proteins are components of β carboxysomes, but this nomenclature is widely ignored in genome annotations; therefore, we have retained the names as they appear in relevant databases. |
| § SYNW0971 is also predicted as ZnuA. |
| This journal is © The Royal Society of Chemistry 2014 |