Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Conditional protein splicing of α-sarcin in live cells
Spencer C.
Alford†
ab,
Connor
O'Sullivan†
ab,
Jon
Obst
b,
Jennifer
Christie
ab and
Perry L.
Howard
*abc
aCentre for Biomedical Research, University of Victoria, P.O. Box 3020 Station CSC Victoria, BC, Canada V8W 3N5. E-mail: phoward@uvic.ca
bDepartment of Biochemistry/Microbiology, University of Victoria, P.O. Box 3020 Station CSC Victoria, BC, Canada V8W 3N5
cDepartment of Biology, University of Victoria, P.O. Box 3020 Station CSC Victoria, BC, Canada V8W 3N5
First published on 23rd January 2014
Abstract
Protein splicing technology harnesses the ability of inteins to ligate protein fragments, forming a mature protein. This report describes our effort to engineer rapamycin-dependent protein splicing of a ribotoxin, called α-sarcin. Engineering this system required the investigation of important splicing parameters, including extein context and splicing temperature. We show α-sarcin splicing is dependent on rapamycin, is inducible with rapid kinetics, and triggers apoptosis in HeLa cells. These findings establish a proof-of-concept for a conditional cell ablation strategy.
Introduction
Selective cell ablation at key stages of development is an important strategy used to address questions related to cell autonomy, lineage, and tissue regeneration.1–4 The strategy is also relevant for therapeutic interventions, such as eliminating tumor and transplanted cells.5–9 In each of these examples, cell ablation can be achieved through regulated expression of a suicide gene in a specific population of cells using tissue-specific or inducible promoters. However, this approach is limited by slow induction of cell ablation, which prevents the investigation of many developmental questions. Greater temporal control can be achieved through the use of pro-drug converting enzymes, such as herpes simplex thymidine kinase or cytosine deaminase.9 These enzymes activate a pro-drug to trigger cell death, and the timing is controlled by addition of the pro-drug. However, the action of many drugs created by these enzymes requires actively dividing target cells.9 Other cell ablation strategies, such as expression of bacterial nitroreductase and pro-drug CB1954/MTZ, are able to kill non-dividing cells, but require high levels of enzyme expression.10 Therefore, an efficient inducible cell ablation method that rapidly eliminates both dividing and non-dividing cells is desirable. In this study, we determined whether conditional protein splicing could be used as a strategy to tightly regulate cell ablation.Conditional protein splicing utilizes inteins, which are self-excising protein sequences that remove themselves from a precursor protein and concurrently splice together flanking protein sequences, called exteins.11 This process can be used to regulate protein function in live cells with rapid kinetic resolution.11 Splicing typically requires two sequence determinants: a cysteine or serine within the intein sequence at the N-terminal extein–intein splice junction, and a histidine–asparagine dipeptide sequence at the C-terminal intein–extein splice junction, immediately followed by a cysteine, serine, or threonine as the first C-terminal extein residue.12 Flanking residues within the N-terminal and C-terminal extein sequences also influence splicing efficiency,13,14 and many inteins can accommodate a variety of extein sequences and still splice effectively.15,16
Intein activity may also be regulated by changes in structure and folding. Some inteins occur naturally as split trans-splicing elements, and alternatively, cis-splicing inteins can be artificially split by recombinant methods.17,18 Splicing mediated by split inteins is contingent on reconstitution of the native intein fold upon re-association of the intein fragments. By fusing split intein fragments with low re-association affinities to protein (hetero) dimerization domains, splicing becomes conditional on the addition of a dimerizing agent and the consequent re-association of intein fragments.11,19 Two such varieties of conditional protein splicing have been described for splicing proteins in cultured mammalian cells: an estrogen receptor-hydroxytamoxifen system20,21 and an FKBP–rapamycin–FRB system.11,19
Examples of spliced functional proteins in physiological systems have largely been limited to reporter proteins (e.g., luciferase or GFP).11,19,22–25 To date, demonstrations of splicing non-reporter proteins in live cells has been limited20,26,27 and the approach has not been adapted for cell ablation studies. Several limitations have slowed the broad applicability of protein splicing, including a minimum requirement for a naturally occurring (or artificially installed) cysteine, serine, or threonine at the C-terminal intein–extein splice junction, as well as the possible requirement of installing splicing-permissive extein mutations that may destabilize or inactivate the spliced target protein.13,26 We investigated each of these limitations while developing our splicing-mediated cell ablation method.
