Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Modulation of alpha-synuclein toxicity in yeast using a novel microfluidic-based gradient generator
João Tiago S.
Fernandes
ab,
Sandra
Tenreiro
b,
Andreia
Gameiro
a,
Virginia
Chu
a,
Tiago F.
Outeiro
*bc and
João P.
Conde
*ad
aINESC Microsistemas e Nanotecnologias and IN – Institute of Nanoscience and Nanotechnology, R. Alves Redol, 9, 1000-029, Lisbon, Portugal
bInstituto de Medicina Molecular, Instituto de Fisiologia, Faculdade de Medicina da Universidade de Lisboa, Av. Professor Egas Moniz, 1649-028, Lisbon, Portugal
cDepartment of Neurodegeneration and Restorative Research, Center for Nanoscale Microscopy and Molecular Physiology of the Brain, University Medical Center Göttingen, Waldweg 33, 37073 Göttingen, Germany. E-mail: touteir@gwdg.de; Tel: +495513913544
dDepartment of Bioengineering, Instituto Superior Técnico, Av. Rovisco Pais, 1, 1049-001, Lisbon, Portugal. E-mail: joao.conde@tecnico.ulisboa.pt; Tel: +351 213 100 237
First published on 14th August 2014
Abstract
Parkinson's disease (PD) is a common age-associated neurodegenerative disorder. The protein α-synuclein (aSyn) is a key factor in PD both due to its association with familial and sporadic cases and because it is the main component of the pathological protein aggregates known as Lewy bodies. However, the precise cellular effects of aSyn aggregation are still elusive. Here, we developed an elastomeric microfluidic device equipped with a chemical gradient generator and 9 chambers containing cell traps to study aSyn production and aggregation in Saccharomyces cerevisiae. This study involved capturing single cells, exposing them to specific chemical environments and imaging the expression of aSyn by means of a GFP fusion (aSyn-GFP). Using a galactose (GAL) gradient we modulated aSyn expression and, surprisingly, by tracking the behavior of single cells, we found that the response of individual cells in a population to a given stimulus can differ widely. To study the combined effect of environmental factors and aSyn expression levels, we exposed cells to a gradient of FeCl3. We found a dramatic increase in the percentage of cells displaying aSyn inclusions from 27% to 96%. Finally, we studied the effects of ascorbic acid, an antioxidant, on aSyn aggregation and found a significant reduction in the percentage of cells bearing aSyn inclusions from 87% to 37%. In summary, the device developed here offers a powerful way of studying aSyn biology with single-cell resolution and high throughput using genetically modified yeast cells.
Introduction
The budding yeast Saccharomyces cerevisiae has been widely explored as a model organism to investigate the molecular underpinnings of several human diseases.1–3 This yeast is the best-characterized eukaryotic cell, is easy to grow under defined conditions, and can easily be genetically manipulated.4,5Parkinson's disease (PD) is a neurodegenerative disorder characterized by the loss of dopaminergic neurons in the substantia nigra in the brain and by the accumulation of intraneuronal protein deposits known as Lewy bodies.6 Although the precise molecular mechanisms underlying PD are still elusive, the identification of genes causing familial forms of the disease enabled significant advances in our understanding of the disease.7 The SNCA gene was the first to be associated with familial forms of PD and encodes for α-synuclein (aSyn), the main protein component of Lewy bodies.6,8,9 However, it is currently unclear whether aggregated forms of aSyn are toxic or, instead, neuroprotective.10,11
To investigate the basic molecular effects of aSyn in the context of a living cell, human aSyn was expressed in yeast and found to induce dose-dependent cytotoxicity.12 Using GFP as a reporter and the galactose (GAL)-inducible promoter, aSyn-GFP fusions were found to initially associate with the plasma membrane and then to accumulate in cytoplasmic inclusions reminiscent of protein aggregates.12 Several additional studies followed, exploiting this yeast model for high-throughput analyses that proved instrumental in informing on the molecular mechanisms involved in aSyn toxicity.13–16
Microfluidics technology, relying on transparent elastomeric materials such as polydimethylsiloxane (PDMS), has made it possible to quickly develop prototypes of devices appropriate for cell culture and live-cell imaging.17 Microfluidic devices offer the possibility of performing biological experiments with small amounts of reagents while still being able to parallel process tens or hundreds of experiments in a very small area.17,18 Since in microfluidic channels liquids flow in the laminar regime19,20 it is possible to generate microenvironments with specific chemical compositions and expose individual cells within a given population to these environments.21
Although classical cell culture protocols enable the study of populations of cells exposed to different chemical stimuli over time, difficulties arise when it is necessary to achieve single-cell resolution. Single-cell studies are important for following the response of individual cells to a static or changing environment over time. Currently used cell culture and cell separation techniques do not allow the continuous tracking of hundreds of individual cells simultaneously in an efficient way. Moreover, single-cell tracking in culture is particularly difficult for non-adherent cells such as yeast. Several methods for trapping cells in microfluidic devices have been described,22 but the most straightforward relies on placing arrays of physical traps in the streamlines of cells in suspension flowing in the device and allows the isolation and immobilization of single cells.23
In addition to the trapping of cells, microenvironments with specific chemical formulations and high spatial resolution can also be created in microfluidics by using gradient generators.24,25 This type of device is able to produce a stable chemical gradient with a predictable profile and offers potential for high parallelization.
