 Open Access Article
Open Access ArticleAn integrated study of the affinities of the Aβ16 peptide for Cu(I) and Cu(II): implications for the catalytic production of reactive oxygen species†
Tessa R.
Young
ab,
Angie
Kirchner‡
ab,
Anthony G.
Wedd
ab and
Zhiguang
Xiao
*ab
aThe Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Victoria 3010, Australia
bSchool of Chemistry, The University of Melbourne, Victoria 3010, Australia. E-mail: z.xiao@unimelb.edu.au
First published on 24th January 2014
Abstract
A new fluorescent probe Aβ16wwa based upon the Aβ16 peptide has been developed with two orders of magnitude greater fluorescence intensity for sensitive detection of interactions with Cu(II). In combination with the Cu(I) probe Ferene S, it is confirmed that the Aβ16 peptide binds either Cu(I) or Cu(II) with comparable affinities at pH 7.4 (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KID = −10.4; log
KID = −10.4; log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIID = −10.0). It follows from this property that the Cu–Aβ16 complex is a robust if slow catalyst for the aerial oxidation of ascorbate with H2O2 as primary product (initial rate, ∼0.63 min−1 for Cu–Aβ16 versus >2.5 min−1 for Cuaq2+). An integrated study of variants of this peptide identifies the major ligands and binding modes involved in its copper complexes in solution. The dependence of KID upon pH is consistent with a two-coordinate Cu(I) site in which dynamic processes exchange Cu(I) between the three available pairs of imidazole sidechains provided by His6, His13 and His14. The N-terminal amine is not involved in Cu(I) binding but is a key ligand for Cu(II). Acetylation of the N-terminus alters the redox thermodynamic gradient for the Cu centre and suppresses its catalytic activity considerably. The data indicate the presence of dynamic processes that exchange Cu(II) between the three His ligands and backbone amide at physiological pH. His6 is identified as a key ligand for catalysis as its presence minimises the pre-organisation energy required for interchange of the two copper redox sites. These new thermodynamic data strengthen structural interpretations for the Cu–Aβ complexes and provide valuable insights into the molecular mechanism by which copper chemistry may induce oxidative stress in Alzheimer's disease.
KIID = −10.0). It follows from this property that the Cu–Aβ16 complex is a robust if slow catalyst for the aerial oxidation of ascorbate with H2O2 as primary product (initial rate, ∼0.63 min−1 for Cu–Aβ16 versus >2.5 min−1 for Cuaq2+). An integrated study of variants of this peptide identifies the major ligands and binding modes involved in its copper complexes in solution. The dependence of KID upon pH is consistent with a two-coordinate Cu(I) site in which dynamic processes exchange Cu(I) between the three available pairs of imidazole sidechains provided by His6, His13 and His14. The N-terminal amine is not involved in Cu(I) binding but is a key ligand for Cu(II). Acetylation of the N-terminus alters the redox thermodynamic gradient for the Cu centre and suppresses its catalytic activity considerably. The data indicate the presence of dynamic processes that exchange Cu(II) between the three His ligands and backbone amide at physiological pH. His6 is identified as a key ligand for catalysis as its presence minimises the pre-organisation energy required for interchange of the two copper redox sites. These new thermodynamic data strengthen structural interpretations for the Cu–Aβ complexes and provide valuable insights into the molecular mechanism by which copper chemistry may induce oxidative stress in Alzheimer's disease.
Introduction
While the underlying molecular causes of Alzheimer's disease remain unknown, mis-handling of the Aβ peptides derived from the amyloid precursor protein during recycling may be an initiating event (‘the amyloid cascade’).1–4 Aberrant metal ion homeostasis appears to contribute by promoting aggregation of the peptides and/or inducing toxic gain of function.5–7 In this context, the potential role of the trace metal copper has been emphasised (see ref. 8 for a review).The affinity of Aβ peptides for Cu(II) remains a topic of discussion (see recent reviews9,10) although a consensus of KD ∼ 10−10 M is building.11–13 On the other hand, the affinity for Cu(I) remains problematical.14,15 Spectroscopic data have provided extensive information on the structure of Cu–Aβ complexes and of those of the truncated form Aβ16(DAEFRHDSGYEVHHQK), in particular. This fragment appears to provide the copper binding sites of highest affinity. The two major coordination forms of monomeric CuII–Aβ are related by a pKa ∼ 7.8,16–22 while a single dominant Cu(I) form has been characterised spectroscopically.23,24 Rapid ligand exchange processes have been detected.21,25 The ability of these complexes to catalyse the production of H2O2 by reduction of O2 by ascorbate (Asc) has been explored as a model for the production of reactive oxygen species that may be primarily responsible for inflammation responses.
The present work defines the dissociation constants of both Cu(I) and Cu(II) for the peptide Aβ16 under the same conditions using chromophoric and fluorescence probes of appropriate affinities. A new probe for Cu(II) has been developed from the Aβ16 peptide itself while those for Cu(I) were defined in recent work.26 Application to variant forms of the Aβ16 peptide has correlated the new thermodynamic data with existing spectroscopic structural data while the ability of the Cu complexes of the peptide and its variants to catalyse production of reactive oxygen species is explored.
Experimental section
Materials and general methods
Materials purchased from Sigma included ligands Ferene S (Fs) and Ferrozine (Fz) (as their sodium salts Na2Fs and NaHFz), reductants NH2OH (as its H2SO4 salt) and ascorbic acid, a copper standard (as a standard solution for calibration of atomic absorption spectroscopy), Amplex Red (also called Ampliflu Red) and horseradish peroxidase (HRP). They were all used as received. Proteins CopK and its variant CopK-H70F were expressed and isolated as reported previously.27 Peptides Aβ16 (sequence: DAEFRHDSGYEVHHQK), Ac-Aβ16 (Ac-DAEFRHDSGYEVHHQK); Aβ16-H13A (DAEFRHDSGYEVAHQK), Aβ16-H6,13A (DAEFRADSGYEVAHQK); Aβ16-H13,14A (DAEFRHDSGYEVAAQK) were purchased from GL Biochem (Shanghai) with purity estimated at >98%. Peptides Aβ16-F4W,Y10W,H14A (denoted as Aβ16wwa, DAEWRHDSGWEVHAQK); Aβ16-H6A (DAEFRADSGYEVHHQK) and Aβ16-H14A (DAEFRHDSGYEVHAQK) were synthesised on site by solid phase peptide techniques. Identity was verified by electrospray ionisation mass spectrometry (ESI-MS) while purity was confirmed to be >98% by HPLC. Peptide concentrations were estimated from absorbance maxima at ∼276 nm using εmax = 1410 M−1 cm−1 for those Aβ16 peptides containing a single tyrosine residue and using ε = 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 M−1 cm−1 at 280 nm for the Aβ16wwa probe peptide containing two tryptophan residues. The concentrations obtained matched those estimated from fluorescence titrations with the copper standard assuming formation of a 1
000 M−1 cm−1 at 280 nm for the Aβ16wwa probe peptide containing two tryptophan residues. The concentrations obtained matched those estimated from fluorescence titrations with the copper standard assuming formation of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex.
1 complex.
All ligand stock solutions, reaction buffers, reductant solutions and protein solutions were prepared in rigorously deoxygenated Milli-Q water and stored in an anaerobic glove-box (<1 ppm O2). The working solution of ligand Fs was prepared freshly from aliquots of frozen stock solution stored in a freezer at temperature −80 °C.26 All reactions involving Cu(I) were performed anaerobically in the glove-box and transported outside in a capped cuvette for spectroscopic characterisation.
UV-Vis and fluorescence titration
UV-visible spectra were recorded on a Varian Cary 300 spectrophotometer in dual beam mode with quartz cuvettes of 1.0 cm path length. All metal titrations were performed in appropriate buffers and corrected for baseline and dilution. Fluorescence emission spectra were obtained on a Varian Cary Eclipse spectrophotometer. The excitation wavelength was 280 nm with a band pass of 20 nm for both excitation and emission spectra. Spectra were recorded between 290–600 nm at a scale rate of 600 nm min−1. The absorbance of solutions was maintained below A280 = 0.1 to minimise resorption effects.Quantification of Cu(I) binding
Data of KID for proteins and peptides P were obtained from the competition reaction of eqn (1) and analysed by eqn (2):26 | (1) |
 | (2) |
The term [CuIL2] is the equilibrium concentration of probe complex [CuIL2]3− in eqn (1) and may be determined directly from the solution absorbance under the condition that this complex is the only absorbing species. The other terms in eqn (2) are the known total concentrations of the relevant species. The dissociation constant KID for CuI–P was derived by curve-fitting of the experimental data to eqn (2) (i.e., plots of [P]tot/[Cu]totversus [CuIL2]) with β2 = 1013.7 and 1015.1 M−2 for L = Fs and Fz, respectively.26 The detailed experimental protocol followed that reported recently.26 The experiments were conducted anaerobically in deoxygenated MOPS buffer (50 mM, pH 7.4) containing reductants NH2OH (0.5 mM) and/or Asc (0.5 mM) (denoted as buffer A) in a glovebox ([O2] < 1 ppm). The samples were transferred in sealed containers for characterisation.
The sidechains of the three histidine residues (His-6,13,14) in Aβ16 peptides are likely to be involved in Cu(I) binding. Their pKa values in an Aβ28 peptide have been estimated to be 6.5, 6.6 and 6.5, respectively.28 Consequently, the apparent dissociation constants  for Cu(I) binding are likely to be sensitive to pH around pH 6.5. A quantitative analysis of the relationship between
for Cu(I) binding are likely to be sensitive to pH around pH 6.5. A quantitative analysis of the relationship between  and solution pH should provide information about the number of His residues being involved in the Cu(I) binding. For two- and three-His binding models, the relationship may be described by eqn (3) and (4), respectively:29
and solution pH should provide information about the number of His residues being involved in the Cu(I) binding. For two- and three-His binding models, the relationship may be described by eqn (3) and (4), respectively:29
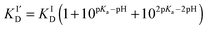 | (3) |
 | (4) |
 and KID refer, respectively, to pH-dependent and pH-independent dissociation constants and pKa is the average proton dissociation constant for the sidechains of the three His residues, taken to be close to 6.5.28 Note that eqn (3) and (4) address protonation equilibria of only those His sidechains that are involved in Cu(I) binding and assume negligible influence from other residues not involved directly in the binding to Cu(I).
and KID refer, respectively, to pH-dependent and pH-independent dissociation constants and pKa is the average proton dissociation constant for the sidechains of the three His residues, taken to be close to 6.5.28 Note that eqn (3) and (4) address protonation equilibria of only those His sidechains that are involved in Cu(I) binding and assume negligible influence from other residues not involved directly in the binding to Cu(I).
The experiments were conducted within the pH range 5.5–7.8 in buffers MES (pKa = 6.1) and MOPS (pKa = 7.2) employing [CuI(Fs)2]3− as a chromophoric probe. The pH of each solution after an experiment was confirmed with a pH micro-electrode to ensure recording of an accurate value for each solution. The  values within this pH range were derived by curve-fitting of the experimental data to eqn (2), taking advantage of the low pKa value of ∼3.2 for the probe ligand Fs. This means that the formation constant β2 of [CuI(Fs)2]3− is essentially pH-independent at pH > 5.5.26 This latter property was confirmed by determination of an essentially identical KID, within experimental error, for control Cu(I) complexes of protein CopK and its variant H70F following the same approach, again using [CuI(Fs)2]3− as the determining probe (see Table S1, ESI†). The Cu(I) sites in both CopK proteins are defined by four methionine sidechains arranged tetrahedrally and so their Cu(I) binding affinities are expected to be pH-independent.27
values within this pH range were derived by curve-fitting of the experimental data to eqn (2), taking advantage of the low pKa value of ∼3.2 for the probe ligand Fs. This means that the formation constant β2 of [CuI(Fs)2]3− is essentially pH-independent at pH > 5.5.26 This latter property was confirmed by determination of an essentially identical KID, within experimental error, for control Cu(I) complexes of protein CopK and its variant H70F following the same approach, again using [CuI(Fs)2]3− as the determining probe (see Table S1, ESI†). The Cu(I) sites in both CopK proteins are defined by four methionine sidechains arranged tetrahedrally and so their Cu(I) binding affinities are expected to be pH-independent.27
Quantification of Cu(II) binding via direct metal ion titration
Direct titration of Cu2+ into a solution of peptide P may induce reaction (5) where the fraction of bound Cu(II) is described by eqn (6):| Cuaq2+ + 2P + ‘CuII–B’ ⇌ 2CuII–P + B | (5) |
 | (6) |
A pre-condition for a meaningful quantitative analysis viaeqn (6) is that KIID(1 + KA[B]) ∼ [P]. To assist such analysis, an Aβ16 variant Aβ16wwa was designed and synthesised. Phe4 and Tyr10 were each replaced with Trp and His14 was replaced with Ala. Upon excitation at 280 nm, the fluorescence emitted by Aβ16wwa at 360 nm was at least 100-fold more intense than that by Aβ16. Binding of paramagnetic Cu(II) to Aβ16wwa quenched this intense fluorescence sensitively. Consequently, the experimental concentration of P = Aβ16wwa could be varied significantly to allow a possible detection of the equilibrium of eqn (5). This, in turn, allowed analysis under the conditions imposed by eqn (6). To minimise potential interference of putative CuII–B complex(es) in eqn (5), the titration was conducted in a minimal concentration of MOPS or HEPES buffers, both of which exhibit a low affinity for Cu(II).
Quantification of Cu(II) binding via ligand competition
Reliable estimation of Cu(II) affinity by direct metal ion titration is restricted by the pre-condition KIID(1 + KA[B]) ∼ [P] of eqn (6) and is subject to further uncertainties associated with the putative term KA[B] that cannot be evaluated reliably for most proton buffers. To lift the restriction (difficult to satisfy for KIID values much less than the experimental [P]) and to suppress the uncertainties, the Cu(II) buffer glycine (Gly; KA1 = 1.17 × 106 M−1 and KA2 = 6.76 × 104 M−1 at pH 7.4)12,30 was used to control the Cu(II) speciation (eqn (7) and (8)). Data was processed according to eqn (9) and (10):12| [CuII(Gly)+] + [CuII(Gly)2] + 2P ⇌ 2CuII–P + 3Gly | (7) |
| [Cu(II)]tot = [CuII(Gly)+] + [CuII(Gly)2] + [CuII–P] | (8) |
 | (9) |
 | (10) |
This approach was applied to determine KIID for the probe Aβ16wwa via quenching of its intense fluorescence upon Cu(II) binding. Typically, a series of solutions in MOPS buffer (10 mM, pH 7.4) were prepared that contained Aβ16wwa (2.0 μM), Cu(II) (1.6 μM; 0.8 equiv.) and increasing concentrations of Gly. The fluorescence intensities at 360 nm (F) of the solutions increased with increasing concentrations of Gly and reached a constant value (Fo) that was >95% that of a control containing Aβ16wwa (2.0 μM) only (see Fig. 6a below). The behaviour indicated that addition of Gly induced the reverse reaction of eqn (7) and that there is little inner filter effect under the conditions of low Cu(II) concentration. Consequently, [CuII–P] = [CuII–Aβ16wwa] may be estimated from eqn (11) and then [Cuaq2+] and KD from eqn (9) and (10):
 | (11) |
An additional estimate of KIID for Aβ16wwa was made by application of an independent Cu(II) competing ligand nitrilotriacetic acid (NTA). The details are given in the ESI† and Fig. S3.
Quantification of Cu(II)-binding to selected Aβ16 peptides with probe Aβ16wwa
The Cu(II) affinities of Aβ16 and its variants were determined conveniently based on competition for Cu(II) with the probe peptide Aβ16wwa:| Aβ16 + CuII–Aβ16wwa ⇌ CuII–Aβ16 + Aβ16wwa | (12) |
 | (13) |
At molar ratios Aβ16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Aβ16wwa < 3, the fluorescence intensity at 360 nm is dominated by that of Aβ16wwa with negligible contribution from Aβ16 (see Fig. 5a below). Consequently, the term [CuII–P] = CuII–Aβ16 may be obtained from eqn (11) and then all other terms in eqn (12) from a mass balance at each known total concentration. This allows definition of KIID for Aβ16 relative to that of Aβ16wwa according to eqn (13).
Aβ16wwa < 3, the fluorescence intensity at 360 nm is dominated by that of Aβ16wwa with negligible contribution from Aβ16 (see Fig. 5a below). Consequently, the term [CuII–P] = CuII–Aβ16 may be obtained from eqn (11) and then all other terms in eqn (12) from a mass balance at each known total concentration. This allows definition of KIID for Aβ16 relative to that of Aβ16wwa according to eqn (13).
The experiments were conducted as two types of complementary titrations. In the first, fluorescence intensity was quenched by titration of aliquots of CuSO4 (10 μL, 80 μM) into a 2.0 mL solution in MOPS (10 mM, pH 7.4) containing either Aβ16wwa (2.0 μM) alone or both Aβ16wwa and the target peptide (each 2.0 μM). In the second, fluorescence intensity was recovered by titration of aliquots of the target peptide (4.0 μL, 500 μM) into a 2.0 mL solution in MOPS (10 mM, pH 7.4) containing Aβ16wwa (2.0 μM) and Cu(II) (1.6 μM). The Cu(II) speciation was analysed viaeqn (11) in both cases.
