Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Differential protein profile in sexed bovine semen: shotgun proteomics investigation†
Michele
De Canio
a,
Alessio
Soggiu
a,
Cristian
Piras
a,
Luigi
Bonizzi
a,
Andrea
Galli
b,
Andrea
Urbani
cd and
Paola
Roncada
*ab
aDipartimento di Scienze Veterinarie e Sanità Pubblica (DIVET), Università degli Studi di Milano, Milano, Italy. E-mail: paola.roncada@guest.unimi.it; Fax: +39 02 50318171; Tel: +39 02 50318138
bIstituto Sperimentale Italiano L. Spallanzani, Milano, Italy
cDipartimento di Medicina Sperimentale e Chirurgia, Università di Roma “Tor Vergata”, Roma, Italy
dFondazione Santa Lucia-IRCCS, Roma, Italy
First published on 10th October 2013
Abstract
The preparation of sexed semen is based on the differential DNA content between the X and Y chromosome bearing sperm cells determined by fluorescence-activated cell sorting. In spite of its intrinsic limitations this represents the only effective method. However, the employment of sexed sperm for breeding food producing animals on a large scale requires additional knowledge in the protein repertoire for the development of improved methods to differentiate X and Y sperm cells maintaining high vitality. In order to address this issue, we performed a comparative shotgun proteomic investigation by nUPLC-MS/MS to characterize sexed bovine semen. The protein profiles of these two types of sperm cells have shown differential expression of proteins that may be directly associated with the main components of cytoskeletal structures of flagellum, as the axoneme, outer dense fibers and fibrous sheath, as well as glycolytic enzymes and calmodulin, involved in the energetic metabolism regulation. Overall these results may provide a base to a better comprehension of the biological features of sperm cells and may be useful to the development of alternative methods of separation.
1. Introduction
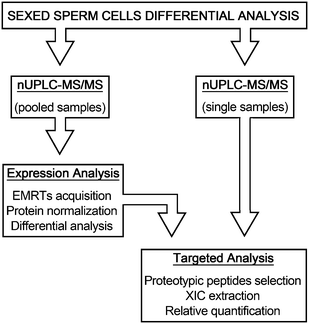
The possibility to control the sex of offspring in food producing mammals is a topic of great interest for researchers of breeding technology. Controlling the ratio between males and females entails direct returns in the livestock industry allowing a better management of food production, faster genetic selection, animal welfare improvement and a decrease of environmental impact.1 The technological development of flow cytometry methods for sorting living X- and Y-chromosome bearing sperm made this possible in Bos taurus species.2 This technology, based on the difference of haploid DNA content between X- and Y-chromosome bearing sperm, evaluated to be 4.05–4.98% depending of cattle breeds,3 provides a tool to separate gametes carrying either the X or Y chromosome with an accuracy of 85–95%.4 But, despite the technological improvements that have succeeded over the years, sorting procedure is still limited by the relatively low number of sperm sorted per unit time (approximately 20 million sperm per hour).1 Moreover, sample dilution diminishes the fertilizing potential of spermatozoa, causing the dilution of protecting and regulating substances of seminal plasma,5 that leads to low pregnancy rates when cryopreserved sexed spermatozoa are employed for the artificial insemination of cows.6 DNA staining methods and UV irradiation also damage spermatozoa and reduce the embryonic viability.7 Although lower pregnancy rates or embryo production using sexed sperm by flow cytometry have been well documented,4,8 the associated mechanisms and the molecular modifications which cause fertility losses and impair embryo production are poorly understood. In spite of these limitations, flow cytometric sorting is the only method to separate X- and Y-chromosome bearing sperm that is suitable for commercial application.2 The relevance of pre-selecting the sex of mammalian offspring pushes for the discovery of faster, cheaper and more efficient methods for the separation of sexed semen. This technological advancement may be reached only by a deeper knowledge of sperm cell biology. In recent years, the application of proteomic methodologies to this field leads to unparalleled progress in the identification of sperm protein constituents, such as transmembrane proteins, kinases and chaperones never previously recognized, providing promising means to answer biological questions related to sperm cells.9 Moreover, specific differential protein content has been correlated with different estimated relative conception rates (ERCR),10 thus associating spermatozoa protein representation with the fertility rates.11 Over the years, several reports engaged the question of diversity between X and Y sperm cells demonstrating significant dissimilarity in structure, morphology, motility and energy metabolism.12,13 However, these differences have not found confirmation in studies on protein expression in sexed sperm cells.14,15 More recently, an extensive proteomic investigation of sexed sperm cells by 2-DE/MS shed light on differentially expressed proteins between bull X and Y spermatozoa including proteins involved in energy metabolism, cellular defense and stress, cytoskeleton and inhibitors of serine proteases.16 In this work, we employed a different proteomic approach based on a nUPLC-MS/MS strategy, with the aim to investigate the global protein expression profiles of these two cell populations. The experimental design was organized in two analytical phases, the discovery phase directed to highlight putative proteome differences between Y- and X-chromosome bearing sperm cells, and the validation phase, where samples were singularly analyzed in order to confirm the findings (Fig. 1). The results reported here provide information on the basic biochemical differences between X and Y sperm cells required for the development of new cell sorting method alternatives to the actual DNA-based procedure.2. Methods
2.1 Sexed sperm cell production