In this study, we developed a device consisting of a microfluidic gradient generator coupled to 9 chambers containing an array of cell traps (Fig. 1A–C). By capturing genetically modified yeast cells expressing aSyn-GFP under the control of a GAL-inducible promoter, we were able to tightly modulate aSyn-GFP expression using a gradient of GAL. We were also able to follow the evolution of aSyn production inside individual immobilized cells. As a proof of concept, we used the same device to determine the effect of FeCl3, a chemical stressor, and of ascorbic acid, an antioxidant, on aSyn inclusion formation. In summary, the device developed here constitutes a powerful platform for high-throughput screens for drug discovery, enabling the simultaneous testing of a range of compound concentrations. Furthermore, the single-cell trapping capabilities offered by this device also enable single-cell behavior analysis within a population when exposed to a given chemical environment.
Experimental
Mold fabrication
The device was fabricated using a variant of the rapid prototyping and soft-lithography techniques, which relies on using a master mold to build PDMS replicas.26,27 Since this device has features on the order of a couple of micrometers, the mold was made directly on top of the physical mask. Briefly, the physical mask was made on a 25 mm × 50 mm sample of glass (Corning, New York, USA) with a 1500–2000 Å layer of AlSiCu deposited by sputtering. Patterning of the metallic layer to the design of the microfluidic mold was performed using standard photolithography and chemical etching techniques. After the patterning of the AlSiCu, a first layer of SU-8 2005 (MicroChem Corp., Newton, MA) was spin-coated on top of the metallic side of the sample to a thickness of about 5 μm and fully exposed. This first layer provides a uniform surface and, given SU-8's poor adhesion to glass, ensures that the small features of the device adhere strongly to the substrate. A second layer of SU-8 2005 with a thickness of about 9 μm was spin-coated on top of the first layer. This SU-8 layer was then exposed through the glass side of the sample with a UV flood lamp, so that only the SU-8 on the patterned AlSiCu-free areas was cross-linked. The second SU-8 layer was then developed in propylene glycol monomethyl ether acetate (Sigma-Aldrich) and hard baked at 150 °C for at least 20 min.Device fabrication
The devices were fabricated with PDMS (Dow Corning, Midland, MI). PDMS with a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (base to curing agent, w/w) was poured on top of the SU-8 mold and cured at 70 °C for 50 min. The molds were placed inside a custom-sized poly(methyl methacrylate) case with 5 mm high internal walls to allow for a consistent thickness of 5 mm between batches. After curing, access ports were punched with a 20 ga syringe tip (Instech, PA, USA). A sealing layer was made by spin-coating PDMS with a 20
1 ratio (base to curing agent, w/w) was poured on top of the SU-8 mold and cured at 70 °C for 50 min. The molds were placed inside a custom-sized poly(methyl methacrylate) case with 5 mm high internal walls to allow for a consistent thickness of 5 mm between batches. After curing, access ports were punched with a 20 ga syringe tip (Instech, PA, USA). A sealing layer was made by spin-coating PDMS with a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio on a glass coverslip (Hirschmann Inc., KY, USA) to a thickness of about 25 μm and curing at 70 °C for 40 min. The PDMS device was then placed on top of the PDMS-covered coverslip and baked for at least 2 h. The partial curing and the difference in curing agent ratios ensure an irreversible, high-pressure-resistant bonding.28 After curing, 20 ga metallic connectors (Instech) were inserted into the access ports and 10
1 ratio on a glass coverslip (Hirschmann Inc., KY, USA) to a thickness of about 25 μm and curing at 70 °C for 40 min. The PDMS device was then placed on top of the PDMS-covered coverslip and baked for at least 2 h. The partial curing and the difference in curing agent ratios ensure an irreversible, high-pressure-resistant bonding.28 After curing, 20 ga metallic connectors (Instech) were inserted into the access ports and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PDMS was poured at the interface using a syringe and cured at 70 °C to seal the connectors and prevent leakage.
1 PDMS was poured at the interface using a syringe and cured at 70 °C to seal the connectors and prevent leakage.
Device operation
Prior to cell insertion, the device is first primed with 1% BSA (w/w) in PBS to minimize the adherence of cells to the PDMS surface along the channels. Cell seeding is performed by flowing cells in medium through the cell inlet at a flow rate of 0.5 μL min−1. The decision of when to stop cell insertion is made independently for each device and takes into account the number of single cells trapped and the number of cell clusters accumulating in the first lines of traps. Our goal is to maximize the former and minimize the latter. During cell insertion, the medium is introduced at a flow rate of 0.4 μL min−1 through the solution inlets to prevent the backflow of cells and their accumulation in the inlets. We found that if such backflow occurs, cells progressively detach from the inlets and enter the cell chambers after the cell insertion phase. This would bias the results since they would have been exposed to a different chemical environment than the previously trapped cells. Before starting the chemical gradient generator, the cell inlet is plugged with a closed metallic connector (Instech) to prevent fluid losses and possible biases in the gradient profile.Syringes containing media are mounted into a syringe pump (New Era Pump Systems, NY, USA) and connected to the left, middle and right solution inlets – from lowest to highest concentration of the chemical species of interest – via polyethylene tubing (Instech).