Catalytic aerobic oxidation of ascorbate and generation of H2O2
Generation of H2O2 by catalytic aerobic oxidation of Asc was monitored by UV-Vis spectroscopy via an assay based upon the dye Amplex Red31 using an experimental procedure described recently.32 The solution conditions for a typical reaction were: [Asc]tot ∼ 50 μM, [Amplex Red]tot ∼ 45 μM; [HRP] ∼ 0.35 U mL−1; [Cu]tot = 5.0 μM (if added) and [ligand]tot = 7.0 μM. The reactions were started by addition of Asc into a solution containing all other components in air-saturated MOPS buffer (50 mM, pH 7.4). Spectral changes were recorded at intervals of 50 s. A control solution without Asc served for baseline correction. The initial absorbance observed at 265 nm was sensitive to the reaction rate and solution composition. Use of Cuaq2+ as catalyst led to considerably lower initial absorbance than the other test solutions due to its higher relative reaction rate.The apparent reduction potentials of the copper centres in various Cu–Aβ16 complexes were estimated from the relative affinities of the peptides for Cu(I) and Cu(II) via the Nernst relationship of eqn (14):
 | (14) |
Results and discussion
Aβ16 binds Cu(I) with sub-nanomolar affinity
The chromogenic ligands Fs (Fig. 1a, inset) and Fz have been developed as quantitative probes for weaker Cu(I) binding.26 When in excess, both react quantitatively to yield well-defined complex anions [CuI(Fs)2]3− (λmax = 484 nm; ε = 6700 cm−1 M−1; β2 = 1013.7 M−2) and [CuI(Fz)2]3− (λmax = 470 nm, ε = 4320 cm−1 M−1; β2 = 1015.1 M−2). This enables them to buffer free Cuaq+ concentrations (expressed hereafter as pCu+ = −log[Cuaq+]) in the respective ranges pCu+ = 8–12 and 10–14. However, the complexes are air-sensitive and subject to substitution by other weak Cu(I) ligands, especially for [CuI(Fs)2]3−. Consequently, all reactions must be performed under anaerobic conditions in MOPS buffer containing reductants NH2OH and/or Asc with exclusion of all other potential Cu(I) ligands such as Cl−.26 | ||
Fig. 1 Quantification of Cu(I) binding to the Aβ16 peptide in buffer A. (a) Changes in absorbance of [CuI(Fs)2]3− solution ([Cu]tot = 30 μM; [Fs]tot = 70 μM) with increasing concentration of Aβ16 peptide. The structure of the Fs ligand is shown in the inset; (b, c) variation of concentration of [CuI(Fs)2]3− with increasing molar ratio Aβ16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu in Cuaq+ buffer of initial p[Cu+] = 8.2 ([Cu]tot = 30 μM; [Fs]tot = 70 μM) in (b) and initial p[Cu+] = 10.3 ([Cu]tot = 31 μM; [Fs]tot = 180 μM) in (c). The experimental data in solid circles were derived from the solutions prepared as described in the Experimental section. The data in empty circles was obtained after 1 Cu in Cuaq+ buffer of initial p[Cu+] = 8.2 ([Cu]tot = 30 μM; [Fs]tot = 70 μM) in (b) and initial p[Cu+] = 10.3 ([Cu]tot = 31 μM; [Fs]tot = 180 μM) in (c). The experimental data in solid circles were derived from the solutions prepared as described in the Experimental section. The data in empty circles was obtained after 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of the original solutions with proton buffer A. The two solid traces in (i, ii) are the fitting of the experimental data to eqn (2). The dotted traces in (ii) are the simple 1 1 dilution of the original solutions with proton buffer A. The two solid traces in (i, ii) are the fitting of the experimental data to eqn (2). The dotted traces in (ii) are the simple 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilutions of the data set (i). The two dashed lines intercepting Aβ16 1 dilutions of the data set (i). The two dashed lines intercepting Aβ16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Cu = 1.0 in (b) allowed estimation of the binding stoichiometry. Cu = 1.0 in (b) allowed estimation of the binding stoichiometry. | ||
A probe solution of [Cu]tot = 30 μM with a minimum molar ratio of Fs/Cu ∼ 2.3 (to ensure the dominance of [CuI(Fs)2]3−) was necessary to observe quantitative transfer of Cu(I) from [CuI(Fs)2]3− to the Aβ16 peptide (i.e., eqn (1) goes to completion; see Experimental section). The peptide has a relatively weak affinity for Cu(I) and, even in this solution buffered at pCu+ ∼ 8.2, it is only able to extract Cu(I) from [CuI(Fs)2]3− quantitatively for Aβ16/Cu < 0.4 (Fig. 1a and b). Linear extrapolation of the data at low Aβ16/Cu ratios demonstrates that Aβ16 possesses a single site of highest affinity for Cu(I).
Increasing [Fs]tot to 180 μM under otherwise identical conditions constrained the free Cuaq+ concentration to pCu+ ∼ 10.3 and imposed an effective competition for Cu(I) according to eqn (1) (Fig. 1c). Dilution of each equilibrium solution resulted in partial transfer of Cu(I) from the complex to the peptide to reach a new equilibrium position, as shown by comparison with the dotted traces in (ii) of Fig. 1b and c, derived from simple 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dilution of data sets (i). Eqn (2) (which assumes a single binding site) was used to fit the four sets of independent experimental data. This process allowed extraction of the equilibrium constant Kex = β2 × KID and estimation of KID = 10−10.4 M at pH 7.4. The estimate was the same for each data set, within experimental error.
1 dilution of data sets (i). Eqn (2) (which assumes a single binding site) was used to fit the four sets of independent experimental data. This process allowed extraction of the equilibrium constant Kex = β2 × KID and estimation of KID = 10−10.4 M at pH 7.4. The estimate was the same for each data set, within experimental error.
Ligand Fz has a higher affinity for Cu(I) and, even at the minimum allowable molar ratio of Fz/Cu ∼ 2.3, imposed an effective competition for Cu(I) between Fz and Aβ16. This allowed estimation of an indistinguishable KID = 10−10.4 M at pH 7.4. This work substantiates preliminary results reported previously,26 but contrasts considerably the other two earlier values (KID ∼ 10−7 M15 and 10−15 M14). The present value was determined relative to β2 = 1015.1 M−2 for [CuI(Fz)2]3− which, in turn, was derived based on β2 = 1017.2 M−2 for [CuI(Bca)2]3− (Bca: bicinchoninic anion).26,34 The latter value was consolidated recently by an independent study.35,36
At least two His are involved in Cu(I) binding to Aβ16 but the N-terminal amine is not
Spectroscopic examination has implicated the His sidechains of the Aβ16 peptide DAEFRHDSGYEVHHQK as Cu(I) ligands.21,23–25 The N-terminal amine and carboxylate sidechains may also be involved.The affinities of a number of variants of Aβ16 were investigated with probe Fs (Fig. 2 and Table 1). Probe Fz is less satisfactory in these systems as its affinity for Cu(I) is too high to allow effective competition. Substitution of any one of the three His residues with non-binding Ala reduced the affinities of the resultant peptides H6A, H13A and H14A by factors of between two and five. Removal of two His residues decreased the affinity of H6,13A and H13,14A by factors of >50 and >250, respectively. On the other hand, acetylation of the N-terminus (Ac-Aβ16) did not alter the affinity of the peptide for Cu(I). The Cu(I) affinity of the triple mutant probe peptide F4W,Y10W,H14A (i.e., Aβ16wwa) is indistinguishable from that of the single mutant H14A (Table 1).
| Peptide | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KID KID![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b b |
K ID/KID(wt) | pKa from curve fitting | Fitting R factor | Fitting curve in Fig. 3 | |
|---|---|---|---|---|---|---|
| Direct det. at pH 7.4 | From fitting pH variation | |||||
| a Unless otherwise indicated, the listed parameters were derived from curve-fitting of the experimental data to eqn (3) based on a CuI(His)2 site model with both KID and pKa as fitting parameters. b Estimated experimental errors ±0.05 unless indicated otherwise. c From curve-fitting to eqn (4) based on a CuI(His)3 site model with both KID and pKa as fitting parameters. d From curve-fitting to eqn (4) (a CuI(His)3 site model) with pKa = 6.5 fixed and KID as the only fitting parameter. e Approximate estimates only at the detection limit of Fs probe with large uncertainty. | ||||||
| Aβ16 | −10.4 | −10.5 | 1.0 | 6.5 | 0.99 | Black trace i |
| −10.4c | 6.1c | 0.97c | Red dotsc | |||
| −10.8d | 6.5 (fixed)d | 0.83d | Green dots | |||
| Ac-Aβ16 | −10.4 | −10.5 | 1.0 | 6.6 | 0.97 | ii |
| H6A | −10.0 | −10.1 | 2.5 | 6.5 | 0.99 | iii |
| H14A | −9.95 | −9.95 | 2.8 | 6.6 | 0.98 | iv |
| Aβ16wwa | −9.98 | — | 2.6 | — | — | — |
| H13A | −9.76 | −9.84 | 4.4 | 6.6 | 0.99 | v |
| H6,13A | >−8e | >250 | ||||
| H13,14A | >−8.7e | >50 | ||||
These experiments demonstrate that (i) at least two His sidechains are required for effective binding of Cu(I) but that all three His ligands contribute significantly; (ii) the N-terminal amine is not a Cu(I) ligand; (iii) substitution of residues not involved directly in Cu(I) binding (e.g., aromatics F4,Y10) has little impact on the affinity, consistent with the structural flexibility of the peptide.
