Open Access Article
Open Access ArticleE88, a new cyclic-di-GMP class I riboswitch aptamer from Clostridium tetani, has a similar fold to the prototypical class I riboswitch, Vc2, but differentially binds to c-di-GMP analogs
Yiling
Luo
,
Bin
Chen
,
Jie
Zhou
,
Herman O.
Sintim
* and
T. Kwaku
Dayie
*
Department of Chemistry and Biochemistry, University of Maryland, College Park, MD 20742, USA. E-mail: dayie@umd.edu; hsintim@umd.edu
First published on 18th December 2013
Abstract
C-di-GMP has emerged as a ubiquitous second messenger, which regulates the transition between sessile and motile lifestyles and virulence factor expression in many pathogenic bacteria using both RNA riboswitches and protein effectors. We recently showed that two additional class I c-di-GMP riboswitch aptamers (Ct-E88 and Cb-17B) bind c-di-GMP with nanomolar affinity, and that Ct-E88 RNA binds 2′-F-c-di-GMP 422 times less tightly than class I Vc2 RNA. Based on sequence comparison, it was concluded that the global folds of Ct-E88 and Vc2 RNAs were similar and that differences in ligand binding were probably due to differences in binding site architectures. Herein, we utilized EMSA, aptamer sensing spinach modules, SAXS and 1D NMR titration to study the conformational transitions of Ct-E88. We conclude that whereas the global folds of the bound states of Vc2 and Ct-E88 RNAs are similar, the unbound states are different and this could explain differences in ligand affinities between these class I c-di-GMP riboswitches.
Introduction
Until recently, the cytoplasmic bacterial second messenger bis-(3′-5′)-cyclic guanosine monophosphate (c-di-GMP) was thought to regulate various physiological processes such as motility reduction, expression of acute virulence genes, formation of adhesins and exopolysaccharides, and cell cycle progression using protein effectors.1–9 High levels of c-di-GMP in the cell are associated with biofilm formation and inhibition of virulent gene expression. Conversely, low levels of c-di-GMP allow bacteria to express virulence factors conducive to motile lifestyle and survival. In response to various extracellular signals, diguanylate cyclases (DGCs) containing Gly-Gly-Asp-Glu-Phe (GGDEF) domains synthesize c-di-GMP from two GTP molecules and phosphodiesterases (PDEs) containing either Glu-Ala-Leu (EAL) or His-Asp-Gly-Tyr-Pro (HDGYP) domains cleave c-di-GMP eventually into two molecules of GMP. Various effector proteins, discovered as potential regulators of c-di-GMP, could not fully explain how the second messenger controls transcription and translation of specific mRNA transcripts important for global cellular regulation.5,7–15 Breaker and co-workers, using computational searches, expanded this effector family to two classes of riboswitch RNAs found in the 5′-untranslated regions of mRNAs associated with DGC and PDE genes involved in c-di-GMP metabolism and regulation.16,17 Appropriately, these riboswitch RNAs fold into an aptamer domain that recognizes metabolites, and a conformational change in the aptamer domain is then relayed to downstream elements that control gene expression at the level of transcription, splicing, or translation. Approximately 500 examples of class I and 45 examples of class II riboswitches have thus far been computationally identified in a variety of bacteria including many pathogens such as Bacillus anthracis, Clostridium tetani, Clostridium difficile, and Vibrio cholera.16,17Out of all these computationally predicted riboswitch families, the 3D structure was solved and the nature of the interaction with c-di-GMP experimentally validated for only one riboswitch RNA from each class: c-di-GMP-I riboswitch associated with the tfoX gene of Vibrio cholera, hereafter referred to as Vc2 RNA-I riboswitch;17 and class II riboswitch aptamer from Clostridium acetobutylicum, hereafter referred to as CdA RNA-II riboswitch.16 Both Vc2 RNA-I and CdA RNA-II riboswitches bind c-di-GMP with dissociation constants ranging from 200 pM to 2 nM.16 These affinities are substantially tighter than those of known protein receptors.18,19
