Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quantitative proteomics identifies unanticipated regulators of nitrogen- and glucose starvation†
Steven V.
Rødkær
,
Dennis
Pultz
,
Michelle
Brusch
,
Martin V.
Bennetzen
,
Lasse G.
Falkenby
,
Jens S.
Andersen
and
Nils J.
Færgeman
*
Villum Center for Bioanalytical Sciences, Department of Biochemistry and Molecular Biology, University of Southern Denmark, Campusvej 55, DK-5230 Odense M, Denmark. E-mail: nils.f@bmb.sdu.dk; Fax: +45 6550 2467; Tel: +45 6550 2453
First published on 23rd May 2014
Abstract
The molecular mechanisms underlying how cells sense, respond, and adapt to alterations in nutrient availability have been studied extensively during the past years. While most of these studies have focused on the linear connections between signaling components, it is increasingly being recognized that signaling pathways are interlinked in molecular circuits and networks such that any metabolic perturbation will induce signaling-wide ripple effects. In the present study, we have used quantitative mass spectrometry (MS) to examine how the yeast Saccharomyces cerevisiae responds to nitrogen- or glucose starvation. We identify nearly 1400 phosphorylation sites of which more than 500 are regulated in a temporal manner in response to glucose- or nitrogen starvation. By bioinformatics and network analyses, we have identified the cyclin-dependent kinase (CDK) inhibitor Sic1, the Hsp90 co-chaperone Cdc37, and the Hsp90 isoform Hsp82 to putatively mediate some of the starvation responses. Consistently, quantitative expression analyses showed that Sic1, Cdc37, and Hsp82 are required for normal expression of nutrient-responsive genes. Collectively, we therefore propose that Sic1, Cdc37, and Hsp82 may orchestrate parts of the cellular starvation response by regulating transcription factor- and kinase activities.
Introduction
One of the most fundamental challenges for all cells and organisms is to sense and respond to the availability of nutrients in the environment. This is achieved by modulating their metabolic pathways in a coordinated manner to maintain cellular homeostasis, achieve balanced growth, and to promote survival. Import, synthesis, and metabolism of a large variety of metabolites and macromolecules must therefore be tightly coordinated to meet the nutritional requirements of cells and organisms. Eukaryotic cells, including the yeast Saccharomyces cerevisiae (S. cerevisiae), have developed numerous signaling pathways for sensing and responding to the availability of carbon sources, nitrogen sources, amino acids, and other nutrients.1 In the yeast S. cerevisiae, the RAS/cAMP-dependent protein kinase A (PKA), AMP-dependent protein kinase (Snf1), Sch9, and Target Of Rapamycin 1 (TOR1) signaling pathways have all been identified as central regulators of nutritional responses to subsequently coordinate metabolic activities. Specifically, the availability of nitrogen affects the activity of the TOR complex 1 (TORC1), which via modulation of Tap42-PP2A phosphatase activity regulates the expression of the nitrogen discrimination pathway- and retrograde signaling genes.2 TORC1 also regulates the expression of stress-responsive- as well as post-diauxic shift/G0 genes via not only PP2A activity but also the yeast PKB homologue, Sch9.3 By regulating the activity of Tap42-PP2A, TORC1 also affects the activity of the transcription factor Gcn4 to modulate transcription of genes involved in amino acid- and purine biosynthesis.4 Finally, inactivation of TORC1 activity induces autophagy, which is further abrogated by PKA- and Sch9 inactivation, arguing that PKA/Sch9 act in parallel to TORC1.5In yeast, the plasma membrane G-protein coupled receptor, Gpr1, and the small GTP-binding proteins Ras1 and Ras2 have previously been proposed to sense the availability of glucose to subsequently modulate cAMP synthesis and hence PKA activity.6,7 In S. cerevisiae TPK1, TPK2, and TPK3 encode closely related catalytic subunits of PKA,8 which when activated inhibits autophagy9 and the expression of stationary phase- and stress-responsive genes.10,11
In response to glucose deprivation, the Snf1 protein kinase, the yeast homologue of the mammalian AMP-activated protein kinase (AMPK), is activated. When activated, Snf1 mediates induction of genes involved in fatty acid β-oxidation and carboxylic acid metabolism via the transcription factor Adr1, as well as genes involved in gluconeogenesis via the transcriptional activator Cat8 and the transcriptional repressor Mig1.12–14 Oppositely, Snf1 inhibits the expression of hexokinase and hexose transporters by promoting phosphorylation of the transcription factor Rgt1.15
Thus, the mechanisms by which S. cerevisiae senses and responds to alterations in nutrient availability and subsequently coordinates the metabolic adaptations to nutrient availability have been widely studied during the past decades. However, the extent to which these mechanisms are dynamically regulated at the molecular level and how these mechanisms are interconnected in molecular circuits and networks still remain elusive. In the present work, we have applied quantitative phosphoproteomics in an effort to understand how S. cerevisiae responds to glucose- and/or nitrogen starvation. In particular, we have directed the present analyses towards the identification of proteins, which are associated to many of the regulatory events and hence potentially involved in proteome-wide regulation. By bioinformatics and downstream biochemical analyses, we identify a number of novel and unanticipated regulators including the co-chaperone Cdc37, chaperone Hsp82, and cyclin-dependent kinase inhibitor Sic1. Our results show that these regulators are required for normal regulation of nutrient-responsive gene expression, suggesting that they potentially serve to coordinate starvation responses.
Materials and methods
Yeast strains and growth conditions
The S. cerevisiae strains used in this study were: the arginine- and lysine auxotrophic strain YAL6b described previously16 was used for the present phosphoproteomic studies. The parental strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) and the corresponding deletion mutants were obtained from Euroscarf (Frankfurt, Germany). The temperature-sensitive cdc37-1/AS146-26b (MATα cdc37-1 ura3-52 leu2-3, 112 lys2) and the corresponding wild-type/WX257-5d were obtained from Mark Winey (University of Colorado at Boulder, Boulder CO, USA) and have been described before.17 Yeast strains were kept on yeast extract peptone dextrose (YPD) or yeast nitrogen base dextrose (YNBD) plates made as described previously.18Sample preparation and mass spectrometry-based analysis
For quantitative phosphoproteomics, the lysine and arginine double auxotrophic strain YAL6B was grown and labeled as described previously.16 Briefly, YAL6B was grown in YPD at 30 °C/200 rpm for 24 hours (ESI,† Fig. S1). Subsequently, cells were diluted 6000-times into YNBD media containing different combinations of arginine- and lysine isotopes; unlabeled arginine/lysine (light), [2H4]lysine/[13C6]arginine (medium), or [13C6,15N2]lysine/[13C6,15N4]arginine (heavy) (Cambridge Isotope Laboratories) and grown to an OD600 of approximately 1. Aliquots of cells (50 OD units) were harvested by gentle centrifugation (1200 × g/5 min/4 °C) and subsequently resuspended in either YNB (for glucose starvation), YNBD without amino acids and NH4(SO2)4 (for nitrogen starvation), or YNB without amino acids and NH4(SO2)4 (for glucose and nitrogen starvation) media in the original volume and incubated for one hour at 30 °C at 200 rpm. Just prior to harvesting, trichloricacid (TCA) was added to 0.5 M and cells were pelleted by centrifugation as described above. An equal number of untreated cells were mixed with starved cells and prepared for MS analysis on a LTQ Orbitrap Velos (Thermo Fisher) as previously described.19 In a similar manner, cells were labeled and starved for glucose for 5 or 15 minutes and subsequently harvested and prepared for mass spectrometry analysis (Fig. S1, ESI†).Data analyses
The MS raw data was processed in the MaxQuant software20 with an initial peptide identification by the MASCOT v. 2.2 search engine.21 A minimum MASCOT score of 7 was required for the identified proteins. Additionally, an estimated false discovery rate of 1% was applied to the search by the decoy database approach.22 Whenever possible, phosphosites were normalized to their own protein level, in order to ensure that changes in the phosphoprofile were caused by phosphorylation events and not due to changes in the level of the protein. Otherwise, the phosphosites were normalized by a standard logarithmic-based procedure.20 Only phosphosites, which had been normalized to the specific protein level, were included in the bioinformatic analyses. Finally, localization probabilities for each phosphosite were calculated as previously described.23 For all processed data, see ESI,† Tables S3–S8. Bioinformatics and statistical analysis were carried out within the R Statistical Computing Environment and several online available bioinformatics software tools described below.The MotifX algorithm24 was used for motif enrichment analyses on the identified- and regulated phosphorylation sites. To ensure that only phosphorylation sites with high-confidence site location assignment were analyzed, the MotifX algorithm was applied only on regulated sites having a localization probability larger than 0.7. The entire yeast proteome was used as background and only motifs that occurred more than 20 times were accepted. A binomial test was used to identify statistically enriched motifs, using a p-value of 1 × 10−06 as significance cut off.
For kinase prediction, we used the NetworKIN software25 on sites having a localization probability of at least 0.9. Only kinases with a NetworKIN score of at least 3 were considered. By applying a Fisher's exact test and a Benjamini–Hochberg-corrected significance threshold, a kinase enrichment analysis was performed with the unregulated phosphosites as background. This was done in order to predict the most pertinent kinase activities after one hour of starvation. Also, only kinases with a frequency count of at least 15 were taken into account.
To create networks of candidate proteins and directly associated proteins, we used STRING 9.0.26 To look for proteins one step further in the network (two nodes away), i.e. proteins indirectly associated to the candidate protein through a direct associated one, we used STRING 8.2.27 For directly associated proteins a confidence value of at least 0.7 was required. To look for indirectly associated proteins, we set the network depth to 2, and a confidence value of at least 0.9 was required.
YEASTRACT was used to identify documented transcriptional regulators of our regulated proteins.28–30 We specified that a transcriptional regulator had to have at least five targets whose abundance changed upon glucose- and/or nitrogen depletion in order to be regarded as relevant. To identify transcriptional regulators with enriched targets among our regulated proteins, we applied a Fisher's exact test with a subsequently Benjamini–Hochberg corrected significance threshold.
RNA extraction, cDNA synthesis, and expression analyses by qPCR
Indicated strains were grown in 5 ml YPD over night at 30 °C/200 rpm, diluted to an OD600 of 0.1, and grown in 50 ml YNBD to an OD600 value of approx. 1. The cdc37-1 temperature-sensitive mutant17 and the parental strain were grown at 24 °C until an OD600 of approx. 0.5, where the cells were placed at 37 °C for two hour. In all cases, upon reaching an OD600 value of approx. 1, cells were harvested in tubes half-filled with crushed ice, spun down (4000 rpm/10 min/4 °C), transferred to a 1.5 ml centrifuge tube with ice-cold water, and spun again (5000 rpm/2 min/4 °C). The supernatant was removed and the pellet was flash-frozen in liquid nitrogen and stored at −80 °C until further use. For the starved population of cells, the medium was changed as described above and starved for one hour. Subsequently, the starved cells were harvested and stored at −80 °C as described for the non-starved cells. RNA extraction, cDNA synthesis, and qPCR analyses were carried out as described previously.18Results and discussion
Both nitrogen- and glucose starvation induce an extreme phosphorylation response
By virtue of its highly characterized proteome, we used S. cerevisiae as a model system to examine how a unicellular organism responds to deprivation of glucose, nitrogen, or both. We used stable isotope labeling by amino acids in cell culture (SILAC) to quantitatively compare starvation-induced changes in the proteome and phosphoproteome of yeast. We grew cultures of an arginine- and lysine auxotroph yeast strain in the presence of light (normal), medium, or heavy isotopes of lysine and arginine to the exponential growth phase. Subsequently, these cultures were harvested and the cells were re-cultured in media without glucose for 5, 15, and 60 minutes, without nitrogen for 60 minutes, or without glucose and nitrogen for 60 minutes (Fig. S1, ESI†). We then harvested and mixed an equal number of cells, and prepared the cells for proteomic- and phosphoproteomic analyses by mass spectrometry.To ensure a proper threshold for assessing 60 minutes starvation-induced changes in the proteome, we determined the 95%- and 5%-percentiles of the global ratio distribution, which corresponded to 1.4-, 1.3-, and 1.5-fold for glucose-, nitrogen-, and glucose and nitrogen starvation, respectively. We identified and quantified 1602, 1604, and 1248 proteins after 60 minutes of starvation of which 224, 148, and 131 changed their expression levels in response to glucose-, nitrogen-, and glucose and nitrogen starvation, respectively (Table 1).
| Starvation condition | ||||||
|---|---|---|---|---|---|---|
| Glucose (5 min) | Glucose (15 min) | Glucose (60 min) | Nitrogen (60 min) | Glucose and nitrogen (60 min) | ||
| a The number of phosphosites represents only those that have been normalized to its own protein level. b The ratio cut of value is indicated in brackets after the number of regulated proteins/sites. | ||||||
| Proteins | Identified | 2042 | 2042 | 2196 | 2196 | 1582 |
| Quantifieda | 1293 | 1293 | 1602 | 1604 | 1248 | |
| Regulatedb | 43 (1.4) | 62 (1.4) | 224 (1.4) | 148 (1.3) | 131 (1.5) | |
| Phospho-proteins | Identified | 2765 | 2765 | 2924 | 2924 | 1896 |
| Quantifieda | 1022 | 1022 | 1336 | 1359 | 874 | |
| Regulatedb | 431 (1.5) | 392 (2.0) | 578 (2.0) | 516 (1.5) | 446 (2.0) | |
In a similar way, we calculated the same percentiles for the ratio distribution of the different phosphorylation sites after 60 minutes of starvation. However, due to an extreme phosphorylation response, resulting in high cut-off values, we calculated two standard deviations of the non-modified proteins instead. Hence, a threshold of 1.5-fold was chosen for nitrogen starvation, whereas 2.0-fold was used for glucose as well as for the combination of glucose and nitrogen starvation (Table 1). In total, 1336, 1359, and 874 phosphosites were quantified, of which 587, 516, and 446 were regarded as being regulated during glucose-, nitrogen-, or glucose and nitrogen starvation, respectively (Table 1). Thus, the rigorous nature of starvation was clearly reflected in the datasets showing a massive phosphorylation- and dephosphorylation response during nitrogen- and in particular glucose deprivation.