We envisioned a strategy in which the cytotoxicity of a protein toxin is regulated by protein splicing, such that addition of a dimerizing agent rapidly activates the toxin. The ability to inducibly trigger toxin activity should be valuable in genetic, developmental, and therapeutic studies where one wants to eliminate specific cells with precise temporal and spatial resolution. To test our concept, we chose a ribonuclease named α-sarcin, which is cytotoxic to many tumor cell lines as well as dividing and non-dividing cells.28–30 Using rational design and molecular biology, we recombinantly split α-sarcin into non-toxic fragments and generated a system to splice the fragments together upon addition of rapamycin. Our demonstration of conditional splicing of α-sarcin adds to the limited number of proteins effectively spliced together in live cells and serves as a proof-of-concept for a conditional cell ablation strategy.
Materials and methods
Molecular biology and construct design
All restriction enzymes, DNA polymerase, and DNA ligase were purchased from New England Biolabs. The α-sarcin gene was initially assembled by annealing complimentary oligonucleotides (Operon) and was subsequently used as a template to PCR amplify its gene fragments. Desired splice junctions were generated by overlap extension PCR and full-length gene fusions, X-VMAN-FKBP and FRB-VMAC-Y were cloned into pcDNA3 plasmids via BamHI/XhoI and HindIII/XhoI, respectively (X = NSar, NGFP, or MBP; Y = CSar, CGFP, or polyHis). All constructs were verified by sequence analysis.Cell culture and transfections
HeLa cells were grown in DMEM supplemented with 10% FBS and penicillin/streptomycin (100 μg ml−1 each) at 37 °C and 5.0% CO2. Cells were seeded at a density of 3.0 × 105 cells per well on 6-well plates. After 24 hours, transfections were performed with designated plasmids using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. Transfected cells were treated with 10 nM rapamycin (unless otherwise stated) or DMSO, 24 hours post transfection. After rapamycin addition, cells were incubated at 30 °C for 12 hours before being processed for induction of apoptosis.Immunoblotting
Whole cell lysates were prepared with 1× SDS-PAGE sample buffer (62.5 mM Tris pH 6.8, 2% (w/v) SDS, 75 mM DTT, 7.5% glycerol, 0.02% bromophenol blue) and heated at 95 °C for ten minutes prior to electrophoresis. Protein samples were resolved through 11% SDS-PAGE gels and transferred to nitrocellulose membranes (0.45 μm) using transfer buffer (25 mM Tris pH 8.3, 192 mM glycine, 20% (w/v) methanol). Anti-GFP (Roche), anti-FLAG (Stratagene), and anti-β-actin antibodies (Sigma) were used at working dilutions of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4000 or 1
4000 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000. Anti-mouse IgG horseradish peroxidase was used as a secondary antibody/detection system (R and D systems) at a working dilution of 1
5000. Anti-mouse IgG horseradish peroxidase was used as a secondary antibody/detection system (R and D systems) at a working dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8000 to 1
8000 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Proteins were detected using an enhanced chemiluminescence immunoblotting detection system (Amersham). Blots were exposed to film and developed using an automated X-ray film developer. Lysate from HeLa cells expressing a non-toxic 3xFLAG-sarcin was used as an electrophoretic mobility standard for spliced 3xFLAG-sarcin.
000. Proteins were detected using an enhanced chemiluminescence immunoblotting detection system (Amersham). Blots were exposed to film and developed using an automated X-ray film developer. Lysate from HeLa cells expressing a non-toxic 3xFLAG-sarcin was used as an electrophoretic mobility standard for spliced 3xFLAG-sarcin.
Cell viability assay
HeLa cells were seeded at 2.0 × 104 cells per well in a 24-well dish and allowed to grow for 24 hours. Transient transfections were performed using Lipofectamine 2000 (Invitrogen). Empty plasmid (pcDNA3.1) was added to the transfection mix to maintain the total DNA concentration per transfection at 500 ng per transfection. 48 hours after transfection, cell viability was assayed. HeLa cells were washed once with phosphate buffered saline and fixed with 10% formalin for 10 minutes at room temperature. Cells were washed two times with dH2O and incubated with 0.1% crystal violet for 30 minutes at room temperature. To remove excess stain, cells were washed with dH2O. Crystal violet was extracted using 10% acetic acid. Extracts were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 and their absorbance was measured at 595 nm using a Victor3V 1420 Multilabel plate reader. Percent viability was determined as the ratio of A595nm of toxin-transfected cells to the A595nm of the pcDNA 3.1 transfected control cells. Experiments were performed in triplicate.
4 and their absorbance was measured at 595 nm using a Victor3V 1420 Multilabel plate reader. Percent viability was determined as the ratio of A595nm of toxin-transfected cells to the A595nm of the pcDNA 3.1 transfected control cells. Experiments were performed in triplicate.