Three bulls were utilized for producing a total of six batches of sexed sperm cells. Ejaculates were collected via an artificial vagina and immediately evaluated for concentration and visual motility using spectrophotometer and phase contrast microscopy, respectively. Ejaculates with concentrations less than 500 million sperm per mL were rejected. A single ejaculate from each bull was sorted in two batches of sexed X- and Y-chromosome bearing sperm cells by Cogent Breeding Ltd (Aldford, Chester, UK). Semen was processed according to the Beltsville sperm sorting technology.17 The procedure is briefly described as follows: an aliquot of fresh semen was stained with Hoechst 33342 for 45 minutes at 34 °C in modified Tyrode's albumin lactate pyruvate buffer18 at approximately 100 × 106 sperm per mL. Fluorescence of dead or moribund cells was quenched by adding the vital-staining red food dye (FD&C 40) so that these cells were removed during the sorting process. Samples were then sorted at a rate of 5500 cells per second and pressure of 40 psi, and collected into conical plastic tubes in TRIS extender without egg yolk. After sorting, sexed sperm samples were centrifuged and cell pellets were re-suspended in TRIS extender and frozen according to the “Harmony frozen” method by Cogent Breeding Ltd protocol. Sexed sperm cells were packaged and frozen in 0.25 mL straws at 5 million total concentration. The percentage of spermatozoa bearing X or Y chromosomes, estimated using a real-time PCR method,19 was always above 90%.2.2 Sperm sample preparation
Three aliquots of sexed X- and three of Y-sperm cells (5 × 106 cells each), originating from different sorting preparations, were washed 5 times with PBS then spermatozoa were re-suspended in 100 mM Tris/HCl pH 7.9 containing 6 M urea and lysed by sonication. Reduction and alkylation of proteins were obtained by adding 100 mM DTT (1 h at 36 °C) and 200 mM iodoacetamide (1 h at RT). Protein samples, at a final concentration of 3 μg μL−1, were digested with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 (w/w) sequence grade trypsin (Promega, Madison, WI, USA) at 36 °C overnight. Reactions were stopped by adding 0.3% (v/v) TFA.
20 (w/w) sequence grade trypsin (Promega, Madison, WI, USA) at 36 °C overnight. Reactions were stopped by adding 0.3% (v/v) TFA.
2.3 Chromatography and mass spectrometry
Tryptic digestions of protein extracts from three different batches of X- or Y-sexed sperm cells were pooled in order to obtain two samples representative of X and Y sperm cells. Pooled samples and single digestions for targeted proteomic analysis were analyzed as previously described.20 Briefly, 0.75 μg of digested proteins were loaded on nanoACQUITY UPLC System (Waters Corp., Milford, MA) coupled to a Q-Tof Premier mass spectrometer (Waters Corp., Manchester, UK). Saccharomyces cerevisiae Enolase (ScEnolase) digestion was added to samples as an internal standard at a final concentration of 100 fmol μL−1. Tryptic peptides were trapped and desalted onto a Symmetry C18 5 μm, 180 μm × 20 mm precolumn (Waters Corp.) and subsequently separated using a NanoEase BEH C18 1.7 μm, 75 μm × 25 cm nanoscale LC column (Waters Corp.) operating at 35 °C. Peptide separation was obtained by a gradient of 3–40% B over 150 min at a flow rate of 250 nL min−1, followed by a gradient of 40–90% B over 5 min and a 15 min rinse with 90% B (phase A: water with 0.1% formic acid; phase B: 0.1% formic acid in acetonitrile). The Q-Tof Premier mass spectrometer operated in “Expression Mode” switching between low (4 eV) and high (15–40 eV) collision energies with a scan time of 0.8 s over 50–1990 m/z mass range. Each X or Y sperm sample was analyzed while performing 3 technical replicates.2.4 Database search
nUPLC-MS/MS data were processed using ProteinLynx GlobalServer v2.4 (PLGS, Waters Corp.). Protein identifications were obtained with the embedded ion accounting algorithm of PLGS software searching into the UniProtKB/Swiss-Prot bovine database release 2012_01 (Bos Taurus; 5879 entries) to which the sequence of ScEnolase was appended. Parameters for the database search were: automatic tolerance for precursor ions, automatic tolerance for product ions, minimum 3 fragment ions matched per peptide, minimum 7 fragment ions matched per protein, minimum 2 peptide matched per protein, 1 missed cleavage, carbamydomethylation of cysteines and oxidation of methionines as fixed and variable modifications. The false positive rate (FPR) of the identification algorithm was set under 4%.2.5 Protein expression profiling
Relative quantitative analysis was performed by the PLGS dedicated tool. Identified proteins were normalized against P00924 entry (ScEnolase) while the most reproducible peptides for retention time and intensity deriving from ScEnolase digestion (m/z 745.43; m/z 814.49; m/z 1288.70; m/z 1416.72; m/z 1755.95) were used to normalize the EMRTs table. The list of normalized proteins was screened according to the following criteria: protein identified in at least 2 out of 3 runs of the same sample; proteins with a 0 < P < 0.05 or 0.95 < P < 1, and proteins with a ratio of expression level ± 0.30 on a natural log scale (about 1.3 on decimal scale).2.6 Targeted label-free quantitation
Targeted analysis was performed in order to assess the relative quantity of the differential proteins in single X and Y cell protein digestions separately run in triplicate. Skyline open source software was employed to build a method for analyzing mass spectrometry data.21 A peptide MS/MS spectral library was created from a csv file generated by PLGS software including peptide and fragment ions information from a pooled sperm sample run on the previously described Waters platform. Duplicate or repeated peptides were automatically removed, as well as peptides without a matching protein in the background proteome, the “Bos taurus” taxonomy-limited sequence list from UniProtKB/Swiss-Prot database. The peptide list was also reduced to the modulated proteins in the PLGS protein expression analysis. Proteotypic peptides were selected following the criteria: uniqueness in the reference background proteome; lack of methionine and tryptophan residues in the amino acid sequence; peptide length ranging from 8 and 25 amino acid residues; evaluation of precursor ion intensity in MS/MS spectra. Raw data from each chromatographic run were processed in order to generate the eXtracted Ion Chromatogram (XIC) of selected peptide masses. Chromatographic traces were inspected for the proper peak picking and peak areas were manually adjusted when necessary. After Savitzky–Golay smoothing, the peaks corresponding to the precursor ion retention times were integrated and the resulting peak areas were normalized to the spiked in ScEnolase peptides and then multiplied by a normalization factor of 100. The normalized peak area was averaged across all X or Y cell acquisitions and a ratio Y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X was generated. A P-value was achieved by a two-tailed, unpaired Mann–Whitney test.