In addition to these binding studies, two crystallographic structures of the aptamer domain of the Vc2 RNA-I riboswitch complexed with c-di-GMP were solved independently by two groups, and the structures revealed an h- or a y-shaped overall architecture with a ligand recognition pocket framed by three paired helical fragments (P1, P2, and P3) held together by interhelical linker regions (J1/2, J2/3, and J3/1).20,21 Here we use the notation and nomenclature suggested by Smith et al. from a well-defined X-ray structure with higher resolution than the other one.20
Building on the insights from the structural and binding studies, the research groups of D'Amare and Rueda used single molecule fluorescence resonance energy transfer (smFRET) experiments and SAXS data to show that, in the absence of c-di-GMP, the Vc2-I RNA does not adopt a side-by-side docking. Rather a key GNRA (G = guanosine, N = any nucleotide, R = purine, and A = adenosine) tetraloop (GT)-tetraloop receptor (TR) interaction and a long-range C44-G83 base pairing become unglued in the presence of physiological Mg2+, and only adopt a bound conformation in the presence of the ligand.22
Despite these extensive studies on the Vc2-I RNA, very little is known about other members of the class I riboswitch RNAs. Using equilibrium microdialysis and gel shift assays, we recently showed that a riboswitch from Clostridium tetani E88 (hereafter Ct-E88 RNA) and Vc2-I RNAs bind c-di-GMP with the same nanomolar affinity but both were unable to bind a c-di-GMP analog with a 2′-methoxy group (2′-OMe-c-di-GMP), suggesting that it belonged to the same class I family as Vc2 (Fig. 1).23 Nonetheless, Vc2-I RNA binds a c-di-GMP analog with a 2′-fluoro group (2′-F-c-di-GMP) with nanomolar affinity, whereas Ct-E88 binds this analog with ∼422 fold loss in affinity.23
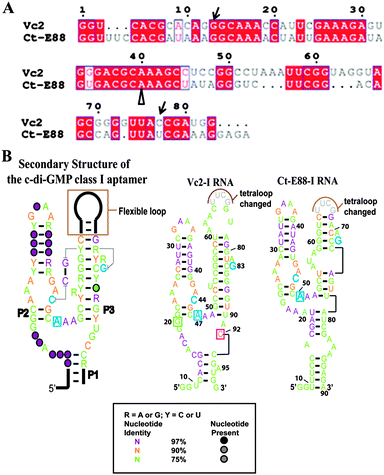 | ||
| Fig. 1 (A) Alignment of predicted c-di-GMP riboswitch sequences of Vc2-I and Ct-E88. Alignment was done using Clustal W224 and ESPript.25 The nucleotides indicated by unfilled arrows below are correlated to the nucleotides in blue in (B) and the nucleotides indicated by arrow are equivalent to G20 and C92 in Vc2-I in (B). (B) Predicted secondary structures of the selected c-di-GMP riboswitch aptamers based on a structure model of the c-di-GMP class I aptamer.20 Cytidine within the P2 stem (in blue) and guanine within the P3 stem (in blue) are expected to form a long range inter-helical Watson–Crick base pairing in the ligand bound structure and adenosine in blue is expected to intercalate between two bases of c-di-GMP in the bound form. In Vc2-I structure, cytidine and guanosine indicated in the boxes are the two important nucleotides that base pair with the two bases of c-di-GMP. | ||
From the alignment (Fig. 1A), it can be observed that Vc2-I and Ct-E88 RNAs share many conserved nucleotides. While such high conservation is suggestive of a similar global architecture, this has not been experimentally verified. Herein, we use various techniques, including native gel electrophoretic mobility shift assay, spinach detection, small-angle X-ray scattering (SAXS) and solution NMR analysis to delineate the nature of the conformational changes that take place during second messenger c-di-GMP induced folding of this newly validated Ct-E88 RNA.
Materials and methods
RNA preparation and purification
The c-di-GMP riboswitch RNAs used in this study are the UUCG-tetraloop modifications of the P3 helix. A pUC57 plasmid containing each target sequence, including the phage T7 RNA polymerase promoter sequence, was constructed using GenScript [Piscataway, NJ] and RNAs were transcribed and purified using methods established in our group.23,26,27Electrophoretic mobility shift assay (EMSA)
To explore the conformational change between the free and bound forms of Ct-E88 RNA, EMSA was performed. 10 μM of RNA was folded by heating to 70 °C for 3 min and slowly cooled in folding buffer (20 mM KCl, 6 mM MgCl2, and 10 mM sodium cacodylate, pH 6.8) in the presence of twice the ligand concentration. The reaction mixtures were incubated overnight at 25 °C, and the samples resolved on a 10% native polyacrylamide gel with running buffer (100 mM Tris–HEPES, pH 7.5, 0.1 mM EDTA) supplemented with 1 mM MgCl2. The gels were stained with ethidium bromide and images were scanned using a STORM phosphoImager (GE Healthcare).Preparation of RNA sensors
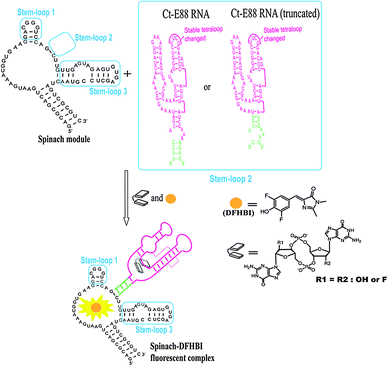
To investigate how the P1 stem transduces signals into adjacent structural elements, we created a c-di-GMP sensor using stem-loop 2 of the spinach RNA replaced with the putative P1 stem of Ct-E88 as reported recently for Vc2-I RNA.28,29 The c-di-GMP-I aptamer was fused to the spinach module by a transducer module (Fig. 2), and secondary structure prediction was performed using Mfold.30 The following conditions were used to test the sensitivity of the fluorescent c-di-GMP riboswitch sensor to sequence variation in the P1 stem of Ct-E88: [c-di-GMP] = 10 μM, [RNA] = 1 μM, and [DFHBI] = 10 μM (0.05% DMSO); and 100 mM HEPES (pH 6.8) buffer containing 100 mM NaCl, 100 mM KCl, and 6 mM MgCl2. RNA in the absence of MgCl2 was heated to 70 °C and kept at 70 °C for 3 min. Then MgCl2 and c-di-GMP were added and cooled to room temperature (24 °C) over ∼10 min. The mixture was then left at 24 °C for 12 h. DFHBI was subsequently added and fluorescence monitored (Ex. 469 nm, Em. 501 nm) at 24 °C until fluorescence plateaued after 6 min.Small angle X-ray scattering experiments and data analysis
To explore Ct-E88 RNA global conformational change as a function of MgCl2 and metabolites, small angle X-ray scattering (SAXS) and wide angle X-ray scattering (WAXS) data were collected at beam line 12-ID of the Advanced Photon Source (APS), Argonne National Laboratory. The wavelength, λ, of the X-ray radiation was set to 1.033 Å. The detailed protocols of SAXS measurements were described previously.26,31 In brief, the Ct-E88 RNA was dissolved in buffer A (100 mM HEPES and 100 mM NaCl, pH 8.0). The sample was heated at 90 °C for 2 min followed by cooling at room temperature for 10 min. MgCl2, or MgCl2 and c-di-GMP were added to buffer A to prepare additional RNA buffer conditions: buffer B (buffer A and 3 mM MgCl2) and buffer C (buffer A, 3 mM MgCl2 and a saturating amount of c-di-GMP [ligand/RNA ≈ 2]). Three concentrations of RNA samples (0.5, 1.0 and 3.0 mg mL−1) were prepared for each of the three buffer conditions, making a total of 9 sample conditions for the SAXS/WAXS data collection.The q-range used for data analysis was between 0.002 and 0.300 Å−1 and the quality of the SAXS data was evaluated by the linearity of a Guinier plot using IGOR PRO (WaveMetrics) software. GNOM software was also used to confirm the radius of gyration (Rg) and to analyze the pair distance distribution function (PDDF) as described earlier.26,32
Low resolution ab initio model reconstructions of the Ct-E88 aptamer
The low resolution ab initio model reconstructions of the Ct-E88 aptamer RNA under the three solution conditions described above were obtained using the ATSAS 2.4 suite of programs including GNOM,32 DAMMIN33 and DAMAVER34 using methods established in our group.26NMR sample preparation and NMR experiments of Ct-E88 RNA
To further corroborate the global changes in structure, simple 1H NMR experiments were performed as described earlier26,27,35 using an NMR sample with a RNA concentration of ∼1 mM and a c-di-GMP concentration of ∼2 mM. NMR experiments were performed on a Bruker Avance 600 MHz spectrometer equipped with an actively shielded z-axis gradient triple resonance probe. NMR spectra were recorded at various temperatures using the WATERGATE module for water suppression.36 All spectra were processed and analyzed using Bruker's TOPSPIN 2.1.Results and discussion
Native gel mobility shift assay
Native PAGE is a useful tool to ascertain RNA folding and conformational changes in response to the presence of metal ions or specific metabolites that trigger the RNA's conformational changes. If alternative states of the RNA are present after interacting with the metal ions or ligands, then bands with different migration rates can be observed provided the exchange between the different states is slow relative to the migration rate of the RNA through the gel. Otherwise, a single band of average mobility is observed.26,37 From our EMSA results, both Vc2-I and Ct-E88 RNAs formed a homogenous band and both RNAs migrate faster on the native PAGE in the presence of c-di-GMP and Mg2+ ions than in their absence (Fig. 3). Thus both RNAs likely adopt a more compact conformation in the presence of the ligand. Intriguingly, 2′-H-c-di-GMP did not cause any band shift for either Vc2-I or Ct-E88 RNA, whereas 2′-F-c-di-GMP induced a band shift for Vc2-I but not Ct-E88 (Fig. 3). These different mobilities on the native gel imply that Vc2-I and Ct-E88 likely undergo different levels of conformational changes with 2′-F-c-di-GMP. These observations are consistent with the results of equilibrium microdialysis and Kd measurements on Vc2-I and Ct-E88 RNA in the presence of the ligand and several different analogs.23 Thus the native gel shift assay provides a hint that Ct-E88 RNA most likely undergoes large conformational changes in the presence of the second messenger but not in the presence of the c-di-GMP analogs modified at the 2′-position.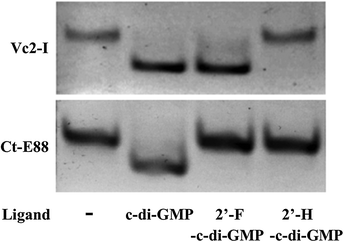 | ||
| Fig. 3 Electrophoretic mobility shift assay of Vc2-I and Ct-E88 RNA in the presence of the ligand c-di-GMP and two different ligand analogs, including 2′-F-c-di-GMP and 2′-H-c-di-GMP. | ||
Spinach
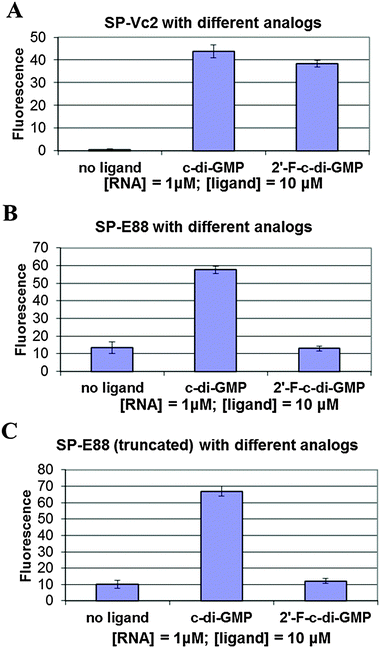
We tested the hypothesis that the stability of the P1 stem is enhanced by ligand binding to the P1 stem attached via a transducer element to a spinach recognition module. As we and others have shown recently, the formation of transducer module stem 2 is critical for the ability of the spinach module to activate the fluorescence of DFHBI.29,38 Therefore transducer elements that are stabilized in the presence of small molecule binding would allosterically regulate the spinach RNA structure. By linking the aptamer domain of either Vc2-I or Ct-E88 RNA to the spinach domain via a transducer linker, the c-di-GMP analog binding to the aptamer is expected to fold up the shared critical stem required for the binding of DFHBI to the spinach RNA, thus leading to fluorescence (Fig. 2).Consistent with our native gel shift data and microdialysis assay, Vc2-I binds both c-di-GMP and 2′-F-c-di-GMP to allosterically regulate the spinach RNA structure leading to enhanced fluorescence (Fig. 4).23 However, Ct-E88 RNA is able to use only c-di-GMP and not 2′-F-c-di-GMP to allosterically regulate the spinach RNA structure inducing increased fluorescence (Fig. 4). Intriguingly, truncation of the P1 stem (by removing residues 10–13 and 87–90 that appear pre-folded in the absence of magnesium ions and metabolite) does not impair the ability of Ct-E88 to transduce fluorescence enhancement (Fig. 4). Thus residues 10–13 and 87–90 are dispensable for the Ct-E88 riboswitch function, and further the P1 stem might actually start with the closing base pair C14–G86.
Following the conformational changes of the Ct-E88 aptamer induced by c-di-GMP and Mg2+ ions with SAXS
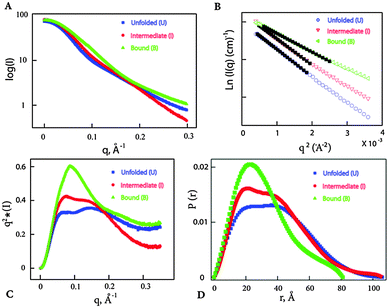
The results from native gel shift assay and microdialysis suggest that both Vc2-I and Ct-E88 RNAs bind c-di-GMP tightly and that Vc2-I but not Ct-E88 is able to recognize a ligand analog with a 2′-fluoro group (2′-F-c-di-GMP) with high affinity. Thus it is not clear if Vc2-I and Ct-E88 RNAs would share a similar global architecture. We therefore tested the nature of the structural response elicited by Mg2+ and c-di-GMP using SAXS, which would provide more information on the global shape of the RNA in the free or bound forms. Solution scattering profiles were collected for the Ct-E88 aptamer in the absence or presence of Mg2+ and c-di-GMP. The three overlaid scattering curves exhibited significant differences, suggesting the existence of at least three possible conformational states along the RNA-folding pathway (U, I and B states) (Fig. 5A). Fig. 5B shows the low angle scattering data extrapolated to zero concentration with straight lines highlighted by the black squares as fitting regions. Radii of gyration, Rg, were obtained from a Guinier analysis from these regions using the linear, first-order Guinier approximation: ln[I(q)] ≅ ln[I(0)] − Rg2q2/3. The data indicate that Rg of the Ct-E88 aptamer changes dramatically only in the presence of both Mg2+ and c-di-GMP (Table 1). In the absence of Mg2+ and c-di-GMP (U state), Ct-E88 aptamer RNA adopts an extended conformation with an Rg of 29.5 Å. In the presence of 3 mM Mg2+ (I state), Rg decreases slightly to 28.9 Å well within the error limit. In the presence of both Mg2+ and c-di-GMP (B state), the RNA is significantly compacted to an Rg of 24.3 Å. To further highlight the degree of RNA compaction, the SAXS data are transformed into the Kratky representation (Fig. 5C), where the scattering intensity is weighted by the square of the momentum transfer (q). The scattering curve of RNA in the U state exhibits a flattened plateau with scattering intensity falling slightly at large angles. Although it is different from an upward rising scattering intensity representative of a completely denatured conformation according to Porod's law, this U state is consistent with a partially ‘folded state’ with an extended conformation.39 Compared to the U and I states, the B state has a pronounced concave peak indicative of a folded structure. To gain further insight into the shape and maximum intramolecular distance of the particles, the pair distance distribution function (PDDF or P(r)) plots were compared (Fig. 5D).40,41 The RNA in U and I states (Table 1) has a relative larger maximum intramolecular distance Dmax than the c-di-GMP bound state of the RNA (B). Thus the RNA undergoes a marked ligand-induced compaction only in the presence of both Mg2+ and c-di-GMP.| States | Conditions | R g (Å) | D max (Å) | NSDc |
|---|---|---|---|---|
| Parameters and quality indicators derived from the scattering data for the Ct-E88 aptamer under different conditions.a The radius of gyration (Rg) values were obtained from Guinier analysis of the low angle scattering data.b The maximum intramolecular distances (Dmax) were obtained using GNOM program.c The normalized spatial discrepancy (NSD) values of three low resolution reconstruction models were generated using DAMAVER program. | ||||
| Unfolded (U) | Buffer | 29.5 ± 0.2 | 106 | 0.95 ± 0.02 |
| Intermediate (I) | Buffer + 3 mM Mg2+ | 28.9 ± 0.1 | 106 | 0.74 ± 0.04 |
| Bound (B) | Buffer + Mg2+ + c-di-GMP | 24.3 ± 0.1 | 81 | 0.60 ± 0.03 |
Low resolution ab initio modeling of Ct-E88 RNA
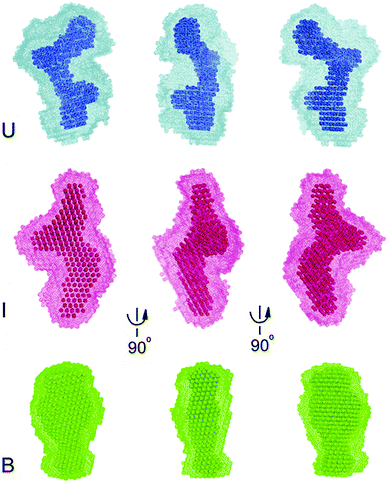
To gain further insight into the nature of the Mg2+ and c-di-GMP induced conformational change, we constructed 20 independent low resolution ab initio models of Ct-E88 RNA based on the SAXS data using the program DAMMIN.33 The program package DAMAVER34 was used to average the resulting 20 models for each sample condition. The final low-resolution reconstruction obtained was the average molecular envelope of the ensemble (Fig. 6). The normalized spatial discrepancy (NSD), which provides a quantitative estimate of similarity between models, was also computed. Identical models give NSD values at zero, similar models give NSD values around one and models with different shapes give NSD values greater than one (Table 1).42 Here the average NSDs for the three reconstructions were 0.95, 0.74 and 0.60 (each NSD was <1) (Table 1), indicating good convergences in both individual DAMMIN fits and overall bead model ensembles for Ct-E88 RNA. The B state had the lowest average NSD value, consistent with the most rigid and folded state.The low resolution models corresponding to the three states of Ct-E88 RNA indicate that both the unfolded (in the absence of both Mg2+ ions and metabolites) and intermediate (with low Mg2+ ions and in the absence of metabolites) states have the same Dmax of 106 Å, whereas the bound state (in the presence of both Mg2+ ions and metabolites) has a much smaller Dmax of 81 Å. Thus the shape of the envelope of the bound state is distinctly different from the other two states. Given the obvious low resolution limitation of SAXS, detailed interpretation of the envelope for the states can be ambiguous, and information on the location and orientation of helices is not available. Nonetheless, both unfolded and intermediate states exhibit two prominent arms absent in the bound state, suggestive of lack of side-by-side packing of helices P2 and P3, or lack of complete folding of helix P1, or both.
However at this low resolution, the nature of this unbound and intermediate state is still unclear from the SAXS data: is helix P1 partially unfolded, and how are helices P2 and P3 packed? For these and other structural questions detailed high resolution multidimensional NMR spectroscopy will be better suited as we recently showed for the SAM-II riboswitch26 and will be the focus of future studies by our group. However to gain insight into the global conformational changes that are revealed by imino proton NMR we present data for three conditions: free, Mg2+ ion-bound, and Mg2+ ion and metabolite-bound Ct-E88 aptamer RNA (see below).26
NMR study of Ct-E88 RNA with c-di-GMP
NMR chemical shift perturbation experiments are ideally suited for probing the characteristic imino proton chemical shifts found in both canonical and non-canonical Watson–Crick configurations as well as for monitoring the formation and breakage of these base pairs in RNAs to define the conformational change upon interacting with metabolites or metal ions. To corroborate the SAXS data analysis that indicated that the magnesium ion bound form of Ct-E88 aptamer RNA has a conformation similar to the free form, NMR titration experiments were performed to determine whether the Mg2+ ion or the metabolite or both induced structural rearrangements with resultant changes in secondary structural elements.26We first tested the effect of temperature on the NMR experiments and found the optimal temperature to be ∼298 K (data not shown). Titration of the riboswitch with Mg2+ ions in the absence of the metabolite reveals no chemical shift perturbation of the existing imino peaks and no appearance of any strong new signals, indicating that addition of Mg2+ ions does not induce any base pairing interactions within the imino region (Fig. 7). However the titration of Ct-E88 RNA with c-di-GMP in the presence of Mg2+ ions revealed significant chemical shift perturbations of existing imino peaks and the appearance of some strong new peaks marked by arrows (Fig. 7), where the peaks in the region of 12–15 ppm are usually indicative of canonical Watson–Crick base pairs (A:U and G:C base pairings) and peaks in the region of 10–12 ppm are often indicative of non-canonical base pairs (G:U, G:A and G:G base pairings). These results indicate that addition of the ligand indeed triggers changes in the secondary structure and folding of the RNA.
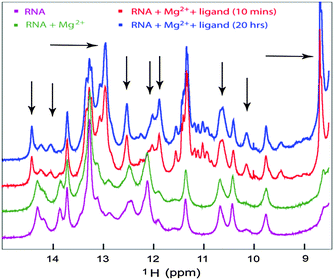 | ||
| Fig. 7 1D NMR titration study of the RNA with the ligand. The imino region of 1D 1H NMR spectra of Ct-E88 RNA, with 3 mM Mg2+ or with both Mg2+ ions and saturating amount of c-di-GMP. | ||
Conclusions
We recently demonstrated that c-di-GMP analogs bind differently to the known class I Vc2 and our new experimentally verified Ct-E88 RNA.23 We now show that even though both Vc2-I and Ct-E88 RNA attain compact global folds in the presence of both Mg2+ ions and c-di-GMP, they do so differently. For example previous work on Vc2-I indicated modest compaction in the presence of physiologic (2.5 mM) Mg2+ (from 32 Å to 28.5 Å) with further compaction to 23.9 Å in the presence of c-di-GMP. Our results indicate that Ct-E88, unlike Vc2, undergoes negligible compaction in the presence of physiologic (2.5 mM) Mg2+. Furthermore SAXS and NMR all further indicated that the c-di-GMP ligand was more critical than Mg2+ for inducing Ct-E88 RNA folding and secondary structure formation. In addition, the fluorescence spinach assay indicated that the last three base pairs in helix P1 of Ct-E88 RNA are not as important for binding affinity and signal transduction as observed for Vc2. In the future, very high resolution structural analysis would be necessary to fully elucidate this intriguing difference between these class I riboswitch RNAs Ct-E88 and Vc2. Insights into understanding these differences might form the basis for the design of small molecules that selectively target different bacteria that harbor different class I c-di-GMP riboswitches in a polymicrobial system, and also enable the design of riboswitches as macromolecular logic gate switches with potential applications in biotechnology.Acknowledgements
We thank Dr Xiaobing Zuo at the Argonne National Laboratory for help with collection of the scattering data. Funding for this work was provided by the National Science Foundation (CHE1307218 to HOS), (CHE1213668 to TKD and HOS), Maryland Biotechnology Award (TKD), American Heart Association (11PRE7890040 fellowship to JZ), UMD Dean's graduate fellowship (JZ), Camille Dreyfus Foundation (HOS) and Jenny T. Sih Endowed Memorial Scholarship (YL).References
- D. Kalia, G. Merey, S. Nakayama, Y. Zheng, J. Zhou, Y. Luo, M. Guo, B. T. Roembke and H. O. Sintim, Chem. Soc. Rev., 2013, 42, 305–341 RSC.
- T. Petters, X. Zhang, J. Nesper, A. Treuner-Lange, N. Gomez-Santos, M. Hoppert, U. Jenal and L. Sogaard-Andersen, Mol. Microbiol., 2012, 84, 147–165 CrossRef CAS PubMed.
- J. R. Tuckerman, G. Gonzalez and M. A. Gilles-Gonzalez, J. Mol. Biol., 2011, 407, 633–639 CrossRef CAS PubMed.
- S. Rakshe, M. Leff and A. M. Spormann, Appl. Environ. Microbiol., 2011, 77, 2196–2198 CrossRef CAS PubMed.
- J. L. Leduc and G. P. Roberts, J. Bacteriol., 2009, 191, 7121–7122 CrossRef CAS PubMed.
- R. Hengge, Nat. Rev. Microbiol., 2009, 7, 263–273 CrossRef CAS PubMed.
- A. Duerig, S. Abel, M. Folcher, M. Nicollier, T. Schwede, N. Amiot, B. Giese and U. Jenal, Genes Dev., 2009, 23, 93–104 CrossRef CAS PubMed.
- J. W. Hickman and C. S. Harwood, Mol. Microbiol., 2008, 69, 376–389 CrossRef CAS PubMed.
- V. T. Lee, J. M. Matewish, J. L. Kessler, M. Hyodo, Y. Hayakawa and S. Lory, Mol. Microbiol., 2007, 65, 1474–1484 CrossRef CAS PubMed.
- D. Srivastava, R. C. Harris and C. M. Waters, J. Bacteriol., 2011, 193, 6331–6341 CrossRef CAS PubMed.
- P. V. Krasteva, J. C. Fong, N. J. Shikuma, S. Beyhan, M. V. Navarro, F. H. Yildiz and H. Sondermann, Science, 2010, 327, 866–868 CrossRef CAS PubMed.
- P. D. Newell, R. D. Monds and G. A. O'Toole, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3461–3466 CrossRef CAS PubMed.
- J. T. Pratt, R. Tamayo, A. D. Tischler and A. Camilli, J. Biol. Chem., 2007, 282, 12860–12870 CrossRef CAS PubMed.
- J. Benach, S. S. Swaminathan, R. Tamayo, S. K. Handelman, E. Folta-Stogniew, J. E. Ramos, F. Forouhar, H. Neely, J. Seetharaman, A. Camilli and J. F. Hunt, EMBO J., 2007, 26, 5153–5166 CrossRef CAS PubMed.
- D. Amikam and M. Y. Galperin, Bioinformatics, 2006, 22, 3–6 CrossRef CAS PubMed.
- E. R. Lee, J. L. Baker, Z. Weinberg, N. Sudarsan and R. R. Breaker, Science, 2010, 329, 845–848 CrossRef CAS PubMed.
- N. Sudarsan, E. R. Lee, Z. Weinberg, R. H. Moy, J. N. Kim, K. H. Link and R. R. Breaker, Science, 2008, 321, 411–413 CrossRef CAS PubMed.
- Y. Luo, J. Zhou, S. K. Watt, V. T. Lee, T. K. Dayie and H. O. Sintim, Mol. Biosyst., 2012, 8, 772–778 RSC.
- P. Ross, R. Mayer, H. Weinhouse, D. Amikam, Y. Huggirat, M. Benziman, E. de Vroom, A. Fidder, P. de Paus and L. A. Sliedregt, et al. , J. Biol. Chem., 1990, 265, 18933–18943 CAS.
- K. D. Smith, S. V. Lipchock, T. D. Ames, J. Wang, R. R. Breaker and S. A. Strobel, Nat. Struct. Mol. Biol., 2009, 16, 1218–1223 CAS.
- N. Kulshina, N. J. Baird and A. R. Ferre-D'Amare, Nat. Struct. Mol. Biol., 2009, 16, 1212–1217 CAS.
- S. Wood, A. R. Ferre-D'Amare and D. Rueda, ACS Chem. Biol., 2012, 7, 920–927 CrossRef CAS PubMed.
- Y. Luo, J. Zhou, J. Wang, T. K. Dayie and H. O. Sintim, Mol. Biosyst, 2013, 9, 1535–1539 RSC.
- M. A. Larkin, G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson and D. G. Higgins, Bioinformatics, 2007, 23, 2947–2948 CrossRef CAS PubMed.
- P. Gouet, E. Courcelle, D. I. Stuart and F. Metoz, Bioinformatics, 1999, 15, 305–308 CrossRef CAS PubMed.
- B. Chen, X. Zuo, Y. X. Wang and T. K. Dayie, Nucleic Acids Res., 2012, 40, 3117–3130 CrossRef CAS PubMed.
- M. Seetharaman, N. V. Eldho, R. A. Padgett and K. T. Dayie, RNA, 2006, 12, 235–247 CrossRef CAS PubMed.
- J. Zhou, D. A. Sayre, J. Wang, N. Pahadi and H. O. Sintim, Molecules, 2012, 17, 13376–13389 CrossRef CAS PubMed.
- S. Nakayama, Y. Luo, J. Zhou, T. K. Dayie and H. O. Sintim, Chem. Commun., 2012, 48, 9059–9061 RSC.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- Y. X. Wang, X. Zuo, J. Wang, P. Yu and S. E. Butcher, Methods, 2010, 52, 180–191 CrossRef CAS PubMed.
- D. I. Svergun, J. Appl. Crystallogr., 1992, 25, 495–503 CrossRef.
- D. I. Svergun, Biophys. J., 1999, 76, 2879–2886 CrossRef CAS.
- V. V. Volkov and D. I. Svergun, J. Appl. Crystallogr., 2003, 36, 860–864 CrossRef CAS.
- K. T. Dayie, J. Biomol. NMR, 2005, 32, 129–139 CrossRef CAS PubMed.
- M. Piotto, V. Saudek and V. Sklenar, J. Biomol. NMR, 1992, 2, 661–665 CrossRef CAS.
- J. R. Cann, Electrophoresis, 1996, 17, 1535–1536 CrossRef CAS PubMed.
- J. S. Paige, T. Nguyen-Duc, W. Song and S. R. Jaffrey, Science, 2012, 335, 1194 CrossRef CAS PubMed.
- J. Lipfert and S. Doniach, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 307–327 CrossRef CAS PubMed.
- X. Zuo, J. Wang, T. R. Foster, C. D. Schwieters, D. M. Tiede, S. E. Butcher and Y. X. Wang, J. Am. Chem. Soc., 2008, 130, 3292–3293 CrossRef CAS PubMed.
- S. Doniach, Chem. Rev., 2001, 101, 1763–1778 CrossRef CAS PubMed.
- M. B. Kozin and D. I. Svergun, J. Appl. Crystallogr., 2001, 34, 33–41 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2014 |