Yeast strains and culture medium
In this work two yeast models of PD were used. The yeast strain VSY72 (can1-100 his3-11 15 leu2-3 112 GAL1pr-SNCA(WT)-GFP::TRP1 GAL1pr-SNCA(WT)-GFP::URA3 ade2-1) carries two copies of the aSyn-GFP fusion inserted in the genome under the control of the GAL1 inducible promoter and was previously described.14 To study the formation of aSyn inclusions we used the strain Y4791 (can1-100 his3-11 15 leu2-3 112 GAL1pr-SNCA(WT)-GFP::TRP1 GAL1pr-SNCA(WT)-GFP::URA3 ade2-1).12 This strain harbors two copies of WT aSyn and displays a stronger phenotype of toxicity associated with the expression of aSyn than that of the VSY72 strain. Cells were grown in synthetic complete (SC) raffinose liquid medium (yeast nitrogen base without amino acids, 6.7 g L−1; raffinose, 10 g L−1; CSM without uracil and tryptophan as indicated by the supplier) for 24 h (doubling time: ~3 h) at 30 °C with orbital agitation (200 rpm). Optical density was measured at 600 nm (OD600 nm) and cells were diluted in the same growth medium in order to be at the exponential growth phase on the following day (with a final OD600 nm around 1). OD600 nm was measured again and the cells in suspension with an OD600 nm of 2 mL−1 were collected by centrifugation at 1800g at 30 °C for 5 min. Cells were then washed in phosphate-buffered saline (PBS) and resuspendend in PBS at a final OD600 nm of 1 mL−1 (~1.5 × 107 cells mL−1). Before loading the cells into the device, vigorous vortexing was applied to minimize cell clusters.In the case of the treatment with ascorbic acid, Y4791 yeast cells were cultured for an additional 24 hours in SC medium without uracil and tryptophan (SC–URA–TRP) and with 1% GAL supplemented with 10 mM FeCl3.
Fluorescence microscopy
The acquisition of fluorescence signals during the tests for the chemical gradient generator was made with a Leica DMLM microscope equipped with a Leica DFC 300 FX digital camera. Live-cell imaging and detection of the aSyn-GFP signal were done using a Zeiss Axiovert 200 M microscope equipped with a motorized stage and a Roper Scientific CoolSnap HQ CCD digital camera. Dry 20× and 40× Zeiss EC Plan-Neofluar and oil 63× Zeiss Plan-Apochromat objectives with numerical aperture values of 0.50, 0.75, and 1.40, respectively, were used. Acquired images were analyzed using ImageJ software (NIH, Bethesda, MD).Results and discussion
Device description
The microfluidic device relies on two connected modules: a chemical gradient generator and a set of 9 chambers each containing hydrodynamic traps for yeast cells (Fig. 1A–C). The chemical gradient generator has three inlets (solution inlets) that allow the insertion of chemical solutions of different compositions. A fourth inlet (cell inlet) branches out from the middle solution inlet and is used to load the device with yeast cells. The addition of this fourth inlet avoids the contamination of solution inlets. The channel connected to the cell inlet is also equipped with a filter made of regularly spaced posts to break or capture cell aggregates that could otherwise clog the device or accumulate in the first line of cell traps.The chemical gradient generator is composed of a multi-level network of 50 μm wide, 9 μm high vertical serpentine-shaped micro-channels where the mixing of chemical solutions occurs. The first level is composed of the three solution inlets followed by an array of four parallel serpentine channels. At each subsequent level, the number of serpentine channels is increased by one until reaching the seventh level, which has nine serpentine channels. At the end of each serpentine channel, the chemical solution is split into two branches. Solutions stemming from adjacent branches will then recombine and mix by flowing side by side in the serpentine channels of the following level.
The micro-channels were shaped as serpentines to increase their length while keeping the structure compact and to maximize the amount of time that the fluids remain in contact with each other to ensure complete diffusive mixing. In this work, all chemical gradients were generated by inserting chemical solutions through the 3 solution inlets simultaneously. By mounting the syringes containing the solutions on a single syringe pump we ensured that the flow rate was the same for the 3 solution inlets. The flow rate used for generating chemical gradients was 0.3 μL min−1, resulting in an average fluid velocity of 11.1 mm s−1 at the inlets, given the dimensions of the channels. The serpentine channels have the same height and width as the inlets and a total length of 11 mm, meaning that the solutions inserted at 0.3 μL min−1 would take about 0.99 s to cross them. However, since the number of channels increases at each level, the flow is split and the fluid velocity in each channel is reduced. Since the serpentine channels only appear at the second level of the structure (the first level being composed of the three solution inlets), fluid flowing in the serpentine channels always takes more than 0.99 s to cross them – and this time increases in proportion to the level the channel is in. Since for small molecules such as FITC, which was used to calibrate the device (diffusion coefficient of 5 × 10−6 cm2 s−1), it takes 1 s to reach 97% diffusive mixing,25 we can say that the dimensions and flow rates used are appropriate to the generation of chemical gradients of small molecules with comparable diffusion coefficients.
At the end of the serpentine channel, the fluid in the micro-channel splits left and right in different proportions. These splitting ratios depend on the level the channel is in and its distance from the edges of the device.25 By multiplying the normalized splitting ratios of two streams converging on a serpentine channel by the concentration of the chemical solutions they carry, we can calculate the chemical composition at the end of that channel. By iterating these calculations to each serpentine in each level, we obtain the expected chemical composition in each of the 9 final chambers.24
The chemical gradient generating network ends in nine final serpentine channels, each of which is connected to a chamber containing an array of hydrodynamic cell traps. In the schematic (Fig. 1A), we consider chamber 1 to be the first from the bottom, and chamber 9 to be the last on the top. The goal of connecting each chamber to a single serpentine channel was twofold: (1) the chemical composition within a given chamber is uniform since the chemical solutions are completely mixed at the end of the serpentine channel; and (2) the adjacent chambers have different chemical compositions and represent the final gradient generated by the microfluidic network. Each chamber is roughly 500 μm wide and 1750 μm long and contains 506 traps disposed in a checkerboard-like pattern. The cell traps are custom-sized for yeast cells, and they consist of semicircular receptacles with an internal diameter of 8 μm and a gap of 4 μm in the middle (Fig. 1C). The gap allows liquid to flow when the trap is empty but is blocked when a yeast cell is trapped. The filling of this gap will re-route other incoming cells' streamlines away from the occupied trap and into neighboring traps. The dimensions of the traps must be designed with a specific type of cell in mind in order to accommodate the necessary physical dimensions.
Efficacy of the chemical gradient generator
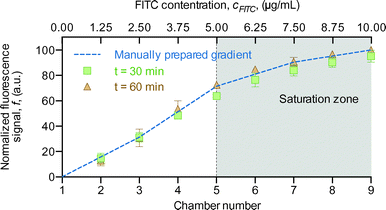
To determine the efficacy of the chemical gradient generator, we started by testing the device using the fluorescent compound fluorescein isothiocyanate (FITC). Aqueous solutions with different concentrations of FITC (0, 0.5 and 10 μg mL−1) were introduced through the solution inlets at a flow rate of 0.3 μL min−1, and the fluorescence signal produced was measured at the chambers located at the end of the generator at t = 30 min and t = 60 min. The experiment was replicated in two individual structures and the average and the standard deviation of the FITC fluorescence signal per chamber to assess the profile shape and stability were calculated. Due to the tendency of PDMS to adsorb small molecules,29 all devices were primed with 1% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) prior to the insertion of the test solutions.30 In parallel, as a control, five vials were prepared with a manually made mixture of 0, 2.5, 5, 7.5 and 10 μg mL−1 FITC. These solutions were sampled onto a microscope slide and fluorescence microscopy images were acquired. By comparing the signal intensity in the different chambers, we verified that the profile of the gradient produced by the generator was consistent with theoretical predictions (Fig. 2). This means that by inserting solutions in the device at concentration ratios of 0%, 50% and 100% for the left, middle and right inlets, respectively, where 100% represents the highest concentration of the chemical species of interest, and 0% represents a solution without the chemical species, we are able to generate a linear gradient with a resolution dependent on the number of channels at the end of the device. This allows us to correlate a given chamber (bottom horizontal axis) with the concentration of the solution (top horizontal axis) (Fig. 2). We also determined that the gradient was already stable after 30 min, since no additional changes were observed even after an additional 30 min (Fig. 2). Finally, we found that the fluorescence signal did not increase linearly with the concentration of FITC in both the microfluidic gradient and the manual gradient. We identified a saturation zone between 5 and 10 μg mL−1 FITC, suggesting that this non-linearity may be due to self-quenching effects in FITC that are unrelated to the device.Cell trapping efficiency
The hydrodynamic traps presented in this device were custom-designed for the particular yeast cells used in this study. Optimization included the shape and the dimensions of the trap (internal diameter, middle gap, horizontal and vertical space between traps and the number of indentation levels). In fact, we found that the spatial disposition of the traps was an essential consideration, since traps that were very densely distributed tended to capture more than a single cell and would generate clustering in and around the traps. This was due to the fact that larger cells or small aggregates did not have enough room to be flushed from the cell chamber, and this clustering could lead to the accumulation of hundreds of cells at the first lines of traps, notwithstanding the cell filter placed after the cell inlet. Since yeast cells also tend to cluster in certain types of media, special consideration was given to the ratio between the trapping of individual cells and clusters. Thus, a compromise had to be found between total trapping efficiency and trapped-single-cell-to-trapped-aggregate ratio when designing the traps and loading cells into the device. Although more densely packed traps enabled the capture of more cells (86% for X = 20 μm, Y = 10 μm compared to 62% for X = 20 μm, Y = 20 μm), the proportion of single cells trapped was higher for more loosely packed traps (39% for X = 20 μm, Y = 10 μm compared to 43% for X = 20 μm, Y = 20 μm) (Fig. 3). Since the goal of this platform was to maximize single-cell analysis, the spacing of traps selected for the following studies was X = 20 μm and Y = 20 μm.Generation and characterization of the galactose gradient
Cells under perfusion in a microfluidic environment are subjected to conditions different from that of a typical cell culture, such as increased shear stress.21 To demonstrate that the microfluidic conditions imposed by this device did not alter the normal physiology of yeast cells, we tested the response of the GAL promoter in a GAL gradient.Cells were seeded in the device in medium with 1% RAF and the gradient was generated by flowing media with (a) 1% RAF, (b) 0.5% RAF plus 0.5% GAL and (c) 1% GAL to the left, middle and right solution inlets, respectively. Cells were imaged by fluorescence microscopy (20× and 40× magnification with an exposure of 100 ms) at sequential time points after turning on the syringe pump, and the average fluorescence signal presented by at least 150 trapped cells in each chamber was evaluated (Fig. 4A). At the 300 min time point we observed that there was a clear increase in the fluorescence signal in all chambers and that, as expected, chambers with higher concentrations of GAL show higher fluorescence signals (Fig. 4A, B). Although we observed a steady increase in the fluorescence signal during the first 3 time points in chambers containing GAL (chambers 2 to 9), we found a sharp increase between t = 180 min and t = 300 min. In fact, in some cases (chamber 8) the signal intensity increased almost threefold between these time points. This occurs because cells need time to adapt and change their metabolic machinery in order to use GAL as a carbon source, resulting in the induction of aSyn-GFP expression. The signal increase was linear between chamber 1 and chamber 6, after which it reached a plateau corresponding to a GAL concentration between 0.6% and 0.8%. This demonstrates the usefulness of this structure, as we are able to identify with very high resolution the GAL concentration at which the production of aSyn saturates.
Furthermore, we also observed budding of yeast cells (Fig. 4B) throughout the course of the experiment (5 hours), indicating that cells were progressing through the cell cycle and demonstrating that normal physiology was maintained under the microfluidic conditions used.
Individual response of cells to galactose
It is virtually impossible to track single cells in a suspension without the use of labels or reporter genes. In the microfluidic device developed in this work, cells remain immobilized in the traps throughout the duration of the experiment. Therefore, it is possible to monitor individual cell responses to stimuli over time without the specific need for using cell labels. To investigate the phenotypic variability of individual cells in the population upon GAL induction, we analyzed the behavior of 35 individual cells trapped in chamber 9 and evaluated the production of aSyn-GFP. We found that individual cells exposed to the same chemical environment displayed a broad range of expression of aSyn-GFP (Fig. 5A). By analyzing the normalized fluorescence signal after 300 min, we observed a normal distribution of the signal with a mean value of 49 ± 18 (SD) (Fig. 5B). This dispersion allowed us to determine not only the minimum and maximum values of the fluorescence signal emitted by the cell population after 300 min (10.64 and 100.00, respectively) but also how it evolved over time (Fig. 5A, grey area). As an example, we show a cell with fluorescence signals of 4.91 at t = 30 min and 18.01 at t = 300 min, and another cell with signal values of 1.43 at t = 30 min and 54.45 at t = 300 min, showing that the production of aSyn can vary greatly from cell to cell within a given population (Fig. 5A). Examples of this phenomenon have already been reported in different types of isogenic cells, from bacteria31 to adherent mammalian cells under microfluidic conditions.32 This highlights the advantage of our device, enabling the study of individual cells in a population over time.By studying the distribution of aSyn production in single cells we are able to further characterize this cell model. In addition, our approach will enable future studies on cells displaying above or below average responses and the assessment of the mechanisms behind increased sensitivity or resistance to the stimuli of choice. Also, by isolating a given cell we could directly observe the increase in the production of aSyn-GFP throughout the duration of the experiment (Fig. 5A), which allowed us to study not only the kinetics of aSyn-GFP formation and degradation but also the effects of changing the chemical environment on a single cell. By using techniques such as 4D imaging, we would also be able to use this device to pinpoint with very high spatial accuracy the appearance and evolution of aSyn accumulation and inclusion formation.
FeCl3 induces the formation of aSyn inclusions in a concentration-dependent manner
After establishing the suitable functioning of the gradient generator and that cells responded to chemical stimuli in the microfluidic environment of our device, we set out to investigate the effects of oxidants and antioxidants in the formation of aSyn inclusions in yeast. FeCl3 was chosen as the chemical stressor since it has been previously reported to induce the formation of aSyn inclusions in yeast.33 For this experiment, yeast cells expressing aSyn-GFP (Y4791 strain) were trapped in the device, exposed to a gradient of FeCl3 with concentrations ranging from 0 to 10 mM for 24 hours and imaged using fluorescence microscopy (63× objective and 50 ms exposure). For that end, a gradient was generated using a medium containing 1% GAL supplemented with 0, 5 and 10 mM FeCl3. Due to the variability in the number of cells available for analysis per chamber across the three tested devices, we chose to analyze the three samples as a whole and not individually, i.e., average and standard deviation with n = 3 for each chamber. As such, for each chamber, we calculated the percentage of cells with inclusions by dividing the sum of cells with inclusions identified across the three devices by the total number of cells analyzed. We observed a linear increase in the percentage of cells displaying aSyn inclusions with increasing concentrations of FeCl3 (from 27% at 0 mM to 96% at 10 mM) (Fig. 6), demonstrating that oxidative stressors can promote the accumulation of aSyn inclusions in yeast. It should be noted that the controlled induction of aSyn inclusions through chemical stressors is an important tool for studying aSyn toxicity.Ascorbic acid reduces the formation of aSyn inclusions
Oxidation is an important mechanism in the aggregation of aSyn.12,15,34,35 Moreover, aSyn expression itself increases oxidative stress by promoting the accumulation of reactive oxygen species (ROS).34,36 Therefore, compounds that reduce oxidative stress could protect cells from aSyn-induced damage. Among other compounds, ascorbic acid (AA) reacts with toxic ROS and renders them non-toxic. AA was reported to counteract the oligomerization of aSyn induced by FeCl2 in neuronal cells overexpressing aSyn.34 Thus, we selected this compound to determine whether it could modulate aSyn inclusion formation in yeast. To test this, yeast cells expressing aSyn-GFP (Y4791 strain) were grown in GAL medium with 10 mM FeCl3 for 24 hours. Cells were then exposed to a gradient of AA and individual cells were followed by fluorescence microscopy and the percentage of cells containing inclusions was calculated, as described above. In the absence of AA (chamber 1), the percentage of cells displaying aSyn inclusions was rather high (~90%) (Fig. 7). However, with increasing concentrations of AA we observed a linear decrease in the percentage of cells with inclusions, decreasing to 32% at 20 mM AA.We hypothesize that the effect of AA on aSyn inclusion formation might be either by preventing the formation of new inclusions or by promoting the clearance of preformed inclusions, but this needs to be further investigated in subsequent studies. Nevertheless, our results clearly demonstrate the usefulness of this microfluidic platform as a tool to test compounds of interest with unparalleled resolution.
Conclusions
This work was aimed at studying aSyn biology using microfluidic technology in order to obtain single-cell resolution within entire cell populations. This was achieved by connecting chambers with arrays of hydrodynamic traps that immobilize cells at the end of a microfluidic chemical gradient generator. The immobilization of non-adherent cells, the control of their microenvironment and the creation of a chemical gradient with a small number of initial solutions are the main features that set this device apart from conventional bulk systems such as Petri dishes and well plates. The trapping of cells is paramount for this type of high-resolution studies, since it guarantees that we can track a single cell's response over time. In this way, we can choose to study either the kinetics of a stimulus in the steady state – in the case of the GAL gradient we were able to pinpoint the moment where aSyn production peaked – or the response of cells to a dynamically changing chemical environment. This approach is not possible in bulk systems since non-adherent cells are usually cultured in suspension and most adherent cells are able to migrate, making it impossible to track a single cell without the use of reporter molecules.The gradient generator, the other main feature of this device, also represents an improvement to classical cell culture techniques as it allows for the creation of a chemical gradient of customizable resolution with just three initial solutions, which is simpler, quicker and less error-prone than the manual mixing of solutions. Moreover, it allows one to dynamically alter the gradient profile by changing either the initial solutions or the relative flow rates in each channel. Whereas in bulk systems the exposure of a cell population to a different chemical stimulus would imply removing the medium, rinsing and inserting a new solution, in this system this change can be made seamlessly by simply changing the solution-containing syringes. Furthermore, the combination of cell trapping and perfusion allows a given cell – or a population of cells – to be exposed to a constant microenvironment, with a constant supply of nutrients and flushing of metabolites, all the while remaining immobilized and available for imaging – something that in conventional cell culture is only possible with bulky and expensive perfusion systems. In the context of cell assays under perfusion conditions, it should also be added that to perform any of the experiments described, we require just three 1 mL syringes containing the chemicals of interest.
We focused on exposing populations of yeast cells expressing aSyn-GFP to highly controlled yet chemically different environments. As previously explained, this device also has the added benefit of allowing the chemical environment to be changed dynamically, allowing cells to be exposed to a sequence of chemical species of interest, such as oxidants and antioxidants. In doing so, we targeted applications where rapid, parallel analysis of aSyn expression levels and location is needed, e.g., for characterizing the effect of the complex and the sequential exposure of the cellular model to solutions with different chemical compositions of interest. To approach these goals, we anticipate further research towards (i) integrating an array of microsensors into the chip at the cell trap locations, able not only to follow the fluorescence of different fluorophores in individual whole cells, but also, desirably, to map the spatial distribution of fluorescence in each cell in real time; (ii) improving cell handling to allow precise on-chip cell collection after sequential exposure for further experiments, namely, genetic expression profiling; and (iii) understanding how the knowledge of single-cell variability can impact our understanding of PD pathology, given that specific populations of neuronal cells appear to show different susceptibility to aSyn or chemical toxins. Eventually, advanced microfluidic cell-chip devices will enable us to develop systems that facilitate the development of novel in vitro disease models that recapitulate the molecular underpinning of the human disease more reliably, potentiating the discovery of novel therapeutic or diagnostic strategies.
Although in this study we explored this device for our specific interest in aSyn pathobiology, it is important to mention that the system developed here is readily adaptable to studies involving other proteins. In fact, yeast cells are unquestionable powerful models to dissect the basic molecular mechanisms underlying PD and other neurodegenerative diseases,3 as well as other human diseases such as mitochondrial diseases,37 cancer,2 and calcium-related diseases,38 among others. Yeast models have also been very useful to identify new compounds with therapeutic interest through high-throughput screening approaches39,40 as well as to identify the molecular basis of the action of pharmacologically active molecules.41,42
This device is also adaptable to studies using other cell types, requiring eventually minor adjustments in the design. There are also advantages for studies employing adherent cells since the trapping allows placing them in pre-determined sites on the chip, prevents cell–cell communication if single cells are captured and avoids potential flushing of cells under perfusion conditions.23 If required, the resolution of the gradient, i.e. the concentration intervals, can be easily increased by adding extra levels and chambers to the device. Finally, bidimensional or more complex gradients, where two different compounds would be inserted through the solution inlets to study synergistic effects, are another interesting and innovative application for this device, with the extra advantage that only very small volumes of the testing compounds would be required. In total, our study not only provides novel information on the biology of aSyn but also expands the toolbox for investigating biological processes of interest with unprecedented resolution.
Acknowledgements
We thank Dr. Paul Muchowski for kindly providing the VSY72 strain, Dr. Susan Lindquist for providing the Y4791 strain and Ana Nascimento and António Temudo of the IMM Bioimaging Unit for their technical support. This work is supported by Fundação para a Ciência e Tecnologia (FCT) through the Associated Laboratories IN and IMM and through project PTDC/BIA-BCM/117975/2010. JTSF and ST were supported by FCT (SFRH/BD/73908/2010 and SFRH/BPD/35767/2007). TFO was supported by an EMBO Installation Grant and by the DFG Center for Nanoscale Microscopy and Molecular Physiology of the Brain.Notes and references
- D. Botstein and G. R. Fink, Genetics, 2011, 189, 695–704 CrossRef CAS PubMed.
- C. Pereira, I. Coutinho, J. Soares, C. Bessa, M. Leão and L. Saraiva, FEBS J., 2012, 279, 697–712 CrossRef CAS PubMed.
- S. Tenreiro, M. C. Munder, S. Alberti and T. F. Outeiro, J. Neurochem., 2013, 127, 438–452 CrossRef CAS PubMed.
- L. Miller-Fleming, F. Giorgini and T. F. Outeiro, Biotechnol. J., 2008, 3, 325–338 CrossRef CAS PubMed.
- S. Tenreiro and T. F. Outeiro, FEMS Yeast Res., 2010, 10, 970–979 CrossRef CAS PubMed.
- M. G. Spillantini, M. L. Schmidt, V. M. Lee, J. Q. Trojanowski, R. Jakes and M. Goedert, Nature, 1997, 388, 839–840 CrossRef CAS PubMed.
- J. M. Shulman, P. L. De Jager and M. B. Feany, Annu. Rev. Pathol.: Mech. Dis., 2011, 6, 193–222 CrossRef CAS PubMed.
- M. Goedert, M. G. Spillantini, K. Del Tredici and H. Braak, Nat. Rev. Neurol., 2012, 9, 13–24 CrossRef PubMed.
- P. Wales, R. Pinho, D. F. Lázaro and T. F. Outeiro, J. Parkinson's Dis., 2013, 3, 415–459 CAS.
- M. R. Cookson, Mol. Neurodegener., 2009, 4, 9 CrossRef PubMed.
- H. A. Lashuel, C. R. Overk, A. Oueslati and E. Masliah, Nat. Rev. Neurosci., 2012, 14, 38–48 CrossRef PubMed.
- T. F. Outeiro and S. Lindquist, Science, 2003, 302, 1772–1775 CrossRef CAS PubMed.
- A. A. Cooper, A. D. Gitler, A. Cashikar, C. M. Haynes, K. J. Hill, B. Bhullar, K. Liu, K. Xu, K. E. Strathearn, F. Liu, S. Cao, K. A. Caldwell, G. A. Caldwell, G. Marsischky, R. D. Kolodner, J. Labaer, J.-C. Rochet, N. M. Bonini and S. Lindquist, Science, 2006, 313, 324–328 CrossRef CAS PubMed.
- V. Sancenon, S.-A. Lee, C. Patrick, J. Griffith, A. Paulino, T. F. Outeiro, F. Reggiori, E. Masliah and P. J. Muchowski, Hum. Mol. Genet., 2012, 21, 2432–2449 CrossRef CAS PubMed.
- L. J. Su, P. K. Auluck, T. F. Outeiro, E. Yeger-Lotem, J. A. Kritzer, D. F. Tardiff, K. E. Strathearn, F. Liu, S. Cao, S. Hamamichi, K. J. Hill, K. A. Caldwell, G. W. Bell, E. Fraenkel, A. A. Cooper, G. A. Caldwell, J. M. McCaffery, J.-C. Rochet and S. Lindquist, Dis. Models & Mech., 2010, 3, 194–208 CAS.
- S. Willingham, T. F. Outeiro, M. J. DeVit, S. L. Lindquist and P. J. Muchowski, Science, 2003, 302, 1769–1772 CrossRef CAS PubMed.
- S. K. Sia and G. M. Whitesides, Electrophoresis, 2003, 24, 3563–3576 CrossRef CAS PubMed.
- J. Melin and S. R. Quake, Annu. Rev. Biophys. Biomol. Struct., 2007, 36, 213–231 CrossRef CAS PubMed.
- D. J. Beebe, G. A. Mensing and G. M. Walker, Annu. Rev. Biomed. Eng., 2002, 4, 261–286 CrossRef CAS PubMed.
- G. M. Whitesides, E. Ostuni, S. Takayama, X. Jiang and D. E. Ingber, Annu. Rev. Biomed. Eng., 2001, 3, 335–373 CrossRef CAS PubMed.
- E. W. K. Young and D. J. Beebe, Chem. Soc. Rev., 2010, 39, 1036–1048 RSC.
- J. Nilsson, M. Evander, B. Hammarström and T. Laurell, Anal. Chim. Acta, 2009, 649, 141–157 CrossRef CAS PubMed.
- D. Di Carlo, L. Y. Wu and L. P. Lee, Lab Chip, 2006, 6, 1445 RSC.
- S. K. W. Dertinger, D. T. Chiu, N. L. Jeon and G. M. Whitesides, Anal. Chem., 2001, 73, 1240–1246 CrossRef CAS.
- N. L. Jeon, S. K. W. Dertinger, D. T. Chiu, I. S. Choi, A. D. Stroock and G. M. Whitesides, Langmuir, 2000, 16, 8311–8316 CrossRef CAS.
- D. C. Duffy, J. C. McDonald, O. J. A. Schueller and G. M. Whitesides, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- M. A. Eddings, M. A. Johnson and B. K. Gale, J. Micromech. Microeng., 2008, 18, 067001 CrossRef.
- M. W. Toepke and D. J. Beebe, Lab Chip, 2006, 6, 1484–1486 RSC.
- G. Shao, J. Wang, Z. Li, L. Saraf, W. Wang and Y. Lin, Sens. Actuators, B, 2011, 159, 44–50 CrossRef CAS PubMed.
- N. Q. Balaban, Science, 2004, 305, 1622–1625 CrossRef CAS PubMed.
- X. R. Bao, I. D. C. Fraser, E. A. Wall, S. R. Quake and M. I. Simon, Biophys. J., 2010, 99, 2414–2422 CrossRef CAS PubMed.
- P. Zabrocki, K. Pellens, T. Vanhelmont, T. Vandebroek, G. Griffioen, S. Wera, F. Van Leuven and J. Winderickx, FEBS J., 2005, 272, 1386–1400 CrossRef CAS PubMed.
- M. Takahashi, L. Ko, J. Kulathingal, P. Jiang, D. Sevlever and S.-H. C. Yen, Eur. J. Neurosci., 2007, 26, 863–874 CrossRef PubMed.
- S. N. Witt and T. R. Flower, FEMS Yeast Res., 2006, 6, 1107–1116 CrossRef CAS PubMed.
- T. R. Flower, L. S. Chesnokova, C. A. Froelich, C. Dixon and S. N. Witt, J. Mol. Biol., 2005, 351, 1081–1100 CrossRef CAS PubMed.
- T. Rinaldi, C. Dallabona, I. Ferrero, L. Frontali and M. Bolotin-Fukuhara, FEMS Yeast Res., 2010, 10, 1006–1022 CrossRef CAS PubMed.
- C. Voisset, N. García-Rodríguez, A. Birkmire, M. Blondel and R. E. Wellinger, Biochim. Biophys. Acta, 2014, 1843, 2315–2321 CrossRef CAS PubMed.
- D. F. Tardiff and S. Lindquist, Drug Discovery Today: Technol., 2013, 10, e121–e128 CrossRef PubMed.
- D. Delneri, FEMS Yeast Res., 2010, 10, 1083–1089 CrossRef CAS PubMed.
- M. Mattiazzi, U. Petrovič and I. Križaj, Toxicon, 2012, 60, 558–571 CrossRef CAS PubMed.
- M. Schenone, V. Dančík, B. K. Wagner and P. A. Clemons, Nat. Chem. Biol., 2013, 9, 232–240 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |