Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cell patterning with a heptagon acoustic tweezer – application in neurite guidance†
F.
Gesellchen
a,
A. L.
Bernassau
b,
T.
Déjardin
a,
D. R. S.
Cumming
b and
M. O.
Riehle
*a
aCentre for Cell Engineering, College of Medical, Veterinary & Life Sciences, University of Glasgow, Glasgow, UK. E-mail: mathis.riehle@glasgow.ac.uk
bSchool of Engineering, College of Science and Engineering, University of Glasgow, Glasgow, UK
First published on 28th April 2014
Abstract
Accurate control over positioning of cells is a highly desirable feature in tissue engineering applications since it allows, for example, population of substrates in a controlled fashion, rather than relying on random seeding. Current methods to achieve a differential distribution of cells mostly use passive patterning methods to change chemical, mechanical or topographic properties of surfaces, making areas differentially permissive to the adhesion of cells. However, these methods have no ad hoc control over the actual deposition of cells. Direct patterning methods like bioprinting offer good control over cell position, but require sophisticated instrumentation and are often cost- and time-intensive. Here, we present a novel electronically controlled method of generating dynamic cell patterns by acoustic trapping of cells at a user-determined position, with a heptagonal acoustic tweezer device. We demonstrate the capability of the device to create complex patterns of cells using the device's ability to re-position acoustic traps by using a phase shift in the acoustic wave, and by switching the configuration of active piezoelectric transducers. Furthermore, we show that by arranging Schwann cells from neonatal rats in a linear pattern we are able to create Bands of Büngner-like structures on a non-structured surface and demonstrate that these features are able to guide neurite outgrowth from neonatal rat dorsal root ganglia.
Introduction
Spatial control of cell positions is of particular importance in the field of tissue engineering. In complex tissues, cells rely on a variety of cues from their environment, such as cell–cell contacts (homo- or heterotypic), substrate adhesion and mechanical forces or extracellular stimuli such as signalling molecules (auto- or paracrine).1 These factors are dependent on accurate positioning of cells in their microenvironment, which remains a major challenge for replication of a functional histoarchitecture in tissue engineering.A variety of microscale methods for patterning cells on a substrate with μm accuracy have been developed in recent years. They are based either on the direct patterning of cells, or indirect patterning via chemical,2 topographic,3 or mechanical4,5 modification of surfaces to direct differential cell adhesion. Direct cell patterning has been demonstrated with inkjet or laser assisted cell printing,6,7 as well as electrical force (dielectrophoresis8,9), optical force (laser guided direct writing10 and laser guided micropatterning11) and magnetic forces using iron oxide-labelled cells.12 Indirect patterning by selective surface modification is mostly done by microcontact printing (μCP) using poly-di-methyl-siloxane (PDMS) stamps fabricated from a master made using photolithography.13 The PDMS stamp is then used to print self-assembling monolayers of derivatized alkanethiols, on to a solid substrate, that promote matrix protein and thus cell adhesion. Alternatively, extracellular matrix proteins can also be printed directly on a substrate to permit cell adhesion.14 PDMS stamps have also been used to create microfluidic devices for cell and protein patterning.15 Other approaches use dynamic substrates, i.e. “switchable surfaces”, whose cell adhesiveness can be modified by light,16 temperature17 or voltage.18
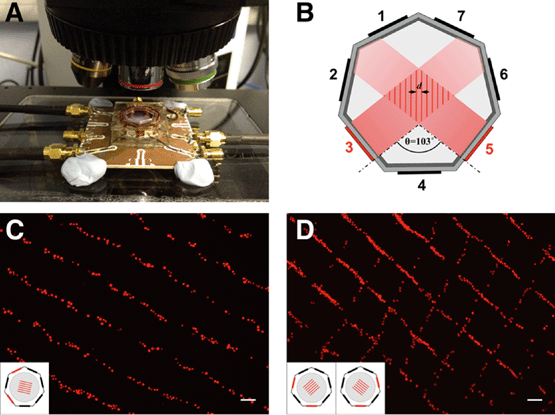
While these methods have been successfully employed to create cell patterns and co-cultures of varying complexity, they often require specialized equipment, and can be cost- and time intensive. Furthermore, many of these methods are inflexible and do not offer dynamic ad hoc geometric control over the pattern; in the case of PDMS stamps, for example, the pattern is limited to that of the lithographic mask used to create the stamp. To address some of these shortcomings we have recently developed a portable device based on acoustic force for spatial manipulation of cells and particles19 (Fig. 1A). The use of acoustic force for immobilization of cells is well established, employing either bulk or surface acoustic waves in a resonant structure that produces a fixed standing wave pattern.20–22 Cells or dense particles agglomerate at nodes with lowest acoustic pressure. The position of these pressure nodes is determined by the geometry of the device and the frequency at which piezoelectric transducers generating the standing acoustic wave are operated. Control over the position of the pressure nodes in conventional transducer-reflector devices is therefore limited. Our device overcomes this limitation by using a heptagonal geometry with transducers that generate travelling waves. The interference pattern produced at the intersection of two or more forward travelling waves creates a nodal pattern that is capable of trapping cells23 (Fig. 1B). Waves are either absorbed at the vertex opposing the transducer, or scattered away from the central region of the device. Because the device operation relies on the use of travelling waves, the precise interference pattern, hence the acoustic pressure nodes, may be electronically controlled by adjusting the phase of the excitation to each transducer.19 The distance between two adjacent pressure nodes is given by d = θ/2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin(λ/2), where θ is the angle formed between the normals to the planes of the active transducers. In a regular heptagonal device with sides numbered 1–7, when transducers placed on two adjacent odd, or equivalently even-numbered, sides and activated at a frequency of 4 MHz (λ = 375 μm in water), θ = 103°. The separation distance between nodes is therefore d = 240 μm. Using the phase shifting capabilities of the device, linear acoustic pressure nodes can then be moved linearly to any desired position in the range d.19
sin(λ/2), where θ is the angle formed between the normals to the planes of the active transducers. In a regular heptagonal device with sides numbered 1–7, when transducers placed on two adjacent odd, or equivalently even-numbered, sides and activated at a frequency of 4 MHz (λ = 375 μm in water), θ = 103°. The separation distance between nodes is therefore d = 240 μm. Using the phase shifting capabilities of the device, linear acoustic pressure nodes can then be moved linearly to any desired position in the range d.19
Our aim in this study was to investigate successive patterning of cells and using acoustic tweezers for an application in tissue engineering. We have previously shown that acoustic tweezers are capable of trapping micron-scale particles, and cells at nodes of minimal acoustic pressure.23 Here, we demonstrate complex multistep cell patterning using phase shifting and varying transducer configurations to arrange cells in six different successive patterns. Furthermore, we show the usefulness of a pattern generated by the device for tissue engineering. In response to peripheral nerve injury, a natural aligned structure termed “Bands of Büngner” develops during peripheral nerve repair.24 These Bands of Büngner consist of aligned Schwann cells and are the natural substrate that regenerating neurons follow and use to reconnect to their peripheral targets. A variety of methods have been developed to create aligned structures that guide Schwann cells in vitro and in vivo with the aim to create an environment conducive to optimal nerve regeneration. Apart from mechanically aligning Schwann cells by directional tension within a collagen gel25 most techniques rely on indirect patterning of cells by proxy of their interaction with a patterned substrate. Substrate features that align neuronal regeneration can be linear chemical micropatterns,26,27 microtopographies,28 and static electrical fields.29 So far direct cell patterning, e.g. with cell printing, optical trapping or magnetic guidance, has not been used to create replicates of Bands of Büngner to the best of our knowledge. Therefore we decided to explore the capability of the heptagon acoustic tweezer device for direct patterning of Schwann cells, obviating the need for prior treatment of the substrate. We show that ultrasonically patterned Schwann cells form linear arrangements resembling Bands of Büngner that serve to guide neurite outgrowth from rat dorsal root ganglia (DRG), in an in vitro model for peripheral nerve regeneration.
Experimental
Sample preparation including cell culture and fluorescent labelling, magnetic labelling and separation of Schwann cells, and image analysis of neurite outgrowth are described in the ESI.†Acoustic tweezer operation
The design and construction of the heptagon acoustic tweezer is described in detail elsewhere.19 The transducers were driven by a 4 MHz sine wave (corresponding to a wavelength of 375 μm in water) at an amplitude of 8 Vpp, generated from an arbitrary waveform generator (TGA12104, Thurlby Thandar Instruments Ltd, UK). The waveform generator enabled synchronisation between channels and independent control of frequency, phase and amplitude to each transducer. Signals were amplified and electronically matched using high-speed buffers (BUF634T, Texas Instruments UK).Prior to the patterning experiments, a layer of 1.5% agar in phosphate buffered saline (PBS) was deposited inside the heptagon cell, filling the cavity up to approximately halfway (~1.2 mL) in order to reduce acoustic streaming that can occur in liquid medium.30
Cell patterning
A 13 mm glass cover slip coated with poly-L-lysine (Sigma-Aldrich) or laminin (AMS Biotechnology, UK), to facilitate cell adhesion, was placed at the centre of the cavity and covered with 0.5 mL of DMEM. The differently labelled cells were introduced to the cavity in successive stages, with different patterns of acoustic excitation for each. 50–100 μl of cells (25–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells) were carefully added to the centre of the device and left to adhere under the influence of the acoustic field for 30–60 min before altering the acoustic excitation for the next round of cell patterning. After patterning was complete and cells had adhered sufficiently strong, the coverslips were transferred to a tissue culture plate for further incubation if required.
000 cells) were carefully added to the centre of the device and left to adhere under the influence of the acoustic field for 30–60 min before altering the acoustic excitation for the next round of cell patterning. After patterning was complete and cells had adhered sufficiently strong, the coverslips were transferred to a tissue culture plate for further incubation if required.