Since short-term glucose deprivation has previously been shown to affect metabolism, transcription as well as translation,31,32 we also examined the phosphoproteomic changes after 5 and 15 minutes of glucose starvation.
For 5 and 15 minutes of glucose starvation, we were able to quantify 1022 phosphorylation sites of which 481 were profiled at all three time points (5, 15, and 60 minutes) (Table S1, ESI†). We found that the changes in the overall phosphorylation profile observed after 60 min of glucose starvation were very similar to the response observed after 5 and 15 min of glucose starvation (Fig. S2A, ESI†). Furthermore, we observed that the phosphorylation pattern of each time point correlated well with that of all the other time points used in this study (Fig. S2B, ESI†). Of the 481 fully profiled sites, 130 were regulated at all times; 95 sites were phosphorylated, and 34 were dephosphorylated. Only one out of the 130 sites did not follow the same phosphorylation pattern throughout all measured time points. For these reasons, we chose to focus the bioinformatics analyses on the 60 minutes starvation results. Assuming that changes in protein expression are slow relative to alterations in phosphorylation pattern, we did not examine the proteomic changes induced after 5 and 15 minutes of glucose deprivation. In support of that, we quantified 1293 proteins of which the level of only 43 and 62 proteins changed after 5 and 15 minutes, respectively.
SP/TP, RxxS, and SxxxL phosphorylation motifs are significantly enriched among regulated phosphorylation sites
In an effort to identify putative kinases involved in regulation of the observed phosphorylation changes after 60 minutes, we first performed sequence motif enrichment analysis using the MotifX algorithm on the identified- and regulated phosphorylation sites.24 Three phosphorylation motifs, the SP, the TP, and the RxxS motifs, were found to be statistical enriched when cells were starved from nitrogen or both glucose and nitrogen. Interestingly, SP/TP and RxxS motifs have previously been enriched as targets, when yeast cells were treated with the TOR1 kinase inhibitor Rapamycin33 and the fatty acid synthase inhibitor Cerulenin.19 Besides these three motifs, we also found the SxxxL motif to be statistically enriched, when cells were starved from glucose (Fig. 1).In order to identify the motifs associated with either phosphorylation- or dephosphorylation events after 60 minutes of starvation, MotifX was applied to all the phosphorylated- and dephosphorylated sites separately (Table S2, ESI†). In addition, we also examined the distribution of the different phosphorylation motifs under the different starvation conditions (Fig. S3, ESI†). We found the TP motif to be primarily phosphorylated, when cells were starved for glucose or both glucose and nitrogen. The SP motif was predominantly dephosphorylated during glucose starvation, while it was almost equally phosphorylated and dephosphorylated upon nitrogen starvation (Fig. S3 and Table S2, ESI†).
When cells were starved from both glucose and nitrogen, the SP motif became heavily dephosphorylated as observed for glucose starvation alone. The RxxS motif was equally distributed among phosphorylation- and dephosphorylation events during glucose starvation, whereas this motif had a minor trend towards dephosphorylated events, when yeast cells were starved from nitrogen.
When cells were starved from both glucose and nitrogen, the RxxS motif was almost equally distributed between phosphorylation- and dephosphorylation events. Finally, we found a profound phosphorylation of SxxxL sites, when cells were deprived from glucose as well as both glucose and nitrogen (Fig. S3 and Table S2, ESI†). Thus, intense phosphorylation of SxxxL sites appears to be a clear signature of glucose starvation.
Several kinases are responsible for the altered phosphoproteome during nitrogen- and glucose starvation
It is well known that the specificity of a kinase is governed not only by the linear motifs surrounding the phosphorylated site of interest but also the contextual networks of the kinases and substrates.34 We therefore applied the NetworKIN algorithm, which incorporates linear motif analysis by position-specific sequence matrices and artificial neural networks35 as well as probabilistic network modeling27 on the regulated high-confident phosphorylation sites. The NetworKIN approach, which has been demonstrated to provide more accurate and meaningful kinase predictions,25 revealed 14 protein kinases as potential global regulators of 60 minutes glucose- and/or nitrogen starvation (Fig. 2). We found the two cyclin-dependent kinases (CDKs) Pho85 and Cdc28, the casein kinases Cka1 and Cka2, and Pkc1 to be associated with phosphorylation events during nitrogen starvation (Fig. 2). Despite that no kinases were found to statistically associate with phosphorylation events during glucose starvation, we found predominantly Snf1 targets to be enriched (1.5-fold) among the identified phosphorylated sites (data not shown). Among the sites we found dephosphorylated upon glucose deprivation, we identified Ste20, Cla4, Skm1, Pho85, Cdc28, Tpk1/2/3, Pkc1, Snf1, Hog1, and Sch9 to potentially phosphorylate these sites (Fig. 2). Except from Sch9, Hog1, Snf1, and Pkc1, the same kinases were predicted to be associated with the dephosphorylation events observed during nitrogen starvation (Fig. 2). Ste20, Cla4, Skm1, Pho85, Cdc28, and Sch9 were also associated with dephosphorylation events during glucose- and nitrogen starvation (Fig. 2).The NetworKIN results suggest that especially the two cell cycle kinases Cdc28 and Pho85 phosphorylate numerous proteins upon starvation, thus linking general starvation responses to cell cycle regulation. Furthermore, our analyses predicted that Cdc28 and Pho85 primarily are associated to the SP/TP phosphorylation motifs (Fig. S4, ESI†). Consistent with this notion, both Cdc28 and Pho85 have been shown to coordinate cell cycle progression with a number of environmental stress signals including changes in inorganic phosphate levels.36,37 Also, in line with previous findings,36 we found that SP phosphorylation sites are increasingly dephosphorylated upon prolonged nutrient deprivation. Besides Cdc28 and Pho85, our analyses indicate that Hog1 also contributes to regulation of SP sites (Fig. S4, ESI†). The RxxS motif was primarily predicted to be controlled by the PAK kinases Ste20, Skm1, and Cla4, and the AGC kinases Sch9, Pkc1, and the cAMP-dependent kinases, Tpk1/2/3 (Fig. S4, ESI†). Generally, we find RxxS sites to be associated with several different kinases and to be equally distributed between phosphorylation- and dephosphorylation events suggesting a less specific role for not only this motif but also for the kinases associated to this site (Fig. 2; Fig. S3 and S4, ESI†). Snf1 was also found to contribute on RxxS sites, and interestingly, Snf1 was the only kinase, which was predicted to regulate SxxxL sites (Fig. S4, ESI†). This finding is consistent with the facts that the SxxxL sites in our study primarily become phosphorylated in response to glucose deprivation (Table S2 and Fig. S3, ESI†), that the Snf1 kinase previously has been shown to recognize not only RxxS motifs but also SxxxL motifs,38 and that Snf1 is activated during glucose scarcity.1 Accordingly, we find that Mig1, a transcriptional repressor, which is inhibited by Snf1-mediated phosphorylation upon glucose deprivation,39 is massively phosphorylated after 5 or 15 minutes of glucose starvation at positions S302 and S314, which are both located within SxxxL motifs (see Table S5, ESI† for details). We therefore propose that Snf1 mediates the extreme phosphorylation response observed on SxxxL sites during glucose starvation.