Variation of KID with pH suggests that only two of the three His residues of Aβ16 are involved simultaneously in Cu(I) binding
The sensitivity of the derived KID values to the availability of the three His residues suggested that this parameter may be pH-dependent. His sidechains exhibit characteristic pKa values of 6.0–6.5 and an average pKa value of the three His sidechains in an Aβ28 peptide was estimated to be 6.53.28 Consequently, at pH < 7, protons may compete with Cu(I) for these sidechains.K ID for Aβ16 is sensitive to pH (black circles; Fig. 3a). With both KID and pKa as fitting parameters, the experimental data were fitted satisfactorily to eqn (3) assuming the availability of two His ligands only (black trace). Fitting to a model that assumes the availability of all three His ligands (eqn (4)) was less adequate (red dotted trace; Table 1). The essential difference is that the average pKa = 6.5 derived from the two-His model is the same as that determined independently by NMR28 while the value of 6.1 derived from the three-His model is significantly smaller (Table 1). However, when the parameter pKa was constrained to the experimental value 6.5 so that KID was the only fitting parameter, the curve fitting remained optimal for the two-His model but was unsatisfactory for the three-His model (Fig. 3a: black trace versus green dotted trace; Table 1).
 | ||
Fig. 3 Variation of pKID (=−log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KID) with solution pH: (a) comparison of curve-fittings of the experimental data for Aβ16-wt to eqn (3) (black solid trace) or eqn (4) (red dotted trace) or to eqn (4) with fixed input of pKa = 6.5 (green dotted trace); (b) curve-fittings of the experimental data to eqn (3) for: (i) Aβ16 (black dots & trace); (ii) Ac-Aβ16 (purple crosses & trace); (iii) H6A (red dots & trace); (iv) H14A (green dots & trace); (v) H13A (blue dots & trace). The fitting parameters in each case are given in Table 1. KID) with solution pH: (a) comparison of curve-fittings of the experimental data for Aβ16-wt to eqn (3) (black solid trace) or eqn (4) (red dotted trace) or to eqn (4) with fixed input of pKa = 6.5 (green dotted trace); (b) curve-fittings of the experimental data to eqn (3) for: (i) Aβ16 (black dots & trace); (ii) Ac-Aβ16 (purple crosses & trace); (iii) H6A (red dots & trace); (iv) H14A (green dots & trace); (v) H13A (blue dots & trace). The fitting parameters in each case are given in Table 1. | ||
Further support for the two-His model is provided by the equivalent experiments for the three Aβ16 variants H6A, H13A and H14A (Fig. 3b(iii)–(v)). Their KID values are also pH sensitive and the relationship in each case was described satisfactorily by the two-His model (eqn (3)) with both KID and pKa values derived from the curve-fitting matching those experimental values (Fig. 2 and Table 1). Each variant peptide features two His residues only and so can contribute a maximum of two His sidechains for Cu(I) binding.
The equivalent experiment for Ac-Aβ16 (acetylated at the N-terminus) generated similar results to those for Aβ16 itself (Fig. 3b, (i) vs. (ii); Table 1), although the former carries one less positive charge than does the native peptide. This supports the condition set previously for derivation of pH dependence viaeqn (3) and (4): protonation of other non-metal-binding sites has minimal impact on the observed KID. It also re-affirms that the N-terminus is not involved in Cu(I) binding.
Taken together, these experiments provide strong evidence that the Cu(I) site in Aβ16 includes two only of the three available His ligands. This conclusion is supported by X-ray absorption spectroscopy and theory.21,23,24,37 On the other hand, the data of Fig. 2 and 3 and Table 1 demonstrate that replacement of any one of the three His residues in Aβ16 led to a marginal decrease only in Cu(I) binding affinity (i.e., any two will do) but that replacement of any two His ligands disabled the binding site. When combined with the NMR study,21 the data are consistent with the presence of solution dynamic processes that exchange Cu(I) between the three available pairs of His ligands (Fig. 4). This dynamic nature proves to be important for redox cycling in the catalytic production of H2O2 (vide infra).
 | ||
| Fig. 4 Model of Cu(I) binding in Aβ16 peptide derived from the experimental data in Fig. 2 and 3. | ||
Development of a highly fluorescent peptide probe for estimation of Cu(II) affinities
The well-characterised Cu(II) ligand Gly has been used as an affinity standard to determine the Cu(II) affinities of Aβ peptides. Spectroscopic approaches have relied on the fluorescence emission of the single Tyr residue in Aβ peptides as the detection probe. However, both detection sensitivity and specificity are compromised by the relatively weak emission intensity of Tyr and interference from secondary Cu(II) binding sites.13A probe based upon the Aβ peptide was designed by replacement of aromatic residues Phe4 and Tyr10 with Trp to increase detection sensitivity and of His14 by Ala to suppress secondary Cu(II) binding. The resultant peptide Aβ16wwa exhibited excellent properties as a probe for quantification of Cu(II) binding to other Aβ peptides: (i) it binds either Cu(I) or Cu(II) with affinities indistinguishable from those of H14A (see Tables 1 and 2); (ii) it emits fluorescence (λex, 280 nm; λem 360 nm) that is an order of magnitude more intense than that of the Aβ16 peptide at 310 nm or more than two orders of magnitude more intense than that at 360 nm (Fig. 5a); (iii) at a concentration of 20 μM, it responds sensitively to Cu(II) binding with a distinct turning point at one equivalent of Cu(II) with evidence of further binding to a second equivalent (Fig. 5b, blue trace). In contrast, the response of Aβ16 to Cu(II) binding is much less sensitive and poorly defined with up to three equivalents of Cu(II) binding detectable under the same conditions (Fig. 5b, red trace).13
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIID for CuII–Aβ complexes estimated via ligand competitiona
KIID for CuII–Aβ complexes estimated via ligand competitiona
| Peptide | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIID KIID |
Affinity std. | Det. probe | Ref. |
|---|---|---|---|---|
| a Refer to ref. 13 for a summary of more extensive literature values estimated via various approaches. b Estimated at pH 7.6. c Estimated using N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) as both proton buffer and Cu(II) affinity standard. | ||||
| Aβ16wwa | −9.8 | Gly | Aβ16wwa | This work |
| −9.8 | NTA | Aβ16wwa | This work | |
| Aβ16 | −10.0 | Gly | Aβ16wwa | This work |
| Aβ16 | −10.0 | Gly | Tyr in Aβ | 13 |
| Aβ28 | −10.0b | Gly | Tyr in Aβ | 11 |
| Aβ42 | −10.2 | Gly | Tyr in Aβ | 11 |
| Aβ16/40 | −9.6 | Gly | ITC | 41 |
| Aβ16 | −9.0c | ACES | ITC | 42 |
| Aβ28 | −8.8c | ACES | ITC | 42 |
Quantification of Cu(II) binding to Aβ16wwa probe
Addition of bis(2-hydroxyethyl)amino-tris(hydroxymethyl)-methane (BisTris) (1.0 mM) into the MOPS buffer (50 mM; pH 7.4) for the Cu2+ titration eliminated Cu(II) binding to the weaker site in Aβ16wwa (20 μM; Fig. S1, ESI†) and induced competition between BisTris and the stronger peptide binding site for Cu(II) according to eqn (5) (P = Aβ16wwa; B = BisTris; KD = 10−5.2 M−1 for CuII-BisTris at pH 7.438 while MOPS has little Cu(II) affinity). After addition of one equivalent of Cu(II), >90% of total added Cu(II) was bound by the peptide. Consequently, an affinity of KIID < 10−9 M may be estimated from eqn (6) based on the above KD for CuII-BisTris.38 On the other hand, the high emission intensity of the Aβ16wwa probe allows its experimental concentration to be reduced significantly. At concentrations between 0.2 and 2.0 μM,39 Cu(II) binding to the weaker site was suppressed while that to the stronger site remained dominant (Fig. 5c). Even at the lowest peptide concentration of 0.20 μM, ≥94% of total added Cu(II) was estimated to be bound by the peptide after addition of one equivalent of Cu(II) titration. Consequently, an affinity of KIID ≤ 10−9.1 M may be estimated from eqn (6) without consideration of the possible contribution of Cu(II) binding to the MOPS buffer at 1.0 mM. However, in either case, the degree of complex formation was too high (>90%) to allow a reliable estimation of KIID.40These experiments demonstrate that: (i) the second binding site for Cu(II) in Aβ16wwa is relatively weak (KIID ≥ 10−6 M); (ii) direct metal ion titration defines the binding stoichiometry of Cu(II)![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Aβ16wwa = 1.0 for the stronger site but can provide an approximate value only for the binding affinity: KIID ≤ 10−9.1 M at its lowest experimental concentration of 0.2 μM. The detection sensitivity for the parent Aβ16 peptide is lower by a factor of ∼100, thereby setting estimation limit of its affinity to KIID < 10−7 M with high uncertainty. This provides an answer to the puzzle of why the Cu(II) affinities acquired in the past for Aβ peptides via direct metal ion titration were scattered so widely around KIID ∼ 10−7 M.11,13,20
Aβ16wwa = 1.0 for the stronger site but can provide an approximate value only for the binding affinity: KIID ≤ 10−9.1 M at its lowest experimental concentration of 0.2 μM. The detection sensitivity for the parent Aβ16 peptide is lower by a factor of ∼100, thereby setting estimation limit of its affinity to KIID < 10−7 M with high uncertainty. This provides an answer to the puzzle of why the Cu(II) affinities acquired in the past for Aβ peptides via direct metal ion titration were scattered so widely around KIID ∼ 10−7 M.11,13,20
It is apparent that the Cu(II) affinity of the Aβ16wwa probe is too high (i.e., KIID is too small) to be determined by direct metal ion titration and a ligand competition approach is required for reliable estimation.12 Glycine (Gly) and nitrilotriacetic acid (NTA) are two suitable competing ligands. Experimental results with Gly are described below and those with NTA are given in the ESI† and Fig. S3.
Gly binds Cu(II) to yield 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 complexes with formation constants KA1 = 106.07 and KA2 = 104.77 M−1 at pH 7.4.43 The fluorescence intensity of an Aβ16wwa solution (2.0 μM) in MOPS buffer (10 mM, pH 7.4) was quenched markedly by addition of Cu2+ (0.80 equiv.) but was recovered almost quantitatively (>95%) by titration of a large excess of Gly (>10 mM; Fig. 6a). This demonstrates that: (i) Cu(II) bound to the peptide can be removed reversibly; (ii) Gly imposes no discernable inner-filter effect; (iii) the inner-filter effect of CuII–Gly complex(es) at low concentrations (≤1.6 μM) is negligible. Consequently, at each point of the titration, the occupancy [CuII–P]/[P]tot can be estimated viaeqn (11) and the corresponding free Cuaq2+ concentration analysed viaeqn (7)–(9). Curve-fitting to eqn (10) led to KD = 10−9.8 M at pH 7.4 for CuII–Aβ16wwa. This data is supported by equivalent and independent experiment with NTA as a competing ligand that provided the same KIID = 10−9.8 M at pH 7.4 within the experimental error (see Table 2 and Fig. S3, ESI†).
2 complexes with formation constants KA1 = 106.07 and KA2 = 104.77 M−1 at pH 7.4.43 The fluorescence intensity of an Aβ16wwa solution (2.0 μM) in MOPS buffer (10 mM, pH 7.4) was quenched markedly by addition of Cu2+ (0.80 equiv.) but was recovered almost quantitatively (>95%) by titration of a large excess of Gly (>10 mM; Fig. 6a). This demonstrates that: (i) Cu(II) bound to the peptide can be removed reversibly; (ii) Gly imposes no discernable inner-filter effect; (iii) the inner-filter effect of CuII–Gly complex(es) at low concentrations (≤1.6 μM) is negligible. Consequently, at each point of the titration, the occupancy [CuII–P]/[P]tot can be estimated viaeqn (11) and the corresponding free Cuaq2+ concentration analysed viaeqn (7)–(9). Curve-fitting to eqn (10) led to KD = 10−9.8 M at pH 7.4 for CuII–Aβ16wwa. This data is supported by equivalent and independent experiment with NTA as a competing ligand that provided the same KIID = 10−9.8 M at pH 7.4 within the experimental error (see Table 2 and Fig. S3, ESI†).
 | ||
| Fig. 6 Determination of Cu(II) affinity of Aβ16wwa in MOPS buffer (10 mM, pH 7.4): (a) fluorescence spectra of Aβ16wwa (2.0 μM; blue trace); Aβ16wwa (2.0 μM) plus Cu(II) (1.6 μM) (green trace); Aβ16wwa (2.0 μM) plus Cu(II) (1.6 μM) plus ≥ 10 mM Gly (red trace); (b) recovery of fluorescence intensity for CuII0.8–Aβ16wwa (2.0 μM) with increasing Gly concentration; (c) curve fitting of [CuII–P]/[P]totversus log[Cuaq2+] to eqn (10) derived an estimate of KIID = 10−9.8 M for CuII–Aβ16wwa. | ||
Estimation of the affinities of Aβ16 peptides for Cu(II) using the Aβ16wwa probe
As the affinity of the probe peptide Aβ16wwa for Cu(II) is comparable to those of many other Aβ peptides, two complementary approaches based on eqn (12) and (13) were employed: (i) monitoring of fluorescence quenching by direct titration of Cuaq2+ into a solution containing Aβ16wwa and the target peptide in equimolar concentrations (2.0 μM) relative to a control that contained Aβ16wwa only (Fig. S4, ESI†); (ii) monitoring the fluorescence recovery of the probe by titration of the target peptide into a solution containing Aβ16wwa (2.0 μM) and 0.8 equiv. of Cu(II) (1.6 μM) (Fig. 7a and b). The Cu(II) speciation in eqn (12) and (13) may be analysed reliably viaeqn (11) as the fluorescence intensity of the probe at 360 nm is more than 100 fold greater than those Aβ peptides that contain a single Tyr residue only (Fig. 5a, Fig. S5a, ESI†). Approach (ii) may be compromised in cases where a large excess of target peptide is required to impose competition. Then, despite their low extinction coefficient around the excitation position (ε276 1410 M−1 cm−1) and low emission intensity at the detection position (360 nm), the target peptides may still impact on the observed fluorescence intensity when in large excess (Fig. S5b, ESI†). This follows from a combination of inner-filter and fluorescence effects of the target peptides. However, in the present systems, the impact was negligible when the target peptide was restricted to no more than two equivalents relative to Aβ16wwa (Fig. 5a, Fig. S5a, ESI†). | ||
| Fig. 7 Determination and comparison of Cu(II) dissociation constants for Aβ16 peptides using Aβ16wwa as a probe at pH 7.4 (a) and 9.0 (b): (i) Aβ16wt; (ii) H14A or H13A; (iii) H6A; (iv) H13,14A; (v) H6,13A; (vi) Ac-Aβ16. All experiments were conducted by titration of target peptide (4.0 μL, 500 μM) into a solution (2.0 mL) containing Aβ16wwa (2.0 μM) and Cu(II) (1.6 μM) in either MOPS buffer (10 mM, pH 7.4) or CHES buffer (10 mM, pH 9.0). The complete set of derived KIID values is given in Table 3. | ||
At pH 7.4, both approaches estimated KIID = 10−10.0 M for Aβ16 based on KIID = 10−9.8 M for Aβ16wwa. This value is in an excellent agreement with the recent consensus value of KD ∼ 10−10 M (Table 2).11,13 Substitution of any one of the three His residues by Ala decreased the affinity for Cu(II) marginally (by 2–3 fold; Fig. 7a and b, Fig. S4, ESI;†Table 3). In addition, KIID values for H14A and Aβ16wwa were the same within experimental error, confirming that removal of the aromatic residues has little impact on the copper binding chemistry. It appears that, as for the case of Cu(I), any two of the three His ligands can contribute to binding of Cu(II) and that a dynamic exchange equilibrium is present. However, the KIID value of H13,14A is identical to that of H6A but is three-fold smaller than that of H6,13A (Table 3). This is consistent with His6 playing a more important role than either His13 or His14 in Cu(II) binding. These observations are in agreement with a current Cu(II) binding model that suggests that His6 is an essential equatorial ligand while a second equatorial ligand is provided interchangeably by either His13 or His14 (Fig. 8).8 On the other hand, acetylation of the N-terminal nitrogen leads to a dramatic decrease of affinity by more than an order of magnitude (Fig. 7a and b, Fig. S4, ESI;†Table 3), demonstrating that the N-terminal nitrogen is another key Cu(II) ligand (Fig. 8). These data support previous analysis of relative affinities,11,42 but provide a sensitive and reliable basis for detection of these differences experimentally (Table 3).
| Aβ16 | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIID KIID |
K IID/KIID(wt) | Aβ28 | K IID/KIID(wt) | ||
|---|---|---|---|---|---|---|
| pH 7.4 | pH 7.4 | pH 9.0 | pH 7.4a | pH 7.8 | ||
| a Estimated using N-(2-acetamido)-2-aminoethanesulfonic acid (ACES) as both proton buffer (20 mM, pH 7.4) and Cu(II) affinity standard and ITC as detection probe. | ||||||
| wt | −10.0 | 1.0 | 1.0 | wt | 1.0 | 1.0 |
| Ac-Aβ16 | −8.3 | 50 | 10 | Ac-Aβ28 | 10.6 | 7.9 |
| H6A | −9.5 | 3.2 | 3.2 | H6A | 1.3 | 2.5 |
| H13A | −9.7 | 2.0 | 1.6 | H13A | 0.8 | 1.3 |
| H14A | −9.7 | 2.0 | 1.3 | H14A | 0.9 | 1.3 |
| Aβ16wwa | −9.8 | 1.6 | — | — | — | |
| H6,13A | −9.0 | 10 | 3.2 | — | — | |
| H13,14A | −9.5 | 3.2 | 1.6 | — | — | |
| Ref. | This work | This work | This work | 42 | 11 | |
 | ||
| Fig. 8 Equilibrium of Cu(II) binding modes in solution (pKa ∼ 7.8) without considering possible axial coordination.8 | ||
Analysis of the relative affinities for Cu(II) in CHES ((cyclohexylamino)ethanesulfonic acid) buffer at pH 9.0 provides a somewhat different story (Fig. 7b). Overall, His ligands and the N-terminal nitrogen appear to make lesser contributions to the Cu(II) binding than those at pH 7.4 although the influence of His6 remains unchanged (Fig. 7 and Table 3). This suggests that increasing pH promotes deprotonation of the Ala2 backbone amide for Cu(II) coordination and formation of the so-called component II (Fig. 8).8 Consistent with this model, variant peptides H13A, H14A and H13,14A all display affinities for CuII that are only marginally weaker than that of the wild-type peptide (Fig. 7b and Table 3). It appears that neither His13 nor His14 are crucial ligands in component II. In contrast, both H6A and H6,13A variants show a significant reduction in affinity (Fig. 7b and Table 3). Current models require only one His ligand for component II,8 and our data suggests that the identity of this ligand is His6 – perhaps due to its proximity to the N-terminal chelate ring. The significant loss of affinity upon acetylation indicates that the N-terminal nitrogen remains as an essential Cu(II) ligand at this pH. Thus the observed changes in the relative affinities of Aβ16 variants at pH 9.0 (Table 3) supports the proposed change in coordination environment in component II (Fig. 8), as observed by a variety of spectroscopic techniques44 including EPR, CD and NMR.16,45
Catalytic aerobic oxidation of ascorbate and generation of H2O2
A common feature in Alzheimer's disease is oxidative stress caused by reactive oxygen species (ROS). It has been proposed that one source is undesirable redox chemistry imposed by Cu bound to the disease proteins/peptides including, most importantly, the Aβ peptides.46 Ascorbic acid is an abundant physiological reductant in the central nerve system (CNS) and is important as a neuromodulator and/or neuroprotective agent in the brain.47,48 Its oxidation by dioxygen can produce H2O2 that, if uncontrolled, may undergo further reduction via Haber–Weiss and related reactions to generate the hydroxyl radical OH, a likely source of oxidative stress and inflammation.49 However, the oxidation is intrinsically slow and must be catalysed by redox-active couples such as CuII/CuI. The Cu ion bound in Aβ peptides has been demonstrated capable of assuming such a catalytic role.32,50 The present work has characterised the thermodynamic properties of a range of Cu centres in selected Aβ16 peptides and, in particular, compared their relative Cu(I) and Cu(II) binding affinities reliably under the same conditions. This provides an unprecedented opportunity for an integrated study to correlate these thermodynamic properties with their efficiencies for generation of H2O2via catalytic aerobic oxidation of Asc.The catalytic reaction was followed by UV-visible spectroscopy (Fig. 9).31 While the ‘free Cuaq2+ ion’ is a robust catalyst, the redox-inactive complex [CuII(EDTA)]2− was not (Fig. 9b and c(i) vs. (vii)). All metal-free Aβ16 peptides were catalytically inactive. As observed previously,32,50 binding of ‘free Cuaq2+ ion’ by Aβ16 diminishes but does not silence the catalytic activity of the Cu centre (Fig. 9b and c(v); Table 4). Overall, the catalytic activity decreases in the following order of ligand environments (relative to that for Aβ16-wt taken as unity):