Patterning of Schwann cells for DRG neurite outgrowth
Schwann cells at P0 or P1 were detached from tissue culture flasks with Trypsin/EDTA and resuspended at a concentration of 5 × 105 cells mL−1 in DMEM/F12. 5 × 104 cells were patterned in the heptagon acoustic tweezer using two active transducers for 30 min to generate a linear pattern as described in the main text. After letting the patterned cells adhere for at least another hour, the 13 mm coverslip was carefully removed with forceps from the device, transferred to a 24 well plate and 500 μl of culture medium added, taking care to prevent dislodging of cells. Patterned Schwann cells and randomly seeded controls were incubated for 24 h at 37 °C (5% CO2, 100% humidity) before initiating co-culture with DRGs.Co-culture of DRG and Schwann cells
Thoracic DRGs were isolated from 2 day-old neonatal Sprague-Dawley rats. Rats were euthanized by a Euthatal® injection and then dissected in accordance with Home Office regulation. After removal of muscle and bones dorsally around the spinal cord, the DRGs were extracted with surgical tweezers and processed to remove remaining nerve tissue.The freshly isolated rat neonatal DRGs were placed onto the patterned area at the center of the coverslip in SCGM supplemented with 10 ng mL−1 NGF 2.5S (Life Technologies, UK). The media level was reduced to 250 μl in order to permit attachment of DRGs and incubation continued for 4 days. One half of the media was exchanged daily.
DRGs placed at the center of coverslips with randomly seeded Schwann cells and treated identically throughout served as controls.
Immunofluorescence
Cells were fixed in 4% formaldehyde, permeabilized (10.3 g sucrose, 0.292 g NaCl, 0.06 g MgCl2, 0.476 g HEPES 0.5 ml Triton X-100 in 100 ml PBS) and blocked using 1% bovine serum albumin (BSA) in PBS. Neurons were immunostained with a βIII-tubulin antibody (mouse anti-TU-20, Santa Cruz Biotechnologies, USA), followed by biotinylated anti-mouse IgG and Fluorescein-labelled streptavidin (Vector Laboratories Ltd, UK). All antibodies were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in PBS/1% BSA.
100 in PBS/1% BSA.
Microscopy & image analysis
For microscopic observation the device was placed on the stage of an upright epifluorescence microscope (Olympus BX51, Olympus UK; Fig. 1A) equipped with a tri-pass filter cube (U-M61002, DAPI/FITC/TexasRed) and a motorized stage (H1P1BX, Prior Scientific UK). Micrographs were taken using a 4 or 5× objective with a cooled CCD camera (QImaging) and the ImagePro Plus 7.0 software package (Media Cybernetics, USA). Tiled images of neurite outgrowth from DRGs were taken with the StagePro module of the ImagePro Plus software; the scan area was manually defined to encompass the entire area of interest and images stitched automatically. Image analysis for measuring distances and angles was performed using the corresponding tools in ImageJ 1.44.31 Qualitative and quantitative analysis of the directionality of neurite outgrowth was performed using the ImageJ plug-in OrientationJ32 (see ESI† for more details). A chi-square test was performed to compare the frequency distributions obtained for neurite outgrowth on patterned and randomly seeded Schwann cells.Results
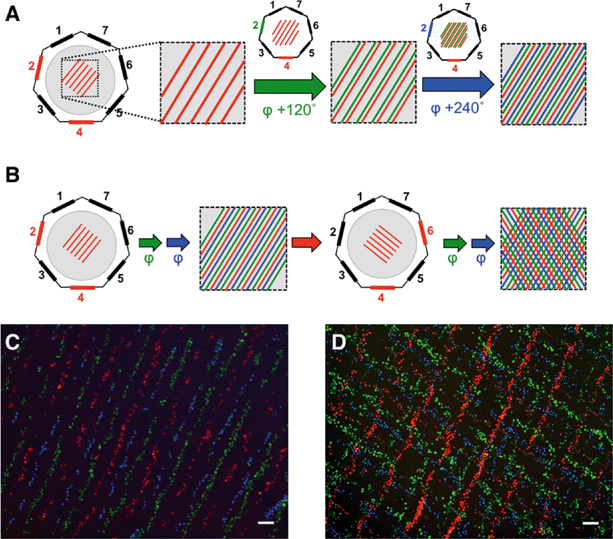
Initial experiments to demonstrate the feasibility of the approach were conducted using fluorescently labelled C2C12 cells (ATCC CRL 1772). Three sets of C2C12 stained with different fluorescent dyes (MitoTracker Green, MitoTracker Red and Hoechst 33342) were used in order to visualize successive patterning events. Once we had demonstrated to our satisfaction that acoustic tweezing could be used to manipulate multiple batches of cells in successive cycles, we went on to demonstrate that manipulated cells could be used to in turn align neurite outgrowth in a peripheral nerve injury model.Acoustic trapping
It was first necessary to show that lines of cells could be formed using the acoustic tweezer, and that they would adhere and begin to culture on a planar surface as determined by the user. The surface was a 13 mm PLL-coated glass coverslip inserted into the centre of the acoustic tweezer device. The cell alignment is illustrated in Fig. 1C where C2C12 that were stained with MitoTracker (MT) red were patterned using transducer pair 2–4.Although 90% of cells remained attached where they had initially settled, some (~10%) were moved to the new position of the pressure node when the phase was shifted, as evident by the presence of green (11.4%) and red cells (9.1%) mixed with the blue cells, which had been patterned last (Fig. 2C).
Schwann cell patterning for neurite guidance
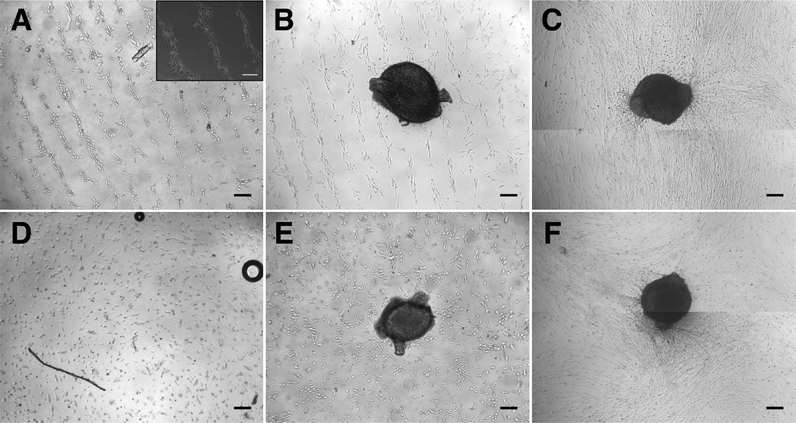
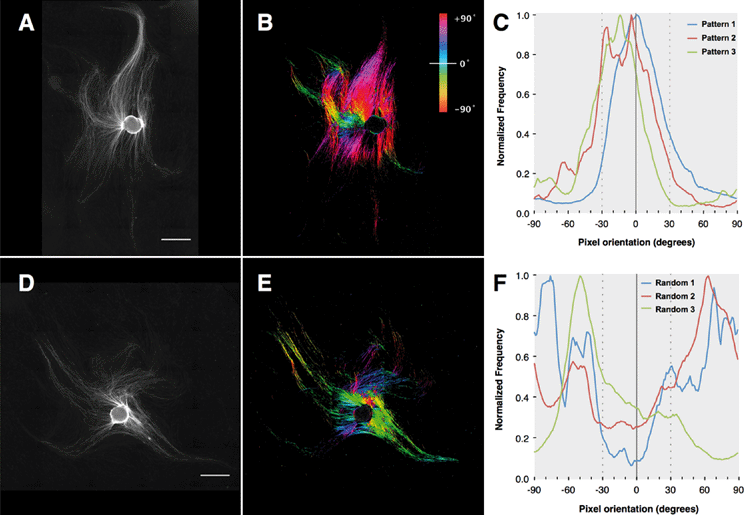
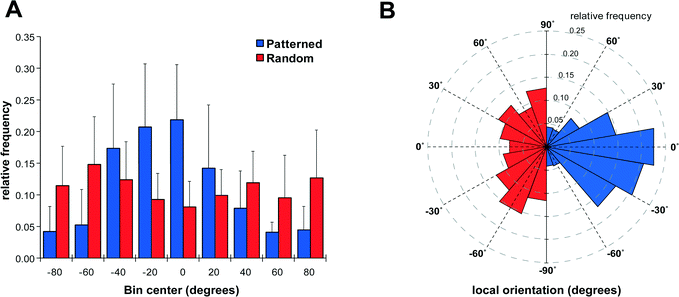
After establishing the capability of the acoustic tweezer for cell patterning, we aimed to test its usefulness in a model system for tissue engineering, specifically in peripheral nerve regeneration. To this end we deposited Schwann cells isolated from neonatal rat sciatic nerves in a linear pattern on PLL-coated cover slips using the heptagon acoustic tweezer device for 30 min, in a 1–3 transducer configuration. After 24 h incubation the cells largely retained the linear pattern and partially arranged in a columnar fashion reminiscent of bands of Büngner (Fig. 3A + inset), formed by denervated Schwann cells during peripheral nerve regeneration. In contrast, randomly seeded Schwann cells displayed no preferential orientation after overnight incubation (Fig. 3D). There was no difference in the number of rounded – non-proliferating cells between the two groups (e.g. in Fig. 3A, D), indicating that acoustic tweezing did not affect cell survival. Neonatal rat DRGs placed on patterned and randomly seeded Schwann cells (Fig. 3B and E, respectively) both had substantial neurite outgrowth after 4 days in culture (Fig. 3C and F).In order to visualize the direction of outgrowing neurites, cells were stained for βIII-tubulin, which revealed an extensive network of neurites on patterned as well as non-patterned Schwann cells (Fig. 4). Neurites growing on a patterned Schwann cell layer exhibited an orientation along a pattern axis, while those on randomly seeded Schwann cells projected networks in several directions (Fig. 4A and D). We used the ImageJ plug-in OrientationJ32 to obtain a qualitative and quantitative measure of neurite orientations. OrientationJ derives the local orientation and isotropic values (coherency and energy) of every pixel in an image and outputs a color-coded representation of local angles and isotropy of features. Fig. 4B and E show the output generated for the images in Fig. 4A and D respectively. For those neurites outgrowing on patterned Schwann cells (Fig. 4B) a clear preference for directions around +90 and −90 degrees, which is in good accordance with the original Schwann cell pattern (Fig. 3B) is evident. The neurites on randomly seeded Schwann cells appear to show a set of two preferential orientations as well, which could be due to a bias e.g. by the position of the nerve stumps, and maybe some neurite associated self-organization of the underlying Schwann cell layer (see difference in cell pattern between 3E and 3F). However, compared to the axon network on patterned Schwann cells, other local angles are just as strongly represented, indicating a more random pathfinding of the axon network. This becomes even more evident when comparing the local angle histograms from three separate experiments (Fig. 4C and F). The local angles of neurites growing on patterned Schwann cells display a preferred direction around the initial angle of the respective Schwann cell pattern (defined as zero degrees) compared to the random controls (where zero denotes the horizontal axis of the image), indicating that patterned Schwann cells provide an efficient guidance cue for neurites growing out from the DRGs. A second analysis using OrientationJ's Measure function, which determines the dominant orientation (as well as coherency and energy) of features within a user defined ROI, allowed us to include faintly stained areas, that were excluded from the measurement of pixel orientation due to the detection threshold being set for the whole image. Using a custom-written macro each image was divided into adjoining 250 × 250 pixel ROIs that were individually analyzed, if their integrated density value exceeded a user-defined threshold. In this way we were able to include even faintly stained neurites in our analysis (Fig. S1†). The relative frequency distributions of local directions, obtained with this analysis are shown in Fig. 5, using 20 degree bins centered on zero degrees (Fig. 5A). Despite experimental variability, neurites growing on patterned Schwann cells show a clustering around the zero degree orientation, with ~60% of the analyzed areas oriented within ±30 degrees of the initial Schwann cell pattern, and angles >50 degrees relative to the pattern clearly underrepresented. In contrast the neurite outgrowth on randomly seeded Schwann cells shows no preferred direction (Fig. 5B). A chi-square test showed a highly significant difference between the two frequency distributions (p ≪ 0.001), indicating that the initial patterning of Schwann cells by acoustic force had a significant influence on the subsequent outgrowth of neurites from explanted DRGs.
Discussion
In the present study we have shown that the acoustic tweezer is capable of trapping cells at predetermined positions and, by using the ability to switch phase, and operate different sets of transducers, we can generate complex cellular patterns. Compared to other methods such as laser guided direct writing, the new device has the advantage of being small, electronically controlled, flexible in the patterning and can be easily integrated with standard microscopy equipment. The device can handle high cell densities and consequently has a relatively good throughput, which is very beneficial for applications in tissue engineering. Additionally it allows handling of different cell types sequentially; if an initially patterned cell type is left to adhere for a sufficient amount of time (usually 30 min) these cells stay in position when the position of the acoustic traps is shifted. Another cell type can then be seeded at the newly positioned pressure nodes, as exemplified by the patterning experiments with fluorescently labelled cells. The technique is made possible because cells, once adhered to a surface, are sufficiently firmly bound that the acoustic forces exerted by the tweezing device cannot move them. Therefore, successive additions of cells can be manipulated independently from previous aliquots of cells. The forces exerted onto individual cells by this acoustic tweezer is in the range 2–10 pN, or 71 kPa.30 However, the forces needed to detach individual cells are of the order of tens, to several hundreds of nN.33–35 Even individual integrin/fibronectin binding events can reach rupture strengths of around 90 pN,36 which exceeds the maximal forces applied by the acoustic tweezer by a factor of about 10–20.While the topologies of the patterns that can be generated with the acoustic tweezer are set by the device geometry, it is possible to actively control the position of any further cell types by adjusting the position of the acoustic pressure nodes by phase shifts, or transducer switching with an accuracy of less than 10 μm, which is smaller than a tissue cell diameter. It should be noted that no treatment of either the cells or the surfaces is necessary to achieve a patterned cell layer. The number of different cell types that can be patterned in this fashion is only limited by space available between the pressure nodes. Using a 1–3 transducer configuration, operated at 4 MHz, results in a spacing of ~240 μm between the nodes, taking the size of cells into account (10–30 μm) this limits side-by-side patterning to 90° phase shifts (~60 μm spacing) and thus four different cell types. Space permitting, cells can then be patterned at an angle to the initial pattern as evidenced by the “cell tartan” (Fig. 2D). Available space being the main limitation, for C2C12 cells, some cells were inevitably stacked on top of previously patterned cells, which prevented them from adhering to the substrate. The other possible transducer combinations (1–2, 1–3–5) were not considered here because of extreme streaming (1–2),30 or because the position of the phase shifted node pattern would in parts overlap with the initial pattern (1–3–5).19 The “cell tartan” shows clearly the versatility of the acoustic tweezer device for complex cell patterning on any flat substrate with any adhesive cell type in normal media, making this technology compatible with a wide array of cell culture techniques.
After demonstrating the versatility of the acoustic tweezer for manipulating cells, we tested its usefulness in a model system for tissue engineering, specifically in peripheral nerve regeneration. Although peripheral nerves have good regenerative properties, the outcome is often less than optimal, highlighting the importance of supporting strategies in nerve repair.37 Schwann cells are instrumental in supporting the repair process, first by initiating clearance of axonal and myelin debris at the injury site and subsequently supporting regrowth of axons sprouting from the proximal stump.38 However, functional outcomes are often suboptimal, limited by misdirection of outgrowing neurites and a slow regeneration rate, leading to chronic denervation of Schwann cells distal to the injury site.39 This in turn causes those Schwann cells to lose their growth supporting phenotype, ultimately leading to a failure to re-innervate the target organ.40 While autologous nerve grafts still represent the gold standards in peripheral nerve repair, tissue engineering approaches to improve the functional outcome have received increasing attention in recent years.41 Several studies utilized Schwann cells to generate a repair-permissive environment in in vitro or animal models of nerve repair.27,28,42–45 While these approaches used micropatterned or microstructured surfaces or polymer scaffolds our goal was to investigate whether it is possible to align Schwann cells for nerve repair without underlying guidance cues in a self-organizing, scaffold-free approach.
A simple, linear pattern of Schwann cells was chosen to assess if we can influence the direction of neurite outgrowth from explanted DRGs in this manner. Interestingly, once patterned, the Schwann cells maintained their linear orientation and formed columnar structures similar to Bands of Büngner in regenerating peripheral nerves (Fig. 3A), a phenomena that has also been observed previously in scaffold based Schwann cell alignment.42 Outgrowth of neurites on the layer of Schwann cells was obvious after 4 days when observed through brightfield microscopy, but directional information could not be extracted due to the background of cells and high density of neurites growing on top of them. We utilized βIII-tubulin staining to visualize the neurites and analyzed area scans taken with a motorized stage. Due to the high density of neurites a tracing of single neurites was not feasible. We therefore used the ImageJ plug-in OrientationJ to obtain quantitative data on directionality. Analyses on a single pixel level (Fig. 4) using the Distribution function of OritentationJ showed that the outgrowth of neurites largely followed the orientation of the initial Schwann cell pattern. While the analysis of pixel orientations provided useful information on the direction of neurite outgrowth, it also had some limitations. In order to restrict the analysis to actual features (i.e. neurites) and omit isotropic areas, the threshold for analysis parameters – coherency and energy – had to be chosen accordingly. Invariably, in some of the images this caused some of the more faintly stained neurites to be omitted from analysis (compare Fig. 4A + B and D + E). In addition, the fact that the distribution histogram is weighted by the coherency parameter makes it difficult to compare the results from different experiments, even after normalization. Analysis of 250 × 250 ROIs (Fig. 5 and S1†) with OrientationJ's Measure function confirmed that the majority of neurites growing on patterned Schwann cells were oriented within ±30° of the pre-patterned direction. This is in good agreement with other studies, which aligned Schwann cells using micropatterned laminin44 or a stretched collagen matrix.46 The fact that in our experimental setup, the Schwann cells had no external guidance cues after the initial patterning by the acoustic stencil suggests that there was a significant degree of self-organization involved. A study by Parrinello et al. found that after peripheral nerve injury, fibroblast are instrumental in organizing Schwann cells into cords, in a cell-sorting process mediated by Sox-2.47 It appears that the acoustic alignment was sufficient to promote a similar process just with Schwann cells. The extending neurites maintain the alignment of their associated Schwann cells, thus stabilizing the patterns48 (Fig. 3C & F). Further studies using the phase-shift capabilities of our acoustic tweezer device could explore if including fibroblasts in the patterning process would improve the neurite guiding properties of the patterned cells.
Conclusions
The engineering of tissues for regenerative medicine sometimes requires accurate positioning of cells. Here we demonstrate the usability of a novel method manipulating cells, using acoustic radiation forces, for the creation of complex, tartan-like patterns of cells. This relies on the fact that the forces acting on the cells are very low, in the pico-Newton range, and that the pattern in which cells are deposited can be switched easily. We test the usefulness of such a cellular pattern, on a model of peripheral nerve injury. In peripheral nerve injury bands of Schwann cells guide axonal regeneration, we create such bands using the acoustic tweezer, and show that outgrowth is guided even though the acoustic forces are no longer present.Acknowledgements
The authors would like to acknowledge support by the Engineering and Physical Sciences Research Council in the UK under the “Sonotweezers” grant (EPSRC ref.: EP/G011494/1), the University of Glasgow for a Lord Kelvin Adam Smith Fellowship in Sensor Systems (AB), and the Stephen Forrest Charitable Trust for funding TD. We also thank the staff of the James Watt Nanofabrication Centre for support in the cleanroom (http://www.jwnc.gla.ac.uk).Notes and references
- Principles of Tissue Engineering, ed. R. Lanza, R. Langer and J. P. Vacanti, Academic Press, Burlington, 2007 Search PubMed.
- C. S. Chen, M. Mrksich, S. Huang, G. M. Whitesides and D. E. Ingber, Science, 1997, 276, 1425–1428 CrossRef CAS.
- L. Csaderova, E. Martines, K. Seunarine, N. Gadegaard, C. D. Wilkinson and M. O. Riehle, Small, 2010, 6, 2755–2761 CrossRef CAS PubMed.
- C. M. Lo, H. B. Wang, M. Dembo and Y. L. Wang, Biophys. J., 2000, 79, 144–152 CrossRef CAS.
- B. Cortese, G. Gigli and M. Riehle, Adv. Funct. Mater., 2009, 19, 2961–2968 CrossRef CAS.
- J. A. Barron, P. Wu, H. D. Ladouceur and B. R. Ringeisen, Biomed. Microdevices, 2004, 6, 139–147 CrossRef CAS.
- T. Xu, J. Jin, C. Gregory, J. J. Hickman and T. Boland, Biomaterials, 2005, 26, 93–99 CrossRef CAS PubMed.
- M. Suzuki, T. Yasukawa, H. Shiku and T. Matsue, Biosens. Bioelectron., 2008, 24, 1049–1053 CrossRef PubMed.
- J. Voldman, Annu. Rev. Biomed. Eng., 2006, 8, 425–454 CrossRef CAS PubMed.
- Y. Nahmias, R. E. Schwartz, C. M. Verfaillie and D. J. Odde, Biotechnol. Bioeng., 2005, 92, 129–136 CrossRef CAS PubMed.
- R. K. Pirlo, Z. Ma, A. Sweeney, H. Liu, J. X. Yun, X. Peng, X. Yuan, G. X. Guo and B. Z. Gao, Rev. Sci. Instrum., 2011, 82, 013708 CrossRef PubMed.
- S. P. Grogan, C. Pauli, P. Chen, J. Du, C. B. Chung, S. D. Kong, C. W. Colwell Jr., M. K. Lotz, S. Jin and D. D. D'Lima, Tissue Eng., Part C, 2012, 18, 496–506 CrossRef CAS PubMed.
- R. Singhvi, A. Kumar, G. P. Lopez, G. N. Stephanopoulos, D. I. Wang, G. M. Whitesides and D. E. Ingber, Science, 1994, 264, 696–698 CAS.
- J. L. Tan, W. Liu, C. M. Nelson, S. Raghavan and C. S. Chen, Tissue Eng., 2004, 10, 865–872 CrossRef CAS PubMed.
- D. T. Chiu, N. L. Jeon, S. Huang, R. S. Kane, C. J. Wargo, I. S. Choi, D. E. Ingber and G. M. Whitesides, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 2408–2413 CrossRef CAS PubMed.
- Y. Nakayama, A. Furumoto, S. Kidoaki and T. Matsuda, Photochem. Photobiol., 2003, 77, 480–486 CrossRef CAS.
- M. Yamato, O. H. Kwon, M. Hirose, A. Kikuchi and T. Okano, J. Biomed. Mater. Res., 2001, 55, 137–140 CrossRef CAS.
- M. N. Yousaf, B. T. Houseman and M. Mrksich, Proc. Natl. Acad. Sci. U. S. A., 2001, 98, 5992–5996 CrossRef CAS PubMed.
- A. L. Bernassau, C. K. Ong, Y. Ma, P. G. MacPherson, C. R. Courtney, M. Riehle, B. W. Drinkwater and D. R. Cumming, IEEE Trans. Ultrason. Ferroelectr. Freq. Control, 2011, 58, 2132–2138 CrossRef PubMed.
- K. A. Garvin, D. C. Hocking and D. Dalecki, Ultrasound Med. Biol., 2010, 36, 1919–1932 CrossRef PubMed.
- L. Gherardini, S. Radel, S. Sielemann, O. Doblhoff-Dier, M. Groschl, E. Benes and A. J. McLoughlin, Bioseparation, 2001, 10, 153–162 CrossRef CAS.
- J. Shi, D. Ahmed, X. Mao, S. C. Lin, A. Lawit and T. J. Huang, Lab Chip, 2009, 9, 2890–2895 RSC.
- A. L. Bernassau, F. Gesellchen, P. G. Macpherson, M. Riehle and D. R. Cumming, Biomed. Microdevices, 2012, 14, 559–564 CrossRef CAS PubMed.
- M. D. Binder, N. Hirokawa and U. Windhorst, Encyclopedia of neuroscience, Springer, Berlin, New York, 2009 Search PubMed.
- V. T. Ribeiro-Resende, B. Koenig, S. Nichterwitz, S. Oberhoffner and B. Schlosshauer, Biomaterials, 2009, 30, 5251–5259 CrossRef CAS PubMed.
- D. M. Thompson and H. M. Buettner, Tissue Eng., 2001, 7, 247–265 CrossRef CAS PubMed.
- K. E. Schmalenberg and K. E. Uhrich, Biomaterials, 2005, 26, 1423–1430 CrossRef CAS PubMed.
- C. Miller, S. Jeftinija and S. Mallapragada, Tissue Eng., 2001, 7, 705–715 CrossRef CAS PubMed.
- J. K. Alexander, B. Fuss and R. J. Colello, Neuron Glia Biology, 2006, 2, 93–103 CrossRef PubMed.
- A. L. Bernassau, P. Glynne-Jones, F. Gesellchen, M. Riehle, M. Hill and D. R. Cumming, Ultrasonics, 2014, 54, 268–274 CrossRef CAS PubMed.
- C. A. Schneider, W. S. Rasband and K. W. Eliceiri, Nat. Methods, 2012, 9, 671–675 CrossRef CAS.
- R. Rezakhaniha, A. Agianniotis, J. T. Schrauwen, A. Griffa, D. Sage, C. V. Bouten, F. N. van de Vosse, M. Unser and N. Stergiopulos, Biomech. Model. Mechanobiol., 2012, 11, 461–473 CrossRef CAS PubMed.
- E. Lamers, J. te Riet, M. Domanski, R. Luttge, C. G. Figdor, J. G. Gardeniers, X. F. Walboomers and J. A. Jansen, Eur. Cells Mater., 2012, 23, 182–193 CAS ; discussion 184–193.
- E. Potthoff, O. Guillaume-Gentil, D. Ossola, J. Polesel-Maris, S. LeibundGut-Landmann, T. Zambelli and J. A. Vorholt, PLoS One, 2012, 7, e52712 CAS.
- G. Weder, N. Blondiaux, M. Giazzon, N. Matthey, M. Klein, R. Pugin, H. Heinzelmann and M. Liley, Langmuir, 2010, 26, 8180–8186 CrossRef CAS PubMed.
- R. I. Litvinov, H. Shuman, J. S. Bennett and J. W. Weisel, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 7426–7431 CrossRef CAS PubMed.
- H. T. Khuong and R. Midha, Curr. Neurol. Neurosci. Rep., 2013, 13, 322 CrossRef PubMed.
- Y. J. Son and W. J. Thompson, Neuron, 1995, 14, 125–132 CrossRef CAS.
- S. M. Hall, Ann. N. Y. Acad. Sci., 1999, 883, 215–233 CrossRef CAS.
- S. Y. Fu and T. Gordon, J. Neurosci., 1995, 15, 3886–3895 CAS.
- R. Deumens, A. Bozkurt, M. F. Meek, M. A. Marcus, E. A. Joosten, J. Weis and G. A. Brook, Prog. Neurobiol., 2010, 92, 245–276 CrossRef PubMed.
- A. Bozkurt, R. Deumens, C. Beckmann, L. Olde Damink, F. Schugner, I. Heschel, B. Sellhaus, J. Weis, W. Jahnen-Dechent, G. A. Brook and N. Pallua, Biomaterials, 2009, 30, 169–179 CrossRef CAS PubMed.
- J. B. Phillips, S. C. Bunting, S. M. Hall and R. A. Brown, Tissue Eng., 2005, 11, 1611–1617 CAS.
- D. M. Thompson and H. M. Buettner, Ann. Biomed. Eng., 2004, 32, 1120–1130 CrossRef.
- Y. G. Zhang, Q. S. Sheng, F. Y. Qi, X. Y. Hu, W. Zhao, Y. Q. Wang, L. F. Lan, J. H. Huang and Z. J. Luo, J. Mater. Sci.: Mater. Med., 2013, 24, 1767–1780 CrossRef CAS PubMed.
- M. Georgiou, S. C. Bunting, H. A. Davies, A. J. Loughlin, J. P. Golding and J. B. Phillips, Biomaterials, 2013, 34, 7335–7343 CrossRef CAS PubMed.
- S. Parrinello, I. Napoli, S. Ribeiro, P. Wingfield Digby, M. Fedorova, D. B. Parkinson, R. D. Doddrell, M. Nakayama, R. H. Adams and A. C. Lloyd, Cell, 2010, 143, 145–155 CAS.
- C. Fernandez-Valle, D. Gorman, A. M. Gomez and M. B. Bunge, J. Neurosci., 1997, 17, 241–250 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4lc00436a |
| This journal is © The Royal Society of Chemistry 2014 |