X was generated. A P-value was achieved by a two-tailed, unpaired Mann–Whitney test.
3. Results
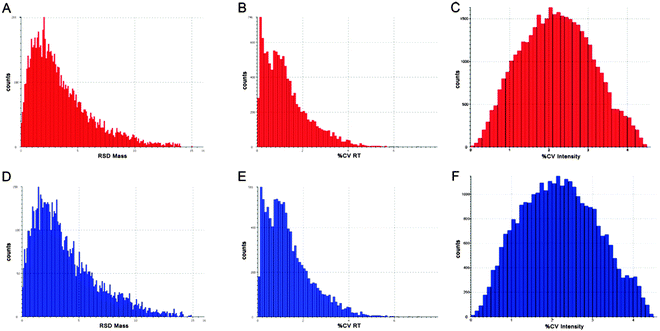
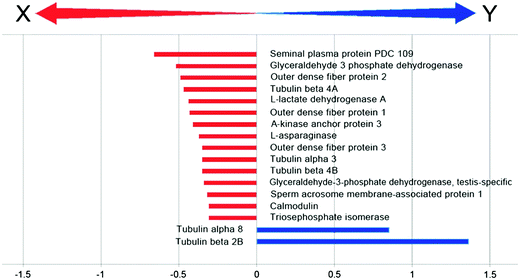
In order to focus on differential expressed proteins between Y- and X-chromosome bearing sperm cells, we performed a comparative analysis of pooled samples of sexed sperm by shotgun nUPLC-MS/MS (Fig. 1). 24.852 EMRTs (Exact Mass Retention Time) peptide clusters were recognized by PLGS software with a RSD (Relative Standard Deviation) mass below 15 ppm (mean 3.2 ppm) and %CV RT (Coefficient of Variation on Retention Time) below 5% (mean 1.2%). The coefficient of variation of EMRTs intensities (%CV Intensity) showed Gaussian distributions centered on mean value of 2.4 %CV in both Y and X sample analyses based on triplicates (Fig. 2). Protein expression profiling was performed after EMRTs and protein normalization. Only proteins identified in at least two of three injections with a fold change higher than 1.3 were taken into consideration. A fold change of 1.3 (0.30 on a natural log scale) indicates expression levels modulated by 30%. Moreover, results were filtered by PLGS statistic filter in order to select only modulated proteins with a confidence level greater than 95%. Applying these stringent filtering criteria 17 differentially expressed proteins were highlighted (Table 1 and Fig. 3), 15 of them were found up-regulated in X-bearing sperm cells, 2 in Y-cells (further details are available as Table S1, ESI†). Most of them were found to be proteins with structural function (five isoforms of Tubulin, three components of outer dense fibers and A-kinase anchor protein 3) while four were glycolytic enzymes (two isoforms of glyceraldehyde 3-phosphate dehydrogenase, triosephosphate isomerase and L-lactate dehydrogenase A). Finally, 4 different proteins were found more expressed in X-cells: L-asparaginase involved in amino acid catabolism, the regulatory protein calmodulin, seminal plasma protein PDC109, a chaperon-like protein which is strongly associated to sperm membranes and sperm acrosome membrane-associated protein 1.| Hit | Accessiona | Description | Mass | Theor. pI | SeqCov (%) | Score | Ratio | ln ratio | StdDev |
|---|---|---|---|---|---|---|---|---|---|
(Y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X) X) |
(Y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X) X) |
[ln ratio (Y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X)] X)] |
|||||||
| a SwissProt/UniprotKB accession number. | |||||||||
| 1 | P02784 | Seminal plasma protein PDC 109 | 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 480.65 480.65 |
4.91 | 39.55 | 1247.4 | 0.52 | −0.66 | 0.31 |
| 2 | P10096 | Glyceraldehyde 3 phosphate dehydrogenase | 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 868.18 868.18 |
8.51 | 20.12 | 504.9 | 0.59 | −0.52 | 0.61 |
| 3 | Q2T9U2 | Outer dense fiber protein 2 | 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 498.34 498.34 |
7.52 | 45.05 | 493.9 | 0.61 | −0.49 | 0.08 |
| 4 | Q3ZBU7 | Tubulin beta 4A | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 585.94 585.94 |
4.78 | 53.83 | 5211.0 | 0.63 | −0.47 | 0.19 |
| 5 | P19858 | L-lactate dehydrogenase A | 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 597.74 597.74 |
8.12 | 25.00 | 370.8 | 0.64 | −0.44 | 0.18 |
| 6 | Q29438 | Outer dense fiber protein 1 | 29![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 435.69 435.69 |
8.40 | 16.41 | 275.9 | 0.65 | −0.43 | 0.16 |
| 7 | O77797 | A kinase anchor protein 3 | 94![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 672.60 672.60 |
6.28 | 40.91 | 421.4 | 0.66 | −0.41 | 0.08 |
| 8 | Q32LE5 | L-asparaginase | 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 050.52 050.52 |
7.00 | 57.47 | 398.8 | 0.69 | −0.37 | 0.11 |
| 9 | Q3MHM5 | Tubulin beta 4B | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 831.18 831.18 |
4.79 | 72.58 | 6746.7 | 0.70 | −0.35 | 0.04 |
| 10 | Q32KN8 | Tubulin alpha 3 | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 925.68 925.68 |
4.98 | 58.22 | 3622.7 | 0.70 | −0.35 | 0.04 |
| 11 | Q2TBH0 | Outer dense fiber protein 3 | 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 633.94 633.94 |
9.89 | 24.41 | 112.4 | 0.70 | −0.35 | 0.20 |
| 12 | Q2KJE5 | Glyceraldehyde 3 phosphate dehydrogenase testis specific | 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 288.08 288.08 |
8.32 | 58.48 | 2376.9 | 0.71 | −0.34 | 0.07 |
| 13 | Q2YDG7 | Sperm acrosome membrane associated protein 1 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 938.92 938.92 |
4.53 | 44.09 | 1611.7 | 0.73 | −0.32 | 0.06 |
| 14 | Q5E956 | Triosephosphate isomerase | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 689.56 689.56 |
6.45 | 68.67 | 697.0 | 0.73 | −0.31 | 0.17 |
| 15 | P62157 | Calmodulin | 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 837.65 837.65 |
4.09 | 71.81 | 4262.9 | 0.73 | −0.31 | 0.05 |
| 16 | Q2HJB8 | Tubulin alpha 8 | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 053.68 053.68 |
5.06 | 32.96 | 1463.4 | 2.34 | 0.85 | 0.14 |
| 17 | Q6B856 | Tubulin beta 2B | 49![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 953.24 953.24 |
4.78 | 52.58 | 4430.9 | 3.90 | 1.36 | 0.36 |
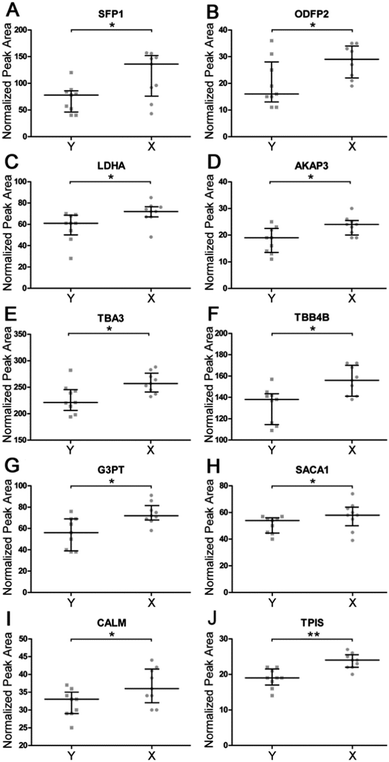
In order to validate shotgun results, we applied a targeted label-free strategy to evaluate the expression profile of the proteins by analyzing the proteotypic peptides. X and Y cell protein digestions were separately run in triplicate. Tryptic digestion of yeast enolase was added to samples as an internal standard. Data were acquired by nUPLC-MS/MS in expression mode. Semi-quantitative analysis was carried on by Skyline software tool, integrating the peak area obtained by the Extracted Ion Chromatograms (XIC) of the MS1 spectra of proteotypic peptides. The normalized peak areas were calculated for each single run and compared (Fig. 4). The significance of the obtained distributions was calculated by applying the Mann–Whitney unpaired test (Table 2). Four of analyzed proteins have been confirmed to be more expressed in X cells with a fold change higher than 30%. These are seminal plasma protein PDC109, outer dense fiber protein 2, A-kinase anchor protein 3 and Glyceraldehyde 3-phosphate dehydrogenase testis-specific. Another six proteins were found to be more abundant in X than Y cells with a fold change calculated by selected peptides ranging from 14% to 23% (L-lactate dehydrogenase A, tubulin alpha-3, tubulin beta-4B, Sperm acrosome membrane-associated protein 1, calmodulin and triosephosphate isomerase).
| Hit | Accessiona | Description | Peptide | m/z | z | RT | Y-cellsb | X-cellsb | Ratio | P-valuec | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean | %CV | Median | IR | Median | IR | (Y![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) X) X) |
|||||||
| a SwissProt/UniprotKB accession number. b Normalized peak area distribution expressed as median and interquartile range (IR). c P-value achieved by two-tailed unpaired Mann–Whitney test. | |||||||||||||
| 1 | P02784 | Seminal plasma protein PDC-109 | CVFPFIYGGK | 594.2995 | 2 | 106.80 | 0.91 | 78 | 52–84 | 136 | 92–148 | 0.57 | 0.0216 |
| 2 | Q2T9U2 | Outer dense fiber protein 2 | VTDLVNQQQTLEEK | 822.9255 | 2 | 64.68 | 1.15 | 16 | 15–25 | 29 | 23–33 | 0.55 | 0.0466 |
| LAECQDQLQGYER | 805.3673 | 2 | 60.66 | 1.29 | |||||||||
| 3 | P19858 | L-lactate dehydrogenase A | LVIITAGAR | 457.2951 | 2 | 70.80 | 1.14 | 61 | 54–68 | 72 | 67–76 | 0.85 | 0.0169 |
| 4 | O77797 | A-kinase anchor protein 3 | ASGTLQSPPNLK | 606.8327 | 2 | 58.00 | 1.24 | 19 | 14–22 | 24 | 21–25 | 0.79 | 0.0263 |
| SPAVSHESSLR | 585.2991 | 2 | 37.79 | 2.99 | |||||||||
| LNECDETGGAFAGLTK | 841.8881 | 2 | 78.28 | 0.91 | |||||||||
| LLQLSAAAVEK | 571.8426 | 2 | 79.06 | 0.95 | |||||||||
| SVGEVLQSVLR | 593.8431 | 2 | 101.01 | 0.77 | |||||||||
| QLDEAVGNVTR | 601.3122 | 2 | 57.96 | 1.47 | |||||||||
| 5 | Q32KN8 | Tubulin alpha-3 | AVFVDLEPTVVDEVR | 844.4487 | 2 | 104.58 | 0.67 | 221 | 214–243 | 257 | 245–270 | 0.86 | 0.0188 |
| 6 | Q3MHM5 | Tubulin beta-4B | INVYYNEATGGK | 664.8276 | 2 | 63.23 | 1.28 | 138 | 116–142 | 156 | 141–168 | 0.88 | 0.0190 |
| 7 | Q2KJE5 | Glyceraldehyde-3-phosphate dehydrogenase, testis-specific | GQLVVDNNEISVFQCK | 925.4592 | 2 | 91.27 | 0.76 | 52 | 40–69 | 72 | 69–77 | 0.77 | 0.0132 |
| GAHQNIIPASTGAAK | 718.3862 | 2 | 51.04 | 1.86 | |||||||||
| VPTPDVSVVDLTCR | 779.4006 | 2 | 89.14 | 0.76 | |||||||||
| LAQPTPYSAIK | 594.8347 | 2 | 65.34 | 1.16 | |||||||||
| AGIALNDNFVK | 581.3167 | 2 | 82.20 | 1.00 | |||||||||
| 8 | Q2YDG7 | Sperm acrosome membrane-associated protein 1 | EVILTSGCPGGESK | 717.3506 | 2 | 57.97 | 1.47 | 54 | 45–55 | 58 | 55–63 | 0.94 | 0.0469 |
| AFECETLDNNEIVASIR | 990.9702 | 2 | 95.12 | 0.83 | |||||||||
| 9 | P62157 | Calmodulin | DTDSEEEIR | 547.2358 | 2 | 40.03 | 2.52 | 33 | 29–34 | 36 | 34–41 | 0.92 | 0.0457 |
| 10 | Q5E956 | Triosephosphate isomerase | VPADTEVVCAPPTAYIDFAR | 1096.5382 | 2 | 101.74 | 0.61 | 19 | 18–21 | 24 | 22–25 | 0.81 | 0.0019 |
| IAVAAQNCYK | 569.2896 | 2 | 49.65 | 1.84 | |||||||||
| IIYGGSVTGATCK | 663.8397 | 2 | 58.00 | 1.45 | |||||||||
| 11 | P00924 | Enolase 1 (yeast) | GNPTVEVELTTEK | 708.8644 | 2 | 71.01 | 1.07 | 100 | 100–100 | 100 | 100–100 | 1 | / |
| NVNDVIAPAFVK | 643.8587 | 2 | 87.44 | 0.83 | |||||||||
| AVDDFLISLDGTANK | 789.9041 | 2 | 106.88 | 0.74 | |||||||||
| TAGIQIVADDLTVTNPK | 878.4779 | 2 | 95.27 | 0.68 | |||||||||
| VNQIGTLSESIK | 644.8589 | 2 | 74.03 | 1.02 | |||||||||
4. Discussion
The application of unsupervised protein profile evaluation to the study of sperm cell biology is a promising tool to study this field that is, from many aspects, still not understood.9 In this paper we approach the relevant question of the differences between Y- and X-chromosome containing sperm cells by a shotgun bottom-up proteomic strategy based on nUPLC-MS/MS investigation. This experimental approach applied to the analysis of peptides obtained by tryptic digestion of cell extracts was employed to give information on the basic structural features of the sperm cells. Collected data showed that significant differences between X and Y cells were related to cytoskeletal proteins which, because of their oligomeric state, are difficult to investigate by proteomic techniques based on protein separation. In the light of these considerations, it was not surprising to find that three tubulin isoforms were more represented in X cells whereas different isotypes were principally observed in Y cells. In a recent issue, Chen et al., performing an extensive differential analysis of sexed bovine sperm cells by 2DE/MS, reported the enhanced levels of the tubulin α3 and β4B isoforms in X cells with respect to Y cells.16 Here, we can complete this experimental frame, demonstrating that two different tubulin isoforms (tubulin α8 and β2B) showed different expression profiles with the inverse fold ratio. Chen et al. suggested that this observation may correlate to the putative cytoskeletal differences influencing the tail shape and movement properties of spermatozoa.16 Indeed, tubulins are the fundamental components of microtubules which, associating in the typical 9 + 2 structure, originate in the central axonemal motor of sperm tail. Moreover, shotgun analysis revealed that not only the tubulins, but also other cytoskeletal components of the sperm tail were more expressed in X cells. An outstanding result indicated that three components of outer dense fibers (ODFs) were found upregulated in X cells. The ODFs are filamentous structures composed of heavily disulfide linked keratin-like proteins,22,23 linked to the external microtubule doublets of the core axoneme. ODFs act to stiffen the axoneme and to anchor it at the basal end of flagellum, where ODFs are united with the connecting piece.24 It was proposed that ODFs are also actively involved in the bending of the sperm tail providing a means to overcome the flexural rigidity imposed by the presence of cytoskeletal accessory structures.25 The role of ODFs in motility of the sperm cells was supported by genetic evidence that highly chimeric mice with ODF2 ± genotype were shown to be infertile, with evident sperm tail defects and impaired motility.26The results of our mass spectrometry investigation arouse interest by considering the involvement of cytoskeletal tail structures in sperm motility. A-kinase anchoring protein 3 (AKAP3), a major component of fibrous sheaths, as well as four glycolytic enzymes, were found more expressed in X-chromosome bearing cells. In addition to the ODFs, the mammalian sperm flagellum is characterized by the presence of a fibrous sheath (FS) that surround the axoneme and ODFs in the principal piece region of the sperm flagellum. Three FS protein constituents are cAMP-dependent protein kinase anchoring proteins (AKAP).27 They function as scaffolds for signaling pathway components that modulate a variety of cell functions.28 Several glycolytic enzymes are associated to the FS including spermatogenic cell-specific hexokinase 1 variants,29,30 testis-specific glyceraldehyde-3-phosphate dehydrogenase,31 enolase,32L-lactate dehydrogenase A and C isotypes33,34 and the A isoform of aldolase 1.34 The glycolytic enzymes arrangement along the principal piece of sperm tail is thought to be important for providing ATP to distal regions of the flagellum. This suggestion was supported by gene targeting studies indicating that testis-specific glyceraldehyde-3-phosphate dehydrogenase is required for sperm motility.35
L-Asparaginase (ALP) is the enzyme which converts the amino acid L-asparagine into L-aspartate. Microscopy studies demonstrated that ALP protein localizes in the mid-piece of rat and human sperm cells, suggesting that ALP is associated with the mitochondria.36 The biological function of this enzyme has still not been clarified. The observation that ALP has been detected in several human tumors37 led to the proposal that ALP activation may be related to the altered metabolic profile of cells characterized by increased glycolysis and reduced flux through the tricarboxylic acid (TCA) cycle.38 Thus, ALP may act to compensate the lack of intermediate metabolites of TCA, converting asparagine to aspartate which, in turn, may be converted into oxaloacetate. This explanation also fits the metabolism of sperm cells which depend on glycolysis to generate most of the energy required for sperm motility.35
Finally, three different proteins were found upregulated in X cells compared to Y cells, sperm acrosome membrane-associated protein 1, calmodulin and seminal plasma protein PDC 109. This third protein, the major component of bovine seminal plasma, is involved in sperm capacitation.39 Due to its properties of molecular chaperone, it is the principal contaminant of protein extracts of sperm cells11 but, in this context, it doesn't seem to have a particular biological meaning. Otherwise, the involvement of calmodulin (CALM) in sperm cell physiology was well documented. CALM mediates the processes triggered by the rinsing of Ca2+ concentration which takes place in the head as well as in the tail of a mammalian sperm cell.40 CALM is involved in sperm capacitation,41 but also regulates the sperm tail beating changes responsible for hyperactivated motility in mammalian oviduct.42,43 Thus, the increased CALM concentration in X cells may be related to a major energetic request necessary to move the spermatozoa, as may be also inferred by the higher levels of glycolytic enzymes. Finally, Sperm acrosome membrane-associated protein 1 (SACA1), a 34 KDa membrane protein is detected in correspondence of acrosome in human sperm cells, with a higher concentration in the equatorial segment.44 In humans, SACA1 was clearly shown to be a differentiation antigen,45 expressed exclusively in germ cells during acrosomal biogenesis. However, its acrosomal localization suggests that this protein may be a potential candidate for an antibody-based separation of sexed sperm cells.
5. Conclusions
In summary, our proteomic analysis pointed out significant differences between bovine X- and Y-chromosome bearing sperm cells principally related to the expression of proteins involved in cytoskeleton organization. These findings suggest the possibility that X and Y cells may differ in structural characteristics concerning the arrangement of the sperm tail. In the past years, the differences in size or shape between X and Y cells have been investigated by different experimental approaches leading to different, even conflicting outcomes. The recent application of atomic force microscopy clearly showed there are no differences in the dimensional parameters of bovine sexed sperm heads,46 even though sperm tails were not investigated. The failure of the previous attempts to separate the X- and Y-chromosome bearing sperm cells on the basis of physical properties2 lead to the conclusion that this way may unlikely lead to results better than the actual method for sperm sexing. Our proteomic analysis highlighted different proteins which could be employed as tentative molecular features to distinguish X and Y cell populations. However, this possibility is linked to the accessibility of specific antibodies to the selected protein targets for detecting and possibly separating a recognized cell type. In this context the sperm acrosome membrane-associated protein 1 should be a potential molecular target by virtue of its antigenic properties and acrosomal membrane localization. The antibodies accessibility of the other differential proteins is not obvious. As this question is relevant for developing alternative methods of sorting, it should be investigated by a direct experimental approach for the selective targeting of sperm surface proteins through antibodies or a selective technique of labeling.47This proteomic investigation also suggested that X cells produce higher levels of glycolytic enzymes, in particular testis-specific glyceraldehyde-3-phosphate dehydrogenase, as well as calmodulin, essential for sperm motility regulation. The analysis of distributions showed a partial overlap of measured values in single samples that induce to exclude a direct employment for distinguishing the two sperm populations. Nevertheless, these emerged differences could be useful to orientate further research toward the development of methods of cell separation based on the differential energetic consumption that may be associated to the motion of sperm cells.
Acknowledgements
The authors are grateful to Dr Innes Drummond of “Cogent Breeding Ltd” (Aldford, Chester, England CH3 6JQ, UK) for sexing the semen. This work was funded by SESSIBOV (MIPAAF- ISILS).References
- D. Rath and L. A. Johnson, Reprod. Domest. Anim., 2008, 43, 338–346 CrossRef PubMed.
- D. L. Garner and G. E. Seidel Jr., Theriogenology, 2008, 69, 886–895 CrossRef CAS PubMed.
- D. L. Garner, B. L. Gledhill, D. Pinkel, S. Lake, D. Stephenson, M. A. Van Dilla and L. A. Johnson, Biol. Reprod., 1983, 28, 312–321 CrossRef CAS.
- G. E. Seidel Jr., J. L. Schenk, L. A. Herickhoff, S. P. Doyle, Z. Brink, R. D. Green and D. G. Cran, Theriogenology, 1999, 52, 1407–1420 CrossRef CAS.
- W. M. Maxwell and L. A. Johnson, Theriogenology, 1999, 52, 1353–1362 CrossRef CAS.
- J. L. Schenk and G. E. Seidel Jr., Soc. Reprod. Fertil. Suppl., 2007, 64, 165–177 CAS.
- L. A. Johnson, J. P. Flook and H. W. Hawk, Biol. Reprod., 1989, 41, 199–203 CrossRef CAS.
- S. L. Underwood, R. Barthgate, M. Ebsworth, W. M. C. Maxwell and G. Evans, Anim. Reprod. Sci., 2010, 118, 7–12 CrossRef CAS PubMed.
- M. A. Baker, Asian J. Androl., 2011, 13, 6–10 CrossRef CAS PubMed.
- J. S. Clay and B. T. McDaniel, J. Dairy Sci., 2001, 84, 1238–1245 CrossRef CAS.
- A. Soggiu, C. Piras, H. A. Hussein, M. De Canio, A. Gaviraghi, A. Galli, A. Urbani, L. Bonizzi and P. Roncada, Mol. BioSyst., 2013, 9, 1188–1195 RSC.
- L. B. Shettles, Nature, 1960, 186, 648–649 CrossRef CAS.
- K. H. Cui and C. D. Matthews, Nature, 1993, 366, 117–118 CrossRef CAS PubMed.
- E. A. Howes, N. G. Miller, C. Dolby, A. Hutchings, G. W. Butcher and R. Jones, Reproduction, 1997, 110, 195–204 CrossRef CAS PubMed.
- P. J. Hendriksen, Theriogenology, 1999, 52, 1295–1307 CrossRef CAS.
- X. Chen, H. Zhu, C. Wu, W. Han, H. Hao, X. Zhao, W. Du, T. Qin, Y. Liu and D. Wang, J. Proteomics, 2012, 77, 59–67 CrossRef CAS PubMed.
- G. E. Seidel Jr. and D. L. Garner, Reproduction, 2002, 124, 733–743 CrossRef PubMed.
- J. L. Schenk, T. K. Suh, D. G. Cran and G. E. Seidel Jr., Theriogenology, 1999, 52, 1375–1391 CrossRef CAS.
- K. Parati, G. Bongioni, R. Aleandri and A. Galli, Theriogenology, 2006, 66, 2202–2209 CrossRef CAS PubMed.
- S. D'Aguanno, I. D'Agnano, M. De Canio, C. Rossi, S. Bernardini, G. Federici and A. Urbani, Mol. BioSyst., 2012, 8, 1068–1077 RSC.
- B. MacLean, D. M. Tomazela, N. Shulman, M. Chambers, G. L. Finney, B. Frewen, R. Kern, D. L. Tabb, D. C. Liebler and M. J. MacCoss, Bioinformatics, 2010, 26, 966–968 CrossRef CAS PubMed.
- J. M. Bedford and H. I. Calvin, J. Exp. Zool., 1974, 187, 181–203 CrossRef CAS PubMed.
- G. E. Olson and D. W. Sammons, Biol. Reprod., 1980, 22, 319–332 CrossRef CAS.
- D. W. Fawcett, Dev. Biol., 1975, 44, 394–436 CrossRef CAS.
- C. B. Lindemann, Cell Motil. Cytoskeleton, 1996, 34, 258–270 CrossRef CAS.
- H. Tarnasky, M. Cheng, Y. Ou, J. C. Thundathil, R. Oko and F. A. van der Hoorn, BMC Dev. Biol., 2010, 10, 67 CrossRef PubMed.
- E. M. Eddy, K. Toshimori and D. A. O'Brien, Microsc. Res. Tech., 2003, 61, 103–115 CrossRef CAS PubMed.
- A. S. Edwards and J. D. Scott, Curr. Opin. Cell Biol., 2000, 12, 217–221 CrossRef CAS.
- C. Mori, N. Nakamura, J. Welch, H. Gotoh, E. Goulding, M. Fujioka and M. Eddy, Mol. Reprod. Dev., 1998, 49, 374–385 CrossRef CAS.
- A. J. Travis, J. A. Foster, N. A. Rosenbaum, P. E. Visconti, G. L. Gerton and S. B. Moss, Mol. Biol. Cell, 1998, 9, 263–276 CrossRef CAS.
- D. Westhoff and G. Kamp, J. Cell Sci., 1997, 110, 1821–1829 CAS.
- V. M. Gitlits, B. H. Toh, K. L. Loveland and J. W. Sentry, Eur. J. Cell Biol., 2000, 79, 104–111 CrossRef CAS.
- S. A. Beyler, T. E. Wheat and E. Goldberg, Biol. Reprod., 1985, 32, 1201–1210 CrossRef CAS.
- M. Krisfalusi, K. Miki, P. L. Magyar and D. A. O'Brien, Biol. Reprod., 2006, 75, 270–278 CrossRef CAS PubMed.
- K. Miki, W. Qu, E. H. Goulding, W. D. Willis, D. O. Bunch, L. F. Strader, S. D. Perreault, E. M. Eddy and D. A. O'Brien, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 16501–16506 CrossRef CAS PubMed.
- L. A. Bush, J. C. Herr, M. Wolkowicz, N. E. Sherman, A. Shore and C. J. Flickinger, Mol. Reprod. Dev., 2002, 62, 233–247 CrossRef CAS PubMed.
- U. H. Weidle, V. Evtimova, S. Alberti, E. Guerra, N. Fersis and S. Kaul, Anticancer Res., 2009, 29, 951–963 CAS.
- Y. Su, C. S. Karamitros, J. Nomme, T. McSorley, M. Konrad and A. Lavie, Chem. Biol., 2013, 20, 533–540 CrossRef CAS PubMed.
- N. Srivastava, A. Jerome, S. K. Srivastava, S. K. Ghosh and A. Kumar, Anim. Reprod. Sci., 2013, 138, 1–13 CrossRef CAS PubMed.
- H. P. Jones, R. W. Lenz, B. A. Palevitz and M. J. Cormier, Proc. Natl. Acad. Sci. U. S. A., 1980, 77, 2772–2776 CrossRef CAS.
- Y. Si and P. Olds-Clarke, Biol. Reprod., 2000, 62, 1231–1239 CrossRef CAS.
- R. Yanagimachi, in The Physiology of Reproduction, ed. E. Knobil and J. Neill, Raven Press, New York, 1994, pp. 189–317 Search PubMed.
- D. F. Katz and R. Yanagimachi, Biol. Reprod., 1980, 22, 759–764 CrossRef CAS.
- Z. Hao, M. J. Wolkowicz, J. Shetty, K. Klotz, L. Bolling, B. Sen, V. A. Westbrook, S. Coonrod, C. J. Flickinger and J. C. Herr, Biol. Reprod., 2002, 66, 735–744 CrossRef CAS.
- E. Boyse and L. Old, Annu. Rev. Genet., 1969, 3, 269–290 CrossRef.
- J. O. Carvalho, L. P. Silva, R. Sartori and M. A. Dode, PLoS One, 2013, 8, e59387 CAS.
- C. Belleannee, M. Belghazi, V. Labas, A. P. Teixeira-Gomes, J. L. Gatti, J. L. Dacheux and F. Dacheux, Proteomics, 2011, 11, 1952–1964 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1. Protein identification by nUPLC-MS/MS. See DOI: 10.1039/c3mb70306a |
| This journal is © The Royal Society of Chemistry 2014 |