| H2O (>4) > H6,13A ∼ H13,14A (2.8) > H13A ∼ H14A (1.5) > wt (1.0) > H6A (0.8) > Ac-Aβ16 (0.3) > EDTA (0) | (15) |
 | ||
Fig. 9 Catalytic aerobic oxidation of Asc and production of H2O2. (a) UV-Vis monitoring of Asc consumption and resorufin formation that monitors H2O2 production. The spectrum of initial solution containing all components except Asc was subtracted from each recorded spectrum; (b) monitoring of Asc consumption at 265 nm and (c) resorufin production at 571 nm (proportional to H2O2 production), in the presence of Cu (5.0 μM) and Cu ligand (7.0 μM). Ligands are: (i) EDTA, (ii) Ac-Aβ16; (iii) Aβ16wt plus Ac-Aβ16 (in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio); (iv) H6A; (v) Aβ16wt; (vi) H13A (indistinguishable from H14A); (vii) free Cu2+ (note: the lower end absorbance and thus less final resorufin production in (c) was due to more extensive extra consumption of Asc by the rapidly produced Amplex Red radicals; see ref. 31). Other initial reaction conditions: [Amplex Red] = 45 μM, [HRP] = 0.35 U mL−1, [Asc] = 50 μM. The reactions were conducted in air-saturated MOPS buffer (20 mM, pH 7.2–7.3) and started by introduction of catalyst. 1 molar ratio); (iv) H6A; (v) Aβ16wt; (vi) H13A (indistinguishable from H14A); (vii) free Cu2+ (note: the lower end absorbance and thus less final resorufin production in (c) was due to more extensive extra consumption of Asc by the rapidly produced Amplex Red radicals; see ref. 31). Other initial reaction conditions: [Amplex Red] = 45 μM, [HRP] = 0.35 U mL−1, [Asc] = 50 μM. The reactions were conducted in air-saturated MOPS buffer (20 mM, pH 7.2–7.3) and started by introduction of catalyst. | ||
Aerobic oxidation of ascorbate may be represented by two redox half-reactions: two-electron oxidation of Asc to dehydroascorbate (D-Asc) coupled to two-electron reduction of O2 to H2O2 (Fig. 10). Their respective reduction potentials at pH 7.0 are about +50 mV51 and +300 mV52 and hence the oxidation is a thermodynamically favored process. However, the reaction is very slow without a catalyst and the catalytic activity of a Cu centre depends on the efficiency of redox cycling between its CuI and CuII forms. This, in turn, is determined by both thermodynamic and kinetic factors. The molecular basis of the order of catalytic activity defined by eqn (15) may be analysed with regard to both factors.
| Aβ16 peptide | Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KID KID |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) KIID KIID |
(mV) (vs. SHE) | Relative catalytic rate | |
|---|---|---|---|---|---|
| Asc consumption | H2O2 production | ||||
| a All data were acquired in MOPS buffer (10–50 mM, pH 7.4). b Calculated from eqn (14) using Eo = +153 mV for the redox couple Cu2+/Cu+. c From ref. 15. d From ref. 14. e The consensus KIID = 10−10.0 M was used for the calculation. f Reactions were too fast to estimate the initial rates reliably. | |||||
| wt | −10.4 | −10.0 | +178 | 1.00 | 1.00 |
| ∼−7c | ∼−24e | ||||
| ∼−15d | ∼+448e | ||||
| Ac-Aβ16 | −10.4 | −8.3 | +277 | 0.37 | 0.34 |
| H6A | −10.0 | −9.5 | +183 | 0.75 | 0.78 |
| H13A | −9.76 | −9.7 | +157 | 1.42 | 1.51 |
| H14A | −9.95 | −9.7 | +168 | 1.42 | 1.52 |
| H6,13A | >−8 | −9.0 | <+94 | ∼2.8 | ∼2.7 |
| H13,14A | >−8.7 | −9.5 | <+106 | ∼2.8 | ∼2.7 |
| Cuaq2+ | +153 | >4f | >5f | ||
Thermodynamically, the reduction potential of a favoured catalyst must fall within the range +50 and +300 mV. ‘Free Cu2+ ion’ is a robust catalyst likely due to its reduction potential (+153 mV vs. SHE) falling about midway in this range and the presence of exchangeable aqua ligands only. The formal reduction potential of a copper centre is linked via the Nernst equation (eqn (14)) to the relative binding affinities of the different oxidation states. Both Cu(I) and Cu(II) affinities in selected Aβ16 peptides have been determined at the same pH = 7.4 in this work and are listed in Table 4. The calculated reduction potential for the copper centre in Aβ16 (Eo′ = +178 mV) closely matches an experimental value (E1/2 = +180 mV)21 determined by direct electrochemistry and predicts it to be a competent catalyst. In contrast, the same calculations based on log KID = −715 or −1514 and the consensus log KIID = −10.0 led to estimates of Eo′ = −24 and +448 mV, respectively. Neither predicts catalytic function. The Cu–Aβ complexes, indeed, catalyse the aerial oxidation of Asc effectively although less actively than does Cuaq2+. The structures of Cu(I) and Cu(II) complexes of the Aβ16 peptide are distinctly different: redox cycling will require energy input for structural reorganisation (Fig. 11). On the other hand, free Cu ions are under tight control in living cells and are present in tightly-bound forms only.
Acetylation of the N-terminal nitrogen has little impact on Cu(I) binding but removes a key Cu(II) ligand and consequently shifts the predicted reduction potential positively to a value (Eo′ = +277 mV) that promotes oxidation of Asc but does not favour reduction of O2. This is consistent with the catalytic activity of Ac-Aβ16 being ∼30% of that of Aβ16-wt (eqn (15)). Notably, addition of Cu2+ into an equimolar mixture of Aβ16 and Ac-Aβ16 produced an activity that was about the average of the combined activities of Aβ16 and Ac-Aβ16 (Fig. 9b and c; compare traces (ii), (iii) and (v)). The affinities of these two peptides for Cu(I) are identical but their affinities for Cu(II) differ considerably. These experiments suggest that the resting state of the catalyst in the presence of Asc is dominated by the Cu(I) form and that the catalytic activity of Cu-Aβ16 is suppressed by Ac-Aβ16. These results are also consistent with the properties of a copper centre bound to the second domain of the amyloid precursor protein (APP-D2): it exhibits similar affinities and catalytic activities to those of the copper centre in Ac-Aβ16.32
Interestingly, APP may be processed in vivo by two enzymes, the α/β secretases, in two different pathways (i.e., so called non-amyloidogenic and amyloidogenic pathways) to secrete two soluble forms of APP N-terminal fragments, sAPPα and sAPPβ. The former cleaved site is within the Aβ sequence between the position 16 and 17 whereas the latter is located right before the N-terminus of the Aβ sequence. Consequently, while both fragments contain Cu site in APP-D2, sAPPα differs from sAPPβ by having a C-terminal 16 amino acid extension equivalent to Ac-Aβ16 in term of Cu binding sites. Our experiments suggest that the Cu sites in APP-D2 and Ac-Aβ16 may be neuroprotective in a sense that ROS generation in the CNS by the Cu-Aβ catalyst may be suppressed partially by competitive Cu(I) binding with APP-D2 and Ac-Aβ16 (Fig. 9b and c, (iii) vs. (v)).32 Notably, it has been reported that sAPPα shows a range of neuroprotective and growth factor properties, including reduction of neuronal injury and improvement in memory performance, in contrast to the generally less potent sAPPβ.53–56
Intriguingly, the predicted reduction potential for the copper centre in each of the three single His variants H6A, H13A and H14A is similar to that in the original Aβ16 peptide. However, mutation on His6 decreased activity by >20% while mutation of either of the other two increased the activity by ∼50%, i.e., the copper centres in the H13A and H14A copper complexes are twice as active as that in H6A (Fig. 9 and Table 4). An electrochemical study has proposed a pre-organisation mechanism for electron exchange between the Cu(I) and Cu(II) forms.57 His6 is an important ligand for both Cu(I) and Cu(II) at pH 7.4 while either His13 and His14 can contribute but play a more important role in binding to Cu(I) than to Cu(II). The data suggests that retention of the His6 ligand is important for optimisation of the rate of electron exchange at pH 7.4 (Fig. 11).
Copper ions in the presence of the double His variants (H6,13A and H13,14) exhibit higher catalytic activities (Table 4). However, these data need to be interpreted cautiously, as the binding affinities of these variants are significantly weaker, in particular for Cu(I) (Table 4). The observed high activities may be related to a significant level of unbound or partially aquated Cu under the conditions. Addition of EDTA (KD = 10−15.9 M at pH 7.4) sequesters the copper into non-redox active CuII-EDTA that inhibits the catalytic activity completely.
Summary and concluding remarks
The Aβ peptides of 40–42 residues are the primary components of the extracellular senile plaques deposited in the AD brain and are proposed as a source of toxicity. The plaques are rich in transition metals Cu, Zn and Fe and the toxicity may be linked to oxidative stress induced by catalytic oxidation mediated via redox-active metal ions and copper ions in particular. The thermodynamic viability of a copper centre as a redox catalyst is linked to its reduction potential that, in turn, is determined by the relative stabilities of the two oxidation states (eqn (14)). However, these stabilities, as measured by dissociation constants KD (affinities) for Cu(I) and Cu(II) bound to Aβ peptides, have remained controversial, primarily due to a lack of reliable detection probes and affinity standards.All essential metal ligands in Aβ peptides are located within the first 16 residues and the fundamental Cu binding properties of Aβ16 peptides have proven to be representative of those of other longer Aβ peptides. This work undertook a systematic quantitative investigation of the Cu(I) and Cu(II) binding properties of various Aβ16 peptides by employing the Fs probe established recently for weaker Cu(I) binding26 and a new highly fluorescent probe Aβ16wwa introduced in this work for weaker Cu(II) binding. The key results are summarised following:
(i) Aβ16 binds Cu(I) in three exchangeable two-coordinate sites defined by two His ligands out of the total of three (Fig. 4). The apparent binding affinity is pH dependant at pH < 7.5 and is estimated to be KID = 10−10.4 M for wild type Aβ16 at pH 7.4. The N-terminal amine and backbone amides are not involved in Cu(I) binding.
(ii) The N-terminal nitrogen is a key Cu(II) ligand and appears to be part of a chelate ring ligand at pH 7.4 (Fig. 8). All three His ligands and at least one backbone amide are involved in Cu(II) binding but not simultaneously and only in several dynamic exchange modes (Fig. 8). His6 appears to play a more important role in Cu(II) binding than does either His13 or His14. The apparent binding affinity for wild type Aβ16 was estimated to be KIID = 10−10.0 M at pH 7.4, consolidating the consensus data reported in several recent studies.
(iii) The dissociation constants KID and KIID allow estimation of the formal reduction potential for the Cu–Aβ16 complex as Eo′ = 178 mV (vs. SHE).36 This value matches E1/2= 180 mV determined directly by cyclic voltammetry.21 Consequently, the complex is predicted to be a robust redox catalyst for oxidation of Asc (∼+50 mV) by dioxygen (∼+300 mV) to generate H2O2 and thus other ROS. Its catalytic activity is about 25% that of ‘free Cuaq2+’ (Eo′ = +153 mV), consistent with the distinct Cu(I) and Cu(II) binding modes present in Aβ16 and the consequent pre-organisation energy required for the redox switching.
These new thermodynamic data consolidate the structural interpretations for the Cu–Aβ complexes deduced previously by spectroscopic investigations and provide molecular insight into the mechanism of ROS production by copper chemistry and of oxidative stress in Alzheimer's disease.
Acknowledgements
We thank the Australian Research Council for financial support under Grant DP130100728.Notes and references
- R. B. Maccioni, J. P. Munoz and L. Barbeito, The molecular bases of Alzheimer's disease and other neurodegenerative disorders, Arch. Med. Res., 2001, 32, 367–381 CrossRef CAS.
- J. Hardy and D. J. Selkoe, The amyloid hypothesis of Alzheimer's disease: progress and problems on the road to therapeutics, Science, 2002, 297, 353–356 CrossRef CAS PubMed.
- R. Cappai and K. J. Barnham, Delineating the mechanism of Alzheimer's disease A beta peptide neurotoxicity, Neurochem. Res., 2008, 33, 526–532 CrossRef CAS PubMed.
- D. M. Holtzman, J. C. Morris and A. M. Goate, Alzheimer's disease: the challenge of the second century, Sci. Transl. Med., 2011, 3, 77sr1 Search PubMed.
- G. Multhaup and C. L. Masters, Metal binding and radical generation of proteins in human neurological diseases and aging, Met. Ions Biol. Syst., 1999, 36, 365–387 CAS.
- A. I. Bush, The metallobiology of Alzheimer's disease, Trends Neurosci., 2003, 26, 207–214 CrossRef CAS.
- D. G. Smith, R. Cappai and K. J. Barnham, The redox chemistry of the Alzheimer's disease amyloid [beta] peptide, Biochim. Biophys. Acta, 2007, 1768, 1976–1990 CrossRef CAS PubMed.
- C. Hureau, Coordination of redox active metal ions to the amyloid precursor protein and to amyloid-β peptides involved in Alzheimer disease. Part 1: An overview, Coord. Chem. Rev., 2012, 256, 2164–2174 CrossRef CAS PubMed.
- I. Zawisza, M. Rózga and W. Bal, Affinity of copper and zinc ions to proteins and peptides related to neurodegenerative conditions (Aβ, APP, α-synuclein, PrP), Coord. Chem. Rev., 2012, 256, 2297–2307 CrossRef CAS PubMed.
- J. H. Viles, Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer's, Parkinson's and prion diseases, Coord. Chem. Rev., 2012, 256, 2271–2284 CrossRef CAS PubMed.
- C. J. Sarell, C. D. Syme, S. E. Rigby and J. H. Viles, Copper(II) binding to amyloid-beta fibrils of Alzheimer's disease reveals a picomolar affinity: stoichiometry and coordination geometry are independent of Abeta oligomeric form, Biochemistry, 2009, 48, 4388–4402 CrossRef CAS PubMed.
- Z. Xiao and A. G. Wedd, The challenges of determining metal-protein affinities, Nat. Prod. Rep., 2010, 27, 768–789 RSC.
- B. Alies, E. Renaglia, M. Rózga, W. Bal, P. Faller and C. Hureau, Cu(II) Affinity for the Alzheimer's Peptide: Tyrosine Fluorescence Studies Revisited, Anal. Chem., 2013, 85, 1501–1508 CrossRef CAS PubMed.
- H. A. Feaga, R. C. Maduka, M. N. Foster and V. A. Szalai, Affinity of Cu(I) for the Copper-Binding Domain of the Amyloid-β Peptide of Alzheimer's Disease, Inorg. Chem., 2011, 50, 1614–1618 CrossRef CAS PubMed.
- B. Alies, B. Badei, P. Faller and C. Hureau, Reevaluation of Copper(I) Affinity for Amyloid-beta Peptides by Competition with Ferrozine-An Unusual Copper(I) Indicator, Chem.–Eur. J., 2012, 18, 1161–1167 CrossRef CAS PubMed.
- C. D. Syme, R. C. Nadal, S. E. Rigby and J. H. Viles, Copper binding to the amyloid-beta (Abeta) peptide associated with Alzheimer's disease: folding, coordination geometry, pH dependence, stoichiometry, and affinity of Abeta-(1-28): insights from a range of complementary spectroscopic techniques, J. Biol. Chem., 2004, 279, 18169–18177 CrossRef CAS PubMed.
- J. W. Karr and V. A. Szalai, Cu(II) binding to monomeric, oligomeric, and fibrillar forms of the Alzheimer's disease amyloid-beta peptide, Biochemistry, 2008, 47, 5006–5016 CrossRef CAS PubMed.
- S. C. Drew, C. L. Masters and K. J. Barnham, Alanine-2 carbonyl is an oxygen ligand in Cu2+ coordination of Alzheimer's disease amyloid-beta peptide – relevance to N-terminally truncated forms, J. Am. Chem. Soc., 2009, 131, 8760–8761 CrossRef CAS PubMed.
- S. C. Drew, C. J. Noble, C. L. Masters, G. R. Hanson and K. J. Barnham, Pleomorphic copper coordination by Alzheimer's disease amyloid-beta peptide, J. Am. Chem. Soc., 2009, 131, 1195–1207 CrossRef CAS PubMed.
- P. Faller and C. Hureau, Bioinorganic chemistry of copper and zinc ions coordinated to amyloid-[small beta] peptide, Dalton Trans., 2009, 1080–1094 RSC.
- C. Hureau, V. Balland, Y. Coppel, P. L. Solari, E. Fonda and P. Faller, Importance of dynamical processes in the coordination chemistry and redox conversion of copper amyloid-beta complexes, JBIC, J. Biol. Inorg. Chem., 2009, 14, 995–1000 CrossRef CAS PubMed.
- P. Dorlet, S. Gambarelli, P. Faller and C. Hureau, Pulse EPR spectroscopy reveals the coordination sphere of copper(II) ions in the 1-16 amyloid-beta peptide: a key role of the first two N-terminus residues, Angew. Chem., Int. Ed., 2009, 48, 9273–9276 CrossRef CAS PubMed.
- R. A. Himes, G. Y. Park, G. S. Siluvai, N. J. Blackburn and K. D. Karlin, Structural Studies of Copper(I) Complexes of Amyloid-β Peptide Fragments: Formation of Two-Coordinate Bis(histidine) Complexes, Angew. Chem., Int. Ed., 2008, 47, 9084–9087 CrossRef CAS PubMed.
- J. Shearer and V. A. Szalai, The Amyloid-β Peptide of Alzheimer's Disease Binds CuI in a Linear Bis-His Coordination Environment: Insight into a Possible Neuroprotective Mechanism for the Amyloid-β Peptide, J. Am. Chem. Soc., 2008, 130, 17826–17835 CrossRef CAS PubMed.
- C. Hureau, Y. Coppel, P. Dorlet, P. L. Solari, S. Sayen, E. Guillon, L. Sabater and P. Faller, Deprotonation of the Asp1-Ala2 peptide bond induces modification of the dynamic copper(II) environment in the amyloid-beta peptide near physiological pH, Angew. Chem., Int. Ed., 2009, 48, 9522–9525 CrossRef CAS PubMed.
- Z. Xiao, L. Gottschlich, R. van der Meulen, S. R. Udagedara and A. G. Wedd, Evaluation of quantitative probes for weaker Cu(i) binding sites completes a set of four capable of detecting Cu(i) affinities from nanomolar to attomolar, Metallomics, 2013, 5, 501–513 RSC.
- M.-R. Ash, L. X. Chong, M. J. Maher, M. G. Hinds, Z. Xiao and A. G. Wedd, Molecular Basis of the Cooperative Binding of Cu(I) and Cu(II) to the CopK Protein from Cupriavidus metallidurans CH34, Biochemistry, 2011, 50, 9237–9247 CrossRef CAS PubMed.
- K. Ma, E. L. Clancy, Y. Zhang, D. G. Ray, K. Wollenberg and M. G. Zagorski, Residue-Specific pKa Measurements of the β-Peptide and Mechanism of pH-Induced Amyloid Formation, J. Am. Chem. Soc., 1999, 121, 8698–8706 CrossRef CAS.
- O. Seneque and J. M. Latour, Coordination properties of zinc finger peptides revisited: ligand competition studies reveal higher affinities for zinc and cobalt, J. Am. Chem. Soc., 2010, 132, 17760–17774 CrossRef CAS PubMed.
- A. Albert and E. P. Serjeant, The Determination of Ionisation Constants- A Laboratory Manual, Cambridge University Press, London, 1984 Search PubMed.
- J. V. Rodrigues and C. M. Gomes, Enhanced superoxide and hydrogen peroxide detection in biological assays, Free Radical Biol. Med., 2010, 49, 61–66 CrossRef CAS PubMed.
- S. L. Leong, T. R. Young, K. J. Barnham, A. G. Wedd, M. G. Hinds, Z. Xiao and R. Cappai, Quantification of Copper Binding to Amyloid Precursor Protein Domain 2 and its Caenorhabditis elegans Ortholog. Implications for Biological Function, Metallomics, 2014, 6, 105–116 RSC.
- G. Milazzo and S. Caroli, Tables of Standard Electrode Potentials, Wiley, New York, 1978 Search PubMed.
- Z. Xiao, J. Brose, S. Schimo, S. M. Ackland, S. La Fontaine and A. G. Wedd, Unification of the copper(I) binding affinities of the metallo-chaperones Atx1, Atox1 and related proteins: detection probes and affinity standards, J. Biol. Chem., 2011, 286, 11047–11055 CrossRef CAS PubMed.
- P. Bagchi, M. T. Morgan, J. Bacsa and C. J. Fahrni, Robust Affinity Standards for Cu(I) Biochemistry, J. Am. Chem. Soc., 2013, 135, 18549–18559 CrossRef CAS PubMed.
- The minor difference in log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) β2 for [CuI(Bca)2]3− determined in ref. 34 and 35 (17.2 versus 17.7) was primarily due to adoption of different standard reduction potentials Eo for the couple Cu2+/Cu+ (+164 mV versus +130 mV). Such minor difference will be passed on to the Cu(I) affinities determined based on different potential standards and thus the reduction potential back-estimated viaeqn (14) from KID and KIID unless the same standard reduction potential (+164 mV or +130 mV) is used. For consistency, we choose to adopt the IUPAC value Eo = +153 mV for all applications and calculations.
β2 for [CuI(Bca)2]3− determined in ref. 34 and 35 (17.2 versus 17.7) was primarily due to adoption of different standard reduction potentials Eo for the couple Cu2+/Cu+ (+164 mV versus +130 mV). Such minor difference will be passed on to the Cu(I) affinities determined based on different potential standards and thus the reduction potential back-estimated viaeqn (14) from KID and KIID unless the same standard reduction potential (+164 mV or +130 mV) is used. For consistency, we choose to adopt the IUPAC value Eo = +153 mV for all applications and calculations. - S. Furlan, C. Hureau, P. Faller and G. La Penna, Modeling the Cu+ binding in the 1-16 region of the amyloid-beta peptide involved in Alzheimer's disease, J. Phys. Chem. B, 2010, 114, 15119–15133 CrossRef CAS PubMed.
- K. H. Scheller, T. H. J. Abel, P. E. Polanyi, P. K. Wenk, B. E. Fischer and H. Sigel, Metal Ion/Buffer Interactions, Eur. J. Biochem., 1980, 107, 455–466 CrossRef CAS.
- A background correction was necessary for Aβ16wwa solutions of 0.20 μM; see Fig. S2, ESI†.
- L. D. Hansen, G. W. Fellingham and D. J. Russell, Simultaneous determination of equilibrium constants and enthalpy changes by titration calorimetry: Methods, instruments, and uncertainties, Anal. Biochem., 2011, 409, 220–229 CrossRef CAS PubMed.
- L. Q. Hatcher, L. Hong, W. D. Bush, T. Carducci and J. D. Simon, Quantification of the binding constant of copper(II) to the amyloid-beta peptide, J. Phys. Chem. B, 2008, 112, 8160–8164 CrossRef CAS PubMed.
- C. Sacco, R. A. Skowronsky, S. Gade, J. M. Kenney and A. M. Spuches, Calorimetric investigation of copper(II) binding to Abeta peptides: thermodynamics of coordination plasticity, JBIC, J. Biol. Inorg. Chem., 2012, 17, 531–541 CrossRef CAS PubMed.
- R. M. Smith, NIST Critically Selected Stability Constants of Metal Complexes, 2007, vol. Version 8.0 Search PubMed.
- C. Hureau and P. Dorlet, Coordination of redox active metal ions to the amyloid precursor protein and to amyloid-β peptides involved in Alzheimer disease. Part 2: Dependence of Cu(II) binding sites with Aβ sequences, Coord. Chem. Rev., 2012, 256, 2175–2187 CrossRef CAS PubMed.
- G. J. Brewer, The Risks of Copper Toxicity Contributing to Cognitive Decline in the Aging Population and to Alzheimer's Disease, J. Am. Coll. Nutr., 2009, 28, 238–242 CrossRef.
- G. Eskici and P. H. Axelsen, Copper and Oxidative Stress in the Pathogenesis of Alzheimer's Disease, Biochemistry, 2012, 51, 6289–6311 CrossRef CAS PubMed.
- R. A. Grünewald, Ascorbic acid in the brain, Brain Res. Rev., 1993, 18, 123–133 CrossRef.
- M. E. Rice, Ascorbate regulation and its neuroprotective role in the brain, Trends Neurosci., 2000, 23, 209–216 CrossRef CAS.
- X. Zhu, B. Su, X. Wang, M. A. Smith and G. Perry, Causes of oxidative stress in Alzheimer disease, Cell. Mol. Life Sci., 2007, 64, 2202–2210 CrossRef CAS PubMed.
- B. Alies, I. Sasaki, O. Proux, S. Sayen, E. Guillon, P. Faller and C. Hureau, Zn impacts Cu coordination to amyloid-[small beta], the Alzheimer's peptide, but not the ROS production and the associated cell toxicity, Chem. Commun., 2013, 49, 1214–1216 RSC.
- B. E. Conway, Electrochemical Data, Greenwood Press, New York, 1969 Search PubMed.
- D. L. Nelson and M. M. Cox, in Lehninger Principles of Biochemistry, ed. W. H. Freeman, New York, 2004 Search PubMed.
- S. W. Barger and M. P. Mattson, Induction of neuroprotective kappa B-dependent transcription by secreted forms of the Alzheimer's beta-amyloid precursor, Mol. Brain Res., 1996, 40, 116–126 CrossRef CAS.
- L. Mucke, C. R. Abraham and E. Masliah, Neurotrophic and Neuroprotective Effects of hAPP in Transgenic Micea, Ann. N. Y. Acad. Sci., 1996, 777, 82–88 CrossRef CAS.
- E. Thornton, R. Vink, P. C. Blumbergs and C. Van Den Heuvel, Soluble amyloid precursor protein α reduces neuronal injury and improves functional outcome following diffuse traumatic brain injury in rats, Brain Res., 2006, 1094, 38–46 CrossRef CAS PubMed.
- C. J. Taylor, D. R. Ireland, I. Ballagh, K. Bourne, N. M. Marechal, P. R. Turner, D. K. Bilkey, W. P. Tate and W. C. Abraham, Endogenous secreted amyloid precursor protein-α regulates hippocampal NMDA receptor function, long-term potentiation and spatial memory, Neurobiol. Dis., 2008, 31, 250–260 CrossRef CAS PubMed.
- V. Balland, C. Hureau and J.-M. Savéant, Electrochemical and homogeneous electron transfers to the Alzheimer amyloid-β copper complex follow a preorganization mechanism, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 17113–17118 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1–S6 and Table S1. See DOI: 10.1039/c4mt00001c |
| ‡ Occupational trainee from Ludwig-Maximilian University, Munich, Germany. |
| This journal is © The Royal Society of Chemistry 2014 |