Apoptosis assay
Apoptosis assays were performed using an annexin V-FITC apoptosis Kit (BD Biosciences) and cytometric analysis was conducted on a BD FACSCalibur cytometer. 2.0 × 104 events per sample were acquired using Cell Quest Software (BD Biosciences) and analyzed using FlowJo Software. Data represents the average annexin V positive population for each of the conditions from 5 independent experiments, each performed in triplicate.Transfection and splicing efficiency
24 hours after transfection with either full length GFP or GFP splice constructs, 2.0 × 104 cells were analyzed on a BD FACSCalibur flow cytometer and GFP intensity was quantified. Full length GFP-transfected cells and cells containing successfully spliced GFP were identified by comparison to untransfected cells.Luciferase assay
8.0 × 103 C2C12 cells per well were seeded on a 96-well plate and 24 hours later, cells were transfected with a plasmid encoding luciferase (pMiR-Luc-Control), and either pcDNA3.1 or sarcin constructs as indicated. 4 hours later, samples were treated with 5 nM rapamycin or DMSO, and incubated at 30 °C for 12 hours. The dual-luciferase reporter assay system (Promega) was used according to the manufacturer's instructions, and luminescence was measured using a Perkin Elmer Victor3V 1420 multilabel plate counter. Percent relative translation inhibition was calculated as [1 − (Lucrap/LucDMSO)] × 100.qRT-PCR
2.0 × 105 C2C12 cells per well were seeded on a 6-well plate and transfected after 24 hours with constructs 2 and 9 as well as pMiR-Luc-Control. 4 hours later, samples were treated with 5 nM rapamycin or DMSO, and incubated at 30 °C for 12 hours. RNA was extracted using direct-zol (Zymo Research), cDNA was synthesized using random primers with the high capacity cDNA reverse transcription kit (Life Technologies), and transcripts were amplified using Ssofast EvaGreen Supermix (Bio-Rad) on a Stratagene MX3000P qPCR system. Firefly luciferase cDNA was amplified using the primers 5′-CTCACTGAGACTACATCAGC-3′ and 5′-TCCAGATCCACAACCTTCGC-3′. GAPDH was amplified using the primers 5′-ACGACCCCTTCATTGACCTC-3′ and 5′-GTCTCGCTCCTGGAAGATGG-3′. Relative mRNA fold change was calculated as 2ΔCtLuc(rap-DMSO)/2ΔCtGAPDH(rap-DMSO).Results
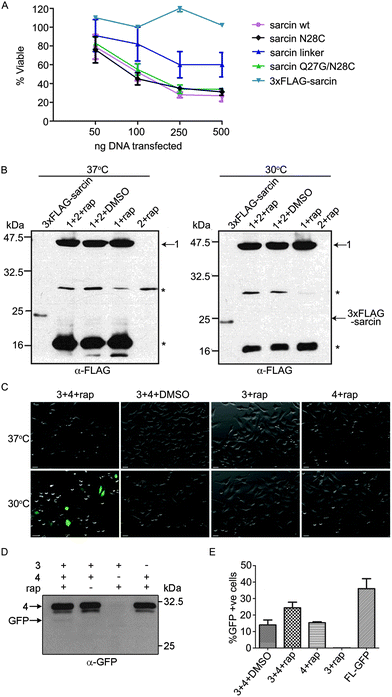
To test whether conditional protein splicing could be used to produce α-sarcin inside mammalian cells, we generated 3-part splicing constructs. First, we cloned the α-sarcin N-terminal fragment (NSar, amino acids 1–27) in frame with the N-terminal VMA intein fragment (VMAN) (Fig. 1A and B). Second, the C-terminal fragment of α-sarcin (CSar, amino acids 28–150) was cloned in frame with the C-terminal VMA intein fragment (VMAC) (Fig. 1A and B). The VMAN and VMAC fragments were, in turn, fused to either the FK506-binding protein (FKBP) or the FK506-rapamycin binding domain of mTor (FRB), respectively11 (Fig. 1B). The splicing constructs were designated NSar-VMAN-FKBP and FRB-VMAC-CSar. In addition, we introduced an N28C mutation into the CSar fragment at the intein splice junction to satisfy the chemical requirements for VMA splicing. Before proceeding, we tested whether the N28C cysteine splicing ‘scar’ would affect α-sarcin toxicity by expressing a full-length N28C α-sarcin mutant in cultured mammalian cells. Expression of the N28C mutant in HeLa cells displayed similar toxicity to wild type α-sarcin, indicating this mutation is tolerated and not detrimental to function (Fig. 2A). Since the N28C product retains cytotoxicity, we next tested whether our designed constructs were able to splice full-length α-sarcin.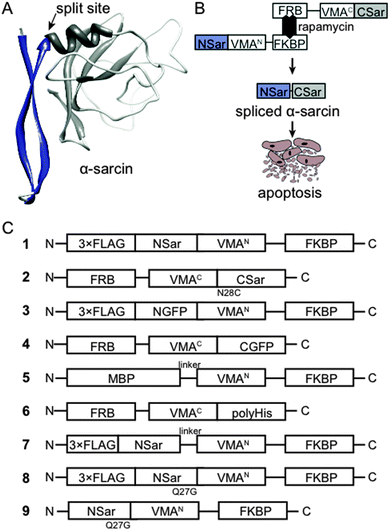 | ||