To further support the potential involvement of Snf1 in the glucose starvation response, we found that phosphorylation of T210 in the Snf1 kinase, which previously has been shown to be crucial for its activity,40 increased dramatically when cells were starved from glucose for 5 and 15 minutes (10.0-fold and 8.3-fold, respectively). T210 phosphorylation is regulated by the three upstream kinases Sak1, Tos3, and Elm1 and a single phosphatase, Reg1-Glc7.41–44 The exact mechanisms by which phosphorylation of T210 is regulated is largely unknown, since neither the kinases nor the phosphatase appear to respond to changes in the availability of glucose.45 However, it has been suggested that its phosphorylation is regulated by the availability of Reg1-Glc7 to T210.45 We found that phosphorylation of numerous residues in Reg1 increases upon glucose deprivation (Tables S3–S5, ESI†), which may impair Reg1 accessibility to T210 in the Snf1 kinase and hence diminish the inactivating dephosphorylation of Snf1. Interestingly, two of these sites, S75 and S346, are located within an RxxSxxxL and an SxxxL site, respectively, indicating that Snf1 itself may phosphorylate and hence regulate the activity of Reg1. In support of that, phosphorylation of the N-terminal part of Reg1 (Reg11–400) has previously been demonstrated to be Snf1-dependent.46 Finally, we also found that S413 in the Snf1 kinase was dephosphorylated (2.4-fold), when cells were starved for glucose for 60 minutes, however, the significance of this is presently unknown.
In addition to Snf1, we found that Cdc28, Hog1, Cla4, Ste20, Pkc1, and Sch9 were all differentially phosphorylated during starvation. We did not identify any sites on the remaining predicted kinases, hence we cannot rule out that their phosphorylation level is regulated as well. Tpk1/2/3 was not identified in this study, however, the Tpk1/2/3-regulatory subunit, Bcy1, was 3.4-fold phosphorylated on S70 one hour after glucose withdrawal. S70 on Bcy1 as well as most of the other identified regulated sites found on our predicted kinases have not previously been reported, and the consequences on kinase activity are therefore presently unknown. In contrast, T723 and S726 on Sch9, which we find to be massively dephosphorylated (more than 20-fold) after 5 and 15 minutes of glucose starvation, have recently been described as TORC1 targets and to be crucial to Sch9 function.47 Similarly, we found that phosphorylation at Y176 of Hog1 was augmented after only 5 minutes of glucose starvation (2.5-fold) and remained elevated when cells were cultured in medium without glucose for 15 and 60 minutes (4.1-fold and 2.0-fold, respectively). Phosphorylation of Y176 has previously been demonstrated to be crucial for Hog1 activity.48 Taken together, we interestingly noticed that all the predicted starvation-associated kinases, if identified, were indeed differentially phosphorylated during starvation.
Protein kinases regulate nutrient-responsive gene expression
To address the role of the predicted kinases during starvation, we monitored the expression of genes known to be induced upon glucose- or nitrogen starvation in wild type and kinase mutants (Fig. S5 and S6, ESI†). We first confirmed that the expression of HXT2, CUP1-1, SUC2, and HSP12 was induced after glucose deprivation as described (Fig. S5, ESI†).49–52 Similarly, we also confirmed that the expression of PEP4, GDH2, GCN4, and GAP1 was induced upon nitrogen starvation (Fig. S6, ESI†) as reported previously.53–56 Next, we examined the expression pattern of these genes after one hour of starvation in strains carrying a deletion of one of the genes encoding one of the predicted kinases.Despite being required for basal expression levels of HXT2, CUP1-1, GDH2, and GCN4 in non-starved cells, we found that the Snf1 kinase, as expected,1 was required for full induction of SUC2 during glucose starvation but also for full induction of HSP12, GDH2, and GAP1 upon nitrogen starvation (Fig. S5 and S6, ESI†). Furthermore, deletion of snf1 resulted in increased expression of GDH2 and PEP4 during glucose starvation and of HSP12 during non-starved conditions. These results confirmed the already well-established role of Snf1 in nutrient regulation.
Cells lacking Cka1 showed increased basal expression of CUP1-1, HSP12, PEP4, GDH2, and GAP1. On the other hand, basal expression of HXT2 under non-starved conditions seemed to depend on Cka1. When starved from glucose, induction of PEP4 and GDH2 expression increased in cka1Δ cells relative to unstarved cells, whereas induction of CUP1-1 expression was reduced. The increased expression of CUP1-1, SUC2, HSP12, and GAP1 during nitrogen starvation depends partly on Cka1.
Based on these observations, we propose that Cka1 is required for prober regulation of many starvation-associated responses.
Functional loss of Pho85 resulted in higher basal expression levels of CUP1-1, HSP12, PEP4, GDH2, and GAP1 but lower basal expression of SUC2 in non-starved cells. Moreover, we found that expression of HXT2 and SUC2 during glucose starvation was further increased in pho85Δ cells compared to wild-type cells. Additionally, when compared to wild-type cells, Pho85 was required for full induction of CUP1-1 expression during glucose- and nitrogen deprivation as well as GDH2 during nitrogen deprivation (Fig. S5 and S6, ESI†). Thus, Pho85, like Snf1 and Cka1, seems to interfere with normal starvation responses of S. cerevisiae.
Cla4 has not previously been associated with starvation responses per se, but its activity is essential in the absence of its redundant and functional homologue Ste20 during vegetative growth.57 Interestingly, Ste20 binding to Cdc42, a Rho-like small GTP-binding protein, is required for filamentous growth in response to nitrogen starvation.57 We found that loss of Cla4 resulted in massive changes in expression of both glucose- and nitrogen sensitive genes. In the non-starved condition, basal expression of CUP1-1 decreased, whereas basal levels of HXT2, HSP12, PEP4, GDH2, and GAP1 increased in cla4Δ cells. Complete induction of HXT2, CUP1-1 and SUC2 upon glucose starvation as well as induction of PEP4, GDH2, GCN4, and GAP1 during nitrogen starvation required functional Cla4. In contrast, during glucose deprivation PEP4, GDH2, GCN4, and GAP1 expression was elevated in cells lacking Cla4 relative to wild-type. Similarly, expression of HXT2, CUP1-1, and SUC2 increased in cla4Δ cells upon nitrogen deprivation compared to wild-type cells. In contrast to Cla4, Ste20 demonstrated only minor effects on the genes tested. Functional loss of Ste20 affected the expression of GCN4 during glucose deprivation resulting in a slightly higher expression. Moreover, the basal expression of GAP1 appeared to depend on Ste20 (Fig. S5 and S6, ESI†). Collectively, these results show that loss of Cla4 leads to an impaired induction of nitrogen- and glucose-responsive gene expression upon nitrogen- and glucose starvation, respectively (Fig. S5 and S6, ESI†). Oppositely, nitrogen-responsive genes were further induced in cla4Δ cells relative to wild-type, when these were starved from glucose and vice versa. However, functional loss of Ste20 had only minor effects on glucose- and nitrogen-responsive gene expression (Fig. S5 and S6, ESI†), suggesting that Cla4 is very important in regulation of starvation-related gene expression and can function independently of Ste20.
Kinase-associated proteins coordinate starvation responses
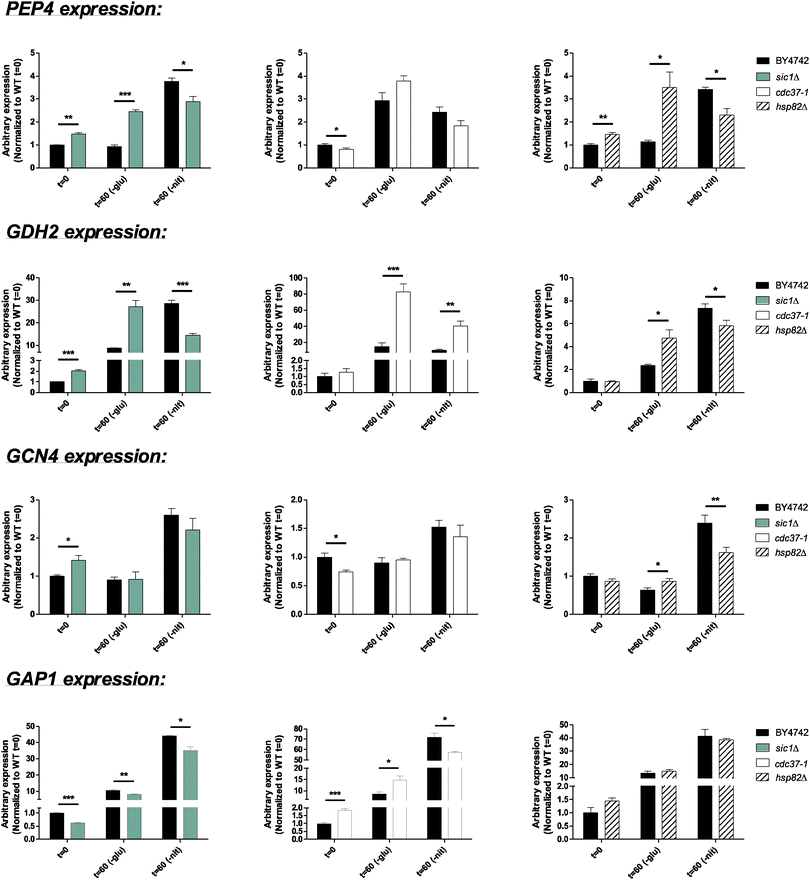
The observations described above suggest that the predicted kinases may possess a crucial regulatory role during starvation from nitrogen and/or glucose. However, how these kinases coordinately regulate the starvation responses is unknown. Therefore, we aimed at identifying proteins, which were capable of linking the different kinases to each other. Such proteins, together with the kinases, would be found in a minor local area of the network surrounded by regulated proteins, and consequently, these interconnecting proteins may as well themselves be regulated and thereby important to the response. Using STRING, we identified kinase-associated proteins, which we found to cluster in 14 different networks, one for each kinase.26,58 Subsequently, we compared these networks in order to identify proteins, which associated with more than just one kinase. The highest associated proteins being associated to more than five kinases were almost exclusively proteins that previously have been described in starvation-related responses. Such proteins included Cdc25, Msn2/4, Ras2, Rim15, Reg1, and Glc7 (Fig. 3A). Cdc25 stimulates the PKA signaling pathway1 and were found in this study to be 1.8-fold dephosphorylated within an SxxxL motif on S423 after 60 minutes of nitrogen deprivation. Interestingly, the same site was 4.2-, 3.2-, and 7.2-fold phosphorylated when cells were starved from glucose for 5, 15, and 60 minutes, respectively. Moreover, we found that S214 in Ras2 becomes 4.0-fold dephosphorylated when cells were deprived from glucose for 60 minutes. This is consistent with a role in starvation responses, which is underlined by previous findings showing that phosphorylation of S214 in Ras2 stimulates cAMP levels and glycogen degradation.59 The protein kinase Rim15 has previously been shown to integrate both PKA- and TOR-mediated signaling and to control gene expression in response to glucose limitation at the diauxic shift in a Gis1- and Msn2/4-dependent manner.60 We identify several sites on Rim15, which all were dephosphorylated during glucose starvation at all time points. One of these sites, S1337, was likewise 1.8-fold dephosphorylated 60 minutes after nitrogen withdrawal. As mentioned above, Reg1-Glc7 comprises a crucial phosphatase, of which Reg1 was identified in this study to be phosphorylated within potential Snf1 motifs. Finally, we identified Msn4 to be phosphorylated on numerous sites upon glucose depletion while dephosphorylated on the same sites during nitrogen starvation. Although the functional role of these modifications remains elusive, they may determine the activity of Msn4 depending on the presence of glucose or nitrogen. Taken together, the identification of known regulators of starvation responses along with the finding of these proteins to be regulated supports the use of our network approach to identify novel players of the yeast starvation response.While most of these kinase-associated proteins (Fig. 3A) have been identified as key regulators of nitrogen- and glucose starvation, the roles of Cdc37 and Sic1 in starvation responses have only been limited described.61,62 This prompted us to examine how inactivation of either Cdc37 or Sic1 affected expression of starvation-responsive genes (Fig. 4 and 5). We found that the basal expression of GAP1 was reduced, whereas the level of HXT2, CUP1-1, HSP12, PEP4, GDH2, and GCN4 increased in sic1Δ under non-starved conditions. Moreover, Sic1 was partly needed for the induction of HXT2, SUC2, and GCN4 during glucose starvation and for PEP4, GDH2, and GCN4 upon nitrogen starvation. In addition, deletion of sic1 further induced the expression of PEP4 and GDH2 in cells growing in glucose-depleted media. In a similar way, HXT2 was induced in sic1Δ cells upon nitrogen deprivation. When growing a cdc37 temperature-sensitive strain at the restrictive temperature, the basal expression level of CUP1-1, HSP12, and GAP1 was elevated, whereas the basal expression of HXT2, PEP4, and GCN4 was reduced under non-starved conditions (Fig. 4 and 5). Furthermore, we found that Cdc37 was required for full induction of HXT2 and GAP1 during nitrogen starvation. Oppositely, inactivation of Cdc37 potentiated the induction of CUP1-1, SUC2, and PEP4 during glucose deprivation as well as GDH2 regardless of the starvation condition (Fig. 4 and 5). Collectively, these results establish a potential role of Sic1 and Cdc37 in coordinating starvation-related responses. Precise regulation during nutrient-scarce conditions may therefore involve the interplay of several regulatory proteins that are closely associated to each other (Fig. 3B). Interestingly, Cdc37 has previously been shown to regulate the activity of multiple protein kinases including Hog1,63 Cdc28,64 PKA,65 and Cka1/2, of which the latter kinase phosphorylates Cdc37 at position S14 and S17 to further promote Cka1/2 activity.66
Regulators of transcription contribute to coordination of starvation responses
It is well known that yeast cells coordinate their metabolism to meet their nutritional needs to sustain growth and survival, not only by changing protein phosphorylation patterns but also by restructuring transcriptional programs.1 Thus, we used YEASTRACT in order to identify transcriptional regulators,28–30 which potentially could contribute to the observed starvation-induced changes in protein levels. This approach enabled us to identify 38 transcriptional regulators having a statistically enriched number of targets among the identified regulated proteins (Fig. 6). The transcriptional regulators Pdr1/3, Ino2, Adr1, Tec1, and Aft1 were all found to have enriched targets among the regulated proteins in each of the three starvation conditions, whereas Rox1, Sok2, Sko1, Gis1, and Cst6 were recognized with enriched targets during nitrogen- as well as glucose starvation. Finally, numerous transcriptional regulators were found with enriched targets in only one of the three different starvation conditions (Fig. 6). Among the identified transcriptional regulators (Fig. 6), we found that the phosphorylation status of sites on Gis1, Sko1, Cst6, Rox1, Mcm1, Ume6, and Msn4 changed in response to starvation (Tables S3–S5, ESI†), suggesting that the activity of these regulators may be controlled by phosphorylation.To further corroborate some of these findings, we addressed the role of Pdr1 and Gis1 in the expression of genes known to be induced upon glucose- or nitrogen deprivation (Fig. S5 and S6, ESI†). Pdr1 was required for full expression of HXT2 and GDH2 during non-starved conditions, of HXT2 and SUC2 after 60 minutes of glucose deprivation, and of CUP1-1 and GAP1 upon 60 minutes of nitrogen starvation. Gis1 was needed for basal expression of HXT2 during non-starved conditions. Additionally, Gis1 was required to fully induce HXT2 (2.1-fold vs. 6.2-fold reduction), CUP1-1, and SUC2 expression after one hour of glucose starvation as well as CUP1-1, HSP12, PEP4, GDH2, and GAP1 after one hour of nitrogen starvation. Oppositely, deletion of pdr1 or gis1 further induced the expression of PEP4, GDH2, and GAP1, when cells were cultured without glucose. When cells were deprived for nitrogen for 60 minutes, expression of PEP4 and GDH2 was further induced in pdr1Δ cells compared to non-starved cells. Furthermore, functional loss of Pdr1 increased the basal expression of HSP12 during the non-starved condition. Similarly, functional loss of Gis1 induced the expression of CUP1-1, HSP12, PEP4, GDH2, and GAP1 during non-starved conditions (Fig. S5 and S6, ESI†). Collectively, these observations clearly suggest that Gis1 and Pdr1 contribute to a normal starvation response.
The chaperone Hsp82 coordinates in part the transcriptional- and kinase responses to starvation
In an effort to identify proteins, which could potentially link all of the above-mentioned transcriptional regulators, we applied a similar network-based approach as described for the kinases. Since our transcriptional regulators were less closely associated, we expanded the analysis by including proteins one step further in the network. We identified seven proteins, which all had a high number of direct- or indirect associations to the transcriptional regulators (Fig. 7). In keeping with this, four of these have previously been associated with starvation-related processes.1 Interestingly, we identified the molecular chaperone Hsp82 to be the most associated protein being directly- or indirectly associated with 20 of the 38 transcriptional regulators (Fig. 7). Hsp82 is less described in starvation responses but is known to be induced during heat treatment and to regulate a number of transcription factors and kinases.67,68 Furthermore, Hsp82 has been described to localize to the nucleus during glucose- as well as nitrogen deprivation.69,70 We did not identify any phosphorylation sites on Hsp82, and its protein level was unaltered in all measured starvation conditions and time points.As described above, we already identified Sic1 and Cdc37 as being highly connected to our candidate kinases. However, as for the transcriptional regulator networks, we also expanded the kinase-associated networks by one step to include indirectly associated proteins. Interestingly, Hsp82 was identified to be associated with 11 of the aforementioned 14 kinases (Fig. S7, ESI†). The large number of closely associated regulatory proteins predicted to be involved in starvation responses suggest that Hsp82 may play a global role in coordinating the starvation response (Fig. 8). This prompted us to examine the expression of nutrient-regulated genes in an hsp82 deletion mutant after 60 minutes of starvation. We found that Hsp82 is required to fully induce the expression of HXT2, CUP1-1, and SUC2 during glucose starvation and CUP1-1, HSP12, PEP4, GDH2, and GCN4 upon nitrogen starvation (Fig. 4 and 5). Under non-starved conditions, deletion of hsp82 increased the expression of CUP1-1, HSP12, and PEP4. In addition, expression of PEP4, GDH2, and GCN4 was further increased in hsp82Δ cells relative to wild-type cells during glucose deprivation (Fig. 4 and 5). These results support our prediction of Hsp82 as being a regulator of starvation responses. Notably, it has previously been shown that Cdc37 and Hsp82 can interact with the same proteins but yet have both converging and diverging functions.71 Accordingly, the expression analyses not only show that both Hsp82 and Cdc37 are required for a normal transcriptional response to either glucose- or nitrogen deprivation, but also show that expression of some of the nutrient-responsive genes are differentially regulated in hsp82Δ and cdc37-1 cells. This suggests that Hsp82 and Cdc37 are both involved in coordinating starvation responses and confirm that they may not always function together.
Conclusions
Our present work suggests in accordance with previous findings that the starvation response is rapidly initiated by a number of kinases and transcriptional regulators organized in complex signaling networks. Based on prior knowledge on protein function, these networks may coordinate metabolism, cell cycle, division, and growth in order to respond and adapt to changes in nutrient availability. The present work identifies several known nutrient-responsive regulatory proteins, but also identifies novel putative regulators of starvation responses. Especially, we aimed our analyses at predicting global regulators that may take part in many starvation-related responses and thus may be important in coordinating the overall outcome. We foresee that our work lays an excellent foundation to further delineate the functions of these regulators and how post-translational protein modifications regulate nutrient sensing- and adaptation pathways.Acknowledgements
This work was supported by funds from The Danish Research Councils (Grant # 23459) and The Novo Nordic Foundation (Grant # 13854). We thank Mark Winey for the Cdc37-1 (AS146-26b) and the wild-type (WX257-5d) strain. We also thank Ida Coordt Elle for proofreading this manuscript.References
- S. Zaman, S. I. Lippman, X. Zhao and J. R. Broach, Annu. Rev. Genet., 2008, 42, 27–81 CrossRef CAS PubMed.
- T. G. Cooper, FEMS Microbiol. Rev., 2002, 26, 223–238 CrossRef CAS PubMed.
- I. Pedruzzi, F. Dubouloz, E. Cameroni, V. Wanke, J. Roosen, J. Winderickx and C. De Virgilio, Mol. Cell, 2003, 12, 1607–1613 CrossRef CAS.
- V. A. Cherkasova and A. G. Hinnebusch, Genes Dev., 2003, 17, 859–872 CrossRef CAS PubMed.
- T. Yorimitsu, S. Zaman, J. R. Broach and D. J. Klionsky, Mol. Biol. Cell, 2007, 18, 4180–4189 CrossRef CAS PubMed.
- L. Kraakman, K. Lemaire, P. Ma, A. W. Teunissen, M. C. Donaton, P. Van Dijck, J. Winderickx, J. H. de Winde and J. M. Thevelein, Mol. Microbiol., 1999, 32, 1002–1012 CrossRef CAS.
- Y. Wang, M. Pierce, L. Schneper, C. G. Guldal, X. Zhang, S. Tavazoie and J. R. Broach, PLoS Biol., 2004, 2, E128 Search PubMed.
- T. Toda, S. Cameron, P. Sass, M. Zoller and M. Wigler, Cell, 1987, 50, 277–287 CrossRef CAS.
- J. S. Stephan, Y. Y. Yeh, V. Ramachandran, S. J. Deminoff and P. K. Herman, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 17049–17054 CrossRef CAS PubMed.
- W. Gorner, E. Durchschlag, J. Wolf, E. L. Brown, G. Ammerer, H. Ruis and C. Schuller, EMBO J., 2002, 21, 135–144 CrossRef CAS PubMed.
- A. Reinders, N. Burckert, T. Boller, A. Wiemken and C. De Virgilio, Genes Dev., 1998, 12, 2943–2955 CrossRef CAS.
- A. Rahner, M. Hiesinger and H. J. Schuller, Mol. Microbiol., 1999, 34, 146–156 CrossRef CAS.
- E. T. Young, K. M. Dombek, C. Tachibana and T. Ideker, J. Biol. Chem., 2003, 278, 26146–26158 CrossRef CAS PubMed.
- S. Ozcan and M. Johnston, Mol. Cell. Biol., 1996, 16, 5536–5545 CAS.
- S. Ozcan, T. Leong and M. Johnston, Mol. Cell. Biol., 1996, 16, 6419–6426 CAS.
- A. Gruhler, J. V. Olsen, S. Mohammed, P. Mortensen, N. J. Faergeman, M. Mann and O. N. Jensen, Mol. Cell. Proteomics, 2005, 4, 310–327 CAS.
- A. R. Schutz, T. H. Giddings, Jr., E. Steiner and M. Winey, J. Cell Biol., 1997, 136, 969–982 CrossRef CAS.
- S. Feddersen, T. B. Neergaard, J. Knudsen and N. J. Faergeman, Biochem. J., 2007, 407, 219–230 CrossRef CAS PubMed.
- D. Pultz, M. V. Bennetzen, S. V. Rødkær, C. Zimmermann, J. M. Enserink, J. S. Andersen and N. J. Færgeman, Mol. BioSyst, 2012, 8, 796–803 RSC.
- J. Cox and M. Mann, Nat. Biotechnol., 2008, 26, 1367–1372 CrossRef CAS PubMed.
- D. N. Perkins, D. J. Pappin, D. M. Creasy and J. S. Cottrell, Electrophoresis, 1999, 20, 3551–3567 CrossRef CAS.
- J. E. Elias and S. P. Gygi, Nat. Methods, 2007, 4, 207–214 CrossRef CAS PubMed.
- J. V. Olsen, B. Blagoev, F. Gnad, B. Macek, C. Kumar, P. Mortensen and M. Mann, Cell, 2006, 127, 635–648 CrossRef CAS PubMed.
- D. Schwartz and S. P. Gygi, Nat. Biotechnol., 2005, 23, 1391–1398 CrossRef CAS PubMed.
- R. Linding, L. J. Jensen, A. Pasculescu, M. Olhovsky, K. Colwill, P. Bork, M. B. Yaffe and T. Pawson, Nucleic Acids Res., 2008, 36, D695–D699 CrossRef CAS PubMed.
- D. Szklarczyk, A. Franceschini, M. Kuhn, M. Simonovic, A. Roth, P. Minguez, T. Doerks, M. Stark, J. Muller, P. Bork, L. J. Jensen and C. von Mering, Nucleic Acids Res., 2011, 39, D561–D568 CrossRef CAS PubMed.
- L. J. Jensen, M. Kuhn, M. Stark, S. Chaffron, C. Creevey, J. Muller, T. Doerks, P. Julien, A. Roth, M. Simonovic, P. Bork and C. von Mering, Nucleic Acids Res., 2009, 37, D412–D416 CrossRef CAS PubMed.
- M. C. Teixeira, P. Monteiro, P. Jain, S. Tenreiro, A. R. Fernandes, N. P. Mira, M. Alenquer, A. T. Freitas, A. L. Oliveira and I. Sa-Correia, Nucleic Acids Res., 2006, 34, D446–D451 CrossRef CAS PubMed.
- P. T. Monteiro, N. D. Mendes, M. C. Teixeira, S. d'Orey, S. Tenreiro, N. P. Mira, H. Pais, A. P. Francisco, A. M. Carvalho, A. B. Lourenco, I. Sa-Correia, A. L. Oliveira and A. T. Freitas, Nucleic Acids Res., 2008, 36, D132–D136 CrossRef CAS PubMed.
- D. Abdulrehman, P. T. Monteiro, M. C. Teixeira, N. P. Mira, A. B. Lourenco, S. C. dos Santos, T. R. Cabrito, A. P. Francisco, S. C. Madeira, R. S. Aires, A. L. Oliveira, I. Sa-Correia and A. T. Freitas, Nucleic Acids Res., 2011, 39, D136–D140 CrossRef CAS PubMed.
- L. M. Castelli, J. Lui, S. G. Campbell, W. Rowe, L. A. Zeef, L. E. Holmes, N. P. Hoyle, J. Bone, J. N. Selley, P. F. Sims and M. P. Ashe, Mol. Biol. Cell, 2011, 22, 3379–3393 CrossRef CAS PubMed.
- M. P. Ashe, S. K. De Long and A. B. Sachs, Mol. Biol. Cell, 2000, 11, 833–848 CrossRef CAS.
- A. Soulard, A. Cremonesi, S. Moes, F. Schutz, P. Jeno and M. N. Hall, Mol. Biol. Cell, 2010, 21, 3475–3486 CrossRef CAS PubMed.
- R. Linding, L. J. Jensen, G. J. Ostheimer, M. A. van Vugt, C. Jorgensen, I. M. Miron, F. Diella, K. Colwill, L. Taylor, K. Elder, P. Metalnikov, V. Nguyen, A. Pasculescu, J. Jin, J. G. Park, L. D. Samson, J. R. Woodgett, R. B. Russell, P. Bork, M. B. Yaffe and T. Pawson, Cell, 2007, 129, 1415–1426 CrossRef CAS PubMed.
- M. L. Miller, L. J. Jensen, F. Diella, C. Jorgensen, M. Tinti, L. Li, M. Hsiung, S. A. Parker, J. Bordeaux, T. Sicheritz-Ponten, M. Olhovsky, A. Pasculescu, J. Alexander, S. Knapp, N. Blom, P. Bork, S. Li, G. Cesareni, T. Pawson, B. E. Turk, M. B. Yaffe, S. Brunak and R. Linding, Sci. Signaling, 2008, 1, ra2 CrossRef PubMed.
- D. Huang, H. Friesen and B. Andrews, Mol. Microbiol., 2007, 66, 303–314 CrossRef CAS PubMed.
- S. Menoyo, N. Ricco, S. Bru, S. Hernandez-Ortega, X. Escote, M. Aldea and J. Clotet, Mol. Cell. Biol., 2013, 33, 1273–1284 CrossRef CAS PubMed.
- S. Dale, W. A. Wilson, A. M. Edelman and D. G. Hardie, FEBS Lett., 1995, 361, 191–195 CrossRef CAS.
- M. A. Treitel, S. Kuchin and M. Carlson, Mol. Cell. Biol., 1998, 18, 6273–6280 CAS.
- R. R. McCartney and M. C. Schmidt, J. Biol. Chem., 2001, 276, 36460–36466 CrossRef CAS PubMed.
- S. P. Hong, F. C. Leiper, A. Woods, D. Carling and M. Carlson, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 8839–8843 CrossRef CAS PubMed.
- J. Tu and M. Carlson, Mol. Cell. Biol., 1994, 14, 6789–6796 CAS.
- J. Tu and M. Carlson, EMBO J., 1995, 14, 5939–5946 CAS.
- S. P. Hong and M. Carlson, J. Biol. Chem., 2007, 282, 16838–16845 CrossRef CAS PubMed.
- E. M. Rubenstein, R. R. McCartney, C. Zhang, K. M. Shokat, M. K. Shirra, K. M. Arndt and M. C. Schmidt, J. Biol. Chem., 2008, 283, 222–230 CrossRef CAS PubMed.
- P. Sanz, G. R. Alms, T. A. Haystead and M. Carlson, Mol. Cell. Biol., 2000, 20, 1321–1328 CrossRef CAS.
- J. Urban, A. Soulard, A. Huber, S. Lippman, D. Mukhopadhyay, O. Deloche, V. Wanke, D. Anrather, G. Ammerer, H. Riezman, J. R. Broach, C. De Virgilio, M. N. Hall and R. Loewith, Mol. Cell, 2007, 26, 663–674 CrossRef CAS PubMed.
- C. Schuller, J. L. Brewster, M. R. Alexander, M. C. Gustin and H. Ruis, EMBO J., 1994, 13, 4382–4389 CAS.
- M. Carlson and D. Botstein, Cell, 1982, 28, 145–154 CrossRef CAS.
- S. Ozcan and M. Johnston, Mol. Cell. Biol., 1995, 15, 1564–1572 CAS.
- K. T. Tamai, X. Liu, P. Silar, T. Sosinowski and D. J. Thiele, Mol. Cell. Biol., 1994, 14, 8155–8165 CAS.
- R. L. Stone, V. Matarese, B. B. Magee, P. T. Magee and D. A. Bernlohr, Gene, 1990, 96, 171–176 CrossRef CAS.
- J. R. Daugherty, R. Rai, H. M. el Berry and T. G. Cooper, J. Bacteriol., 1993, 175, 64–73 CAS.
- J. A. Coffman and T. G. Cooper, J. Bacteriol., 1997, 179, 5609–5613 CAS.
- G. Thireos, M. D. Penn and H. Greer, Proc. Natl. Acad. Sci. U. S. A., 1984, 81, 5096–5100 CrossRef CAS.
- R. Chiva, I. Baiges, A. Mas and J. M. Guillamon, J. Appl. Microbiol., 2009, 107, 235–244 CrossRef CAS PubMed.
- E. Leberer, C. Wu, T. Leeuw, A. Fourest-Lieuvin, J. E. Segall and D. Y. Thomas, EMBO J., 1997, 16, 83–97 CrossRef CAS PubMed.
- B. Snel, G. Lehmann, P. Bork and M. A. Huynen, Nucleic Acids Res., 2000, 28, 3442–3444 CrossRef CAS PubMed.
- B. Xiaojia and D. Jian, FEBS Lett., 2010, 584, 2333–2338 CrossRef PubMed.
- E. Cameroni, N. Hulo, J. Roosen, J. Winderickx and C. De Virgilio, Cell Cycle, 2004, 3, 462–468 CrossRef CAS.
- V. Zinzalla, M. Graziola, A. Mastriani, M. Vanoni and L. Alberghina, Mol. Microbiol., 2007, 63, 1482–1494 CrossRef CAS PubMed.
- O. Donze and D. Picard, Mol. Cell. Biol., 1999, 19, 8422–8432 CAS.
- X. X. Yang, P. Hawle, J. P. Bebelman, A. Meenhuis, M. Siderius and S. M. van der Vies, FEMS Yeast Res., 2007, 7, 796–807 CrossRef CAS PubMed.
- A. Farrell and D. O. Morgan, Mol. Cell. Biol., 2000, 20, 749–754 CrossRef CAS.
- M. Ren, A. Santhanam, P. Lee, A. Caplan and S. Garrett, Eukaryotic Cell, 2007, 6, 1363–1372 CrossRef CAS PubMed.
- S. Bandhakavi, R. O. McCann, D. E. Hanna and C. V. Glover, J. Biol. Chem., 2003, 278, 2829–2836 CrossRef CAS PubMed.
- K. A. Borkovich, F. W. Farrelly, D. B. Finkelstein, J. Taulien and S. Lindquist, Mol. Cell. Biol., 1989, 9, 3919–3930 CAS.
- J. P. Burnie, T. L. Carter, S. J. Hodgetts and R. C. Matthews, FEMS Microbiol. Rev., 2006, 30, 53–88 CrossRef CAS PubMed.
- M. Breker, M. Gymrek and M. Schuldiner, J. Cell Biol., 2013, 200, 839–850 CAS.
- H. Tapia and K. A. Morano, Mol. Biol. Cell, 2010, 21, 63–72 CrossRef CAS PubMed.
- Y. Kimura, S. L. Rutherford, Y. Miyata, I. Yahara, B. C. Freeman, L. Yue, R. I. Morimoto and S. Lindquist, Genes Dev., 1997, 11, 1775–1785 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Fig. S1–S7. See DOI: 10.1039/c4mb00207e |
| This journal is © The Royal Society of Chemistry 2014 |