| Fig. 1 (A) α-Sarcin was split into an N-terminal β-hairpin fragment and a C-terminal catalytic fragment based on its structure.33 (B) General schematic of α-sarcin splicing constructs. Rapamycin drives VMA intein reconstitution and splicing of α-sarcin to induce apoptosis. (C) Schematic of splicing constructs used to design and characterize α-sarcin conditional protein splicing. | ||
We predicted the splicing constructs, NSar-VMAN-FKBP and FRB-VMAC-CSar, would not be toxic when expressed independently or when co-expressed. However, after addition of rapamycin, the VMA intein should reconstitute and splice NSar and CSar fragments together, forming functional α-sarcin (Fig. 1B). We expressed NSar-VMAN-FKBP with an N-terminal 3xFLAG tag (Fig. 1C, construct 1). This enabled immunoblot detection of the precursor as well as any splicing end products. Splicing between construct 1 and FRB-VMAC-CSar (Fig. 1C, construct 2) was thus expected to produce 3xFLAG tagged α-sarcin. Constructs 1 and 2 were expressed in HeLa cells and treated with rapamycin at 37 °C. Construct 1 was readily detected (at 47 kDa) by immunoblot analysis of cell lysates, as were self-cleavage products at 30 and 20 kDa. However, we were unable to detect spliced 3xFLAG α-sarcin in rapamycin-treated cells (Fig. 2B). We speculated the lack of splicing was attributed to a non-optimal incubation temperature.
The VMA intein demonstrates increased splicing activity at temperatures below 37 °C.31 Therefore, we lowered the incubation temperature of our experiments to 30 °C to determine if the elevated temperature of cultured mammalian cells (37 °C) was preventing α-sarcin splicing (Fig. 2B). Even at the reduced temperature we did not detect any spliced product. To confirm the split VMA intein was functional, we generated splicing constructs comprising split green fluorescent protein (GFP) fragments. An N-terminal fragment of GFP (1–128) was fused to VMAN-FKBP (Fig. 1C, construct 3) and the C-terminal fragment (129–238) was fused to FRB-VMAC (Fig. 1C, construct 4). The GFP split site, between residues 128 and 129, falls at the end of β-strand 6 and has previously been shown amenable to intein-mediated splicing in vitro.23,25 To facilitate splicing we introduced I129C and E125I mutations, as previously described.23 Using constructs 3 and 4, GFP fluorescence was readily visible in the presence of rapamycin at the reduced temperature of 30 °C, whereas no fluorescence was detected at 37 °C (Fig. 2C). Splicing of full length GFP was confirmed by immunoblot analysis using an anti-GFP antibody. Although the amount of spliced product compared to the amount of precursor is low, the detection of this band following SDS-PAGE indicates the fluorescence is likely from the spliced GFP protein (Fig. 2D) and not merely protein complementation of the GFP fragments. Analysis of cell lysates showed background fluorescence associated with construct 4, which contains the C-terminal fragment of GFP (Fig. 2E). We detected fluorescence associated with construct 4 by flow cytometry, but it was not visible by fluorescence microscopy (Fig. 2C). Importantly, this background signal does not increase when construct 3 is co-expressed in the absence of rapamycin, indicating fluorescence is dependent on rapamycin and splicing (Fig. 2E). Together, these results indicated the VMA intein (in the context of GFP and α-sarcin) splices poorly at 37 °C, and the intein-α-sarcin splice junctions interfere with splicing.
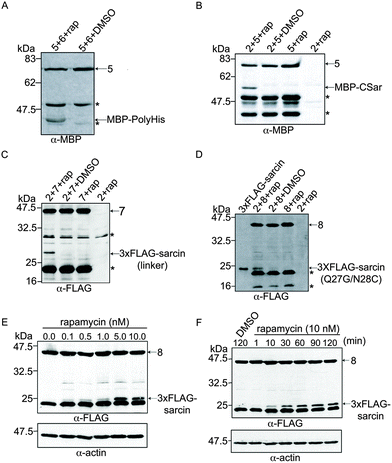
To investigate the α-sarcin extein context at the splice junctions, we used splicing constructs 5 and 6 (Fig. 1C), which are comprised of Maltose binding protein (MBP) fused to VMAN-FKBP and a poly-Histidine (polyHis) sequence fused to FRB-VMAC, respectively. These splicing constructs were used in the first demonstration of conditional protein splicing.11,19 As expected, we readily detected splicing of MBP to polyHis (Fig. 3A). We next attempted to splice MBP to the CSar fragment (using constructs 5 and 2, Fig. 1C) to determine whether the C-terminal splice site within α-sarcin was functional. The CSar fragment readily spliced to MBP, indicating the splicing defect was likely within the NSar extein (Fig. 3B). To test this, we generated construct 7 (Fig. 1C), in which the NSar fragment is fused to the VMAN through a 5 amino acid linker identical in sequence (EFLKG) to the splicing-permissive linker joining MBP and VMAN in construct 5.19 Under these conditions, the 3xFLAG NSar fragment was spliced to the CSar fragment after rapamycin addition, and resulted in a slightly higher migrating band (>25 kDa) as expected from the additional amino acids provided by the linker (Fig. 3C). Therefore, the extein sequence context contributed by the native NSar fragment is not suitable for splicing. Unfortunately, inserting the construct 5 linker sequence into α-sarcin significantly attenuated its toxicity, rendering it unsuitable for our purposes (Fig. 2A).
Based on the above results, we designed a modified NSar extein, in which we eliminated the linker sequence, but introduced a mutation at the NSar residue immediately preceding the intein sequence (designated as the −1 position). We hypothesized the native glutamine found at this position in α-sarcin interferes with splicing and mutating it to glycine, as found in the linker, would improve splicing.15 We first tested whether a full-length α-sarcin Q27G mutant (plus N28C) would retain cytotoxicity as a splicing end product. Introduction of Q27G/N28C into α-sarcin did not affect its cytotoxicity when expressed in HeLa cells (Fig. 2A). Therefore, Q27G was incorporated into the NSar fragment to generate construct 8 (Fig. 1C). In cells transfected with constructs 2 and 8, rapamycin treatment resulted in the production of a spliced product at ∼23 kDa which co-migrated with a full-length 3xFLAG-α-sarcin electrophoretic standard (Fig. 3D). We generated this standard by directly expressing full length 3xFLAG-sarcin from a plasmid in HeLa cells and lysing the cells under identical conditions used in the splicing experiments. Although a faint band at the same size was detected in cells treated with DMSO (Fig. 3D), it was not detected in subsequent experiments (Fig. 3E and F and data not shown), and we conclude it was cross contamination between lanes which occurred during loading. After confirming our system was capable of splicing α-sarcin, we next investigated the inducibility and timescale of splicing.
The extent of conditional protein splicing should depend on concentration of the dimerizing agent (rapamycin in our system).19 To determine whether the spliced α-sarcin product was dependent on rapamycin concentration, constructs 2 and 8 were expressed in HeLa cells and treated with various doses of rapamycin, followed by a two hour incubation. Anti-FLAG immunoblot analysis of cell lysates showed dose-dependent splicing activity; increasing amounts of the spliced product were detected with increasing rapamycin concentrations (Fig. 3E). Splicing was detectable at as low as 0.1 nM rapamycin, and dose-dependence was observed between 0.5 and 10 nM. This compares well to previously reported rapamycin-mediated splicing of MBP to polyHis.11,19 To determine the timescale of α-sarcin splicing, we repeated the experiment using 10 nM rapamycin, and prepared cell lysates at various time points over two hours. Immunoblot analysis showed the spliced product appeared within ten minutes of rapamycin addition and increased steadily over the two-hour period (Fig. 3F). The analyses discussed above highlight features of the α-sarcin splicing system that are advantageous for a conditional cell ablation strategy, but we next investigated if the spliced α-sarcin product exhibited cytotoxicity.
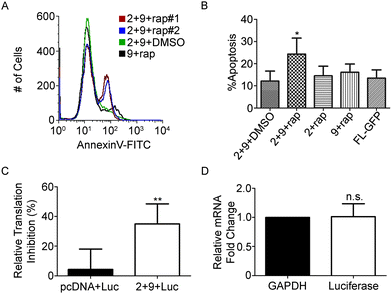
Expression of α-sarcin in the cytoplasm of HeLa cells induces apoptosis.32 However, the addition of an N-terminal 3xFLAG tag on α-sarcin significantly attenuates its cytotoxicity (Fig. 2A).32 This feature permitted immunoblot characterization, but was not amenable for cell ablation. To address this, we generated construct 9, (Fig. 1C) which contained an untagged NSar Q27G fragment, and determined whether the levels of spliced α-sarcin were sufficient to kill cells. HeLa cells were co-transfected with constructs 2 and 9 and were incubated with 10 nM rapamycin, or DMSO. After 12 hours at 30 °C, cell viability was assayed. As shown in Fig. 4A and B, the addition of rapamycin, but not DMSO, induced apoptosis. Approximately 25% of the cells treated with rapamycin were apoptotic. Since the overall transfection efficiency was 36%, as determined by GFP expression (Fig. 2E), an approximate 70% ablation efficiency of transfected cells was obtained. This level of apoptosis was not obtained when either constructs 2 or 9 were expressed individually with rapamycin, or when the constructs were co-expressed in the absence of rapamycin. Nor was the toxicity associated with expression of constructs 2 or 9, individually or together, above the apoptosis seen with the GFP splicing constructs (Fig. 4A and B). The efficiency of apoptosis induction (Fig. 4B) and GFP fluorescence (Fig. 2E) were similar. This suggests that in cells not undergoing apoptosis, the lack of cell death is related to poor folding and stability of the intein constructs and not α-sarcin resistance. To further support the induction of apoptosis we observed was due to production of α-sarcin, we tested whether we could detect inhibition of protein translation that characteristically accompanies α-sarcin activity in cells. The toxicity of α-sarcin is linked to its ribonucleolytic action on 28S rRNA, which stops protein synthesis, and leads to apoptosis. To confirm spliced α-sarcin inhibited protein synthesis, we measured the production of a luciferase reporter after triggering splicing. Co-expression of constructs 2 and 9 inhibited luciferase production after rapamycin treatment (Fig. 4C). No change in mRNA levels of the reporter were detected following rapamycin treatment (Fig. 4D), confirming the decrease in luciferase activity was due to protein synthesis inhibition. In summary, the fact that with a single amino acid change to the NSar extein, a rapamycin-induced FLAG-tagged product was detected by immunoblotting and migrated at precisely the same size as 3xFLAG-sarcin, supports that this was a splicing product and not the result of cleavage and/or degradation. Furthermore, this conclusion is supported by the rapid kinetics and dose dependency of product production.11 The trademark protein synthesis inhibition and accompanying induction of apoptosis seen with constructs 2 and 9 solely in the presence of rapamycin argues the observed cytotoxicity is due to correct splicing of α-sarcin. Taken together, our results show that cytotoxicity of α-sarcin can be regulated by conditional protein splicing.
Discussion
Our report describes the conditional protein splicing of a protein toxin, α-sarcin, in mammalian cells. We showed that by introducing two mutations, which do not affect cytotoxicity, α-sarcin can be split in two non-toxic fragments and quickly reassembled through rapamycin-mediated protein splicing in live cells. This has several advantages for cell ablation studies. First, α-sarcin is toxic in low amounts.28,29 Indeed, although we could not detect the untagged α-sarcin by immunoblot (data not shown), its cytotoxicity was still evident. Therefore, high efficiency splicing is not required. Second, α-sarcin induces apoptosis, in part, by inactivating the ribosome and inhibiting protein synthesis, and is toxic to both dividing and non-dividing cells. Therefore, this ablation strategy is applicable to situations where targeting post-mitotic or quiescent populations is desired. Third, the timescale required for α-sarcin splicing is rapid. Compared to conventional promoter-driven expression, which typically requires 6–8 hours before protein can be detected by immunoblotting, spliced α-sarcin was produced within 10 minutes. Together, the low dose of rapamycin required and the low background splicing should allow researchers to better study kinetic and developmental questions. Although these features are advantageous, our investigation revealed important design considerations and potential obstacles.Fortunately, our study showed these obstacles can be overcome with molecular engineering. Identifying proteins amenable to splicing and choosing appropriate splice sites relies largely on informed guesses based on protein structures and empirical testing of several splice sites. Choosing splice sites that lie between defined domains is not possible when the target protein lacks distinct domains or is comprised of a single domain. Using its solution structure as a guide,33 we chose to split α-sarcin between glutamine-27 and asparagine-28 to generate an N-terminal β-hairpin fragment and a C-terminal catalytic fragment (Fig. 1A). Our rationale to split α-sarcin at this site was that deletion of the N-terminal β-hairpin produces a catalytically active, but non-lethal fragment.34 The fact that the C-terminal fragment retains ribonuclease activity in the absence of the N-terminus implies that it can fold independently.34 Despite presumed autonomous folding of the fragments, our initial attempt at splicing was unsuccessful due to intolerance of the native extein sequence contributed by the NSar fragment. The MBP-VMAN-FKBP and FRB-VMAC-polyHis splicing constructs were critical to identifying the defect as a non-permissive NSar extein residue and provided an easy method to diagnose splicing deficiencies. Although placement of a glycine at the −1 position is not essential and can be substituted with other amino acid residues, there has been some selection for this residue in VMA intein.15 Substitutions at this position are thought to unfavorably shift the equilibrium of the splicing reaction.15 Fortunately, the Q27G/N28C double mutation did not affect the toxicity of α-sarcin when directly expressed in the cytoplasm. Thus, with relatively minor changes it was possible to split α-sarcin into two fragments amenable to protein splicing. Strategies for evolving more promiscuous inteins that tolerate a greater diversity of extein sequences, as has been engineered for the Ssp. DnaB intein, should allow for easier adaptation of this method in the future.16 In addition to the contribution of extein sequence, we also confirmed temperature significantly affects splicing efficiency.31
We chose the VMA split intein for this study because the N- and C-terminal intein fragments show little self-association in the absence of a dimerizing agent.11,19 However, a major limitation to using the VMA intein was poor splicing at 37 °C. This appears to be contextual since the VMA intein/rapamycin-inducible system was previously shown to splice protein fragments at 37 °C in mammalian cells.11 In this instance, intact MBP and polyHis peptide were used as extein fragments and, therefore, similar folding challenges associated with splitting a native protein were not encountered. Evidence of the instability of our intein splicing constructs could be seen in the self-cleavage product of approximately 20 kDa that was detected by anti-FLAG immunoblotting. The presence of this species was decreased at 30 °C (Fig. 2B), suggesting improved stability at the lower temperature may account for the increase in splicing. The inability to splice at 37 °C currently restricts the applicability of our cell ablation strategy to cell culture or to organisms whose ambient temperature is 30 °C or lower. To overcome this limitation in the future, directed evolution could be applied to the VMA intein using NGFP-VMAN, CGFP-VMAC, and GFP fluorescence as a read out. A directed evolution strategy could also be used to improve the efficiency of splicing, which is currently about 70%. Indeed, several studies have already utilized direction evolution to improve splicing of various inteins16,21,35 and we speculate our system can be improved by optimizing the intein sequence as well as the flanking α-sarcin sequences.36–39 Alternatively, using different inteins in the context of the FKBP–FRB system may circumvent this problem.
Conclusions
We have tested whether protein splicing could be exploited to inducibly activate the protein toxin, α-sarcin. After optimizing temperature and extein sequence parameters, we successfully spliced α-sarcin in cultured mammalian cells. Splicing was rapidly induced and dosable with rapamycin concentration. Additionally, translation inhibition and apoptosis were initiated following splicing of α-sarcin. These findings collectively provide a proof-of-concept for a conditional cell ablation strategy with potential applications in developmental biology or other studies requiring the rapid elimination of difficult-to-target quiescent populations of cells.Acknowledgements
The authors thank Dr T. Muir for providing constructs containing the VMA intein fragments conjugated to the FRB and FKBP. This work was supported by a NSERC Discovery Grant awarded to PLH.References
- B. Fraser, M. G. DuVal, H. Wang and W. T. Allison, PLoS One, 2013, 8, e55410 CAS.
- K. Araki, M. Araki and K. Yamamura, J. Biochem., 2006, 140, 793–798 CrossRef CAS PubMed.
- M. Schuldiner, J. Itskovitz-Eldor and N. Benvenisty, Stem Cells, 2003, 21, 257–265 CrossRef CAS PubMed.
- S. Tanoue, P. Krishnan, B. Krishnan, S. E. Dryer and P. E. Hardin, Curr. Biol., 2004, 14, 638–649 CrossRef CAS PubMed.
- M. Malecki, J. Genet. Syndr. Gene Ther., 2012, 3, e114 Search PubMed.
- R. J. Kreitman and I. Pastan, Curr. Drug Targets, 2006, 7, 1301–1311 CrossRef CAS.
- D. Kirn, I. Niculescu-Duvaz, G. Hallden and C. J. Springer, Trends Mol. Med., 2002, 8, S68–S73 CrossRef CAS.
- W. A. Denny, Cancer Invest., 2004, 22, 604–619 CrossRef CAS PubMed.
- W. A. Denny, J. Biomed. Biotechnol., 2003, 1, 48–70 CrossRef PubMed.
- W. Cui, B. Gusterson and A. J. Clark, Gene Ther., 1999, 6, 764–770 CrossRef CAS PubMed.
- H. D. Mootz, E. S. Blum, A. B. Tyszkiewicz and T. W. Muir, J. Am. Chem. Soc., 2003, 125, 10561–10569 CrossRef CAS PubMed.
- S. Elleuche and S. Poggeler, Appl. Microbiol. Biotechnol., 2010, 87, 479–489 CrossRef CAS PubMed.
- G. Amitai, B. P. Callahan, M. J. Stanger, G. Belfort and M. Belfort, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 11005–11010 CrossRef CAS PubMed.
- M. Q. Xu and F. B. Perler, EMBO J., 1996, 15, 5146–5153 CAS.
- S. Chong, K. S. Williams, C. Wotkowicz and M. Q. Xu, J. Biol. Chem., 1998, 273, 10567–10577 CrossRef CAS PubMed.
- J. H. Appleby-Tagoe, I. V. Thiel, Y. Wang, Y. Wang, H. D. Mootz and X. Q. J. Liu, Biol. Chem., 2011, 286, 34440–34447 CrossRef CAS PubMed.
- D. D. Martin, M. Q. Xu and T. C. Evans Jr., Biochemistry, 2001, 40, 1393–1402 CrossRef CAS PubMed.
- T. C. Evans, Jr., D. Martin, R. Kolly, D. Panne, L. Sun, I. Ghosh, L. Chen, J. Benner, X. Q. Liu and M. Q. Xu, J. Biol. Chem., 2000, 275, 9091–9094 CrossRef PubMed.
- H. D. Mootz and T. W. Muir, J. Am. Chem. Soc., 2002, 124, 9044–9045 CrossRef CAS PubMed.
- C. M. Yuen, S. J. Rodda, S. A. Vokes, A. P. McMahon and D. R. Liu, J. Am. Chem. Soc., 2006, 128, 8939–8946 CrossRef CAS PubMed.
- S. H. Peck, I. Chen and D. R. Liu, Chem. Biol., 2011, 18, 619–630 CrossRef CAS PubMed.
- A. Kanno, T. Ozawa and Y. Umezawa, Anal. Chem., 2006, 78, 556–560 CrossRef CAS PubMed.
- T. Ozawa, S. Nogami, M. Sato, Y. Ohya and Y. Umezawa, Anal. Chem., 2000, 72, 5151–5157 CrossRef CAS.
- T. Ozawa, A. Kaihara, M. Sato, K. Tachihara and Y. Umezawa, Anal. Chem., 2001, 73, 2516–2521 CrossRef CAS.
- T. Ozawa, T. M. Takeuchi, A. Kaihara, M. Sato and Y. Umezawa, Anal. Chem., 2001, 73, 5866–5874 CrossRef CAS.
- T. Sonntag and H. D. Mootz, Mol. Biosyst., 2011, 7, 2031–2039 RSC.
- S. S. Wong, I. Kotera, E. Mills, H. Suzuki and K. Truong, Cell Calcium, 2012, 51, 57–64 CrossRef CAS PubMed.
- J. C. Jennings, B. H. Olson, V. Roga, A. J. Junek and D. M. Schuurmans, Appl. Microbiol., 1965, 13, 322–326 CAS.
- Y. Endo, P. W. Huber and I. G. Wool, J. Biol. Chem., 1983, 258, 2662–2667 CAS.
- K. Eiklid, S. Olsnes and A. Pihl, Exp. Cell Res., 1980, 126, 321–326 CrossRef CAS.
- E. C. Schwartz, L. Saez, M. W. Young and T. W. Muir, Nat. Chem. Biol., 2007, 3, 50–54 CrossRef CAS PubMed.
- S. C. Alford, J. D. Pearson, A. Carette, R. J. Ingham and P. L. Howard, BMC Biochem., 2009, 10, 9, DOI:10.1186/1471-2091010-9.
- J. M. Perez-Canadillas, J. Santoro, R. Campos-Olivas, J. Lacadena, P. A. Martinez del, J. G. Gavilanes, M. Rico and M. Bruix, J. Mol. Biol., 2000, 299, 1061–1073 CrossRef CAS PubMed.
- L. Garcia-Ortega, M. Masip, J. M. Mancheno, M. Onaderra, M. A. Lizarbe, M. F. Garcia-Mayoral, M. Bruix, D. P. Martinez and J. G. Gavilanes, J. Biol. Chem., 2002, 277, 18632–18639 CrossRef CAS PubMed.
- A. R. Buskirk, Y. C. Ong, Z. J. Gartner and D. R. Liu, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 10505–10510 CrossRef CAS PubMed.
- M. W. Southworth, K. Amaya, T. C. Evans, M. Q. Xu and F. B. Perler, Biotechniques, 1999, 27, 110–120 CAS.
- S. W. Lockless and T. W. Muir, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10999–11004 CrossRef CAS PubMed.
- S. Chong, G. E. Montello, A. Zhang, E. J. Cantor, W. Liao, M. Q. Xu and J. Benner, Nucleic Acids Res., 1998, 26, 5109–5115 CrossRef CAS PubMed.
- H. Iwai, S. Zuger, J. Jin and P. H. Tam, FEBS Lett., 2006, 580, 1853–1858 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |