Open Access Article
Open Access ArticleDynamically reconfigurable fibre optical spanner†
Thorsten
Kolb
,
Sahradha
Albert
,
Michael
Haug
and
Graeme
Whyte
*
Biophysics Group, Department of Physics, Friedrich-Alexander Universität Erlangen-Nürnberg, Henkestrasse 91, 91052 Erlangen, Germany. E-mail: gwhyte@biomed.uni-erlangen.de
First published on 20th January 2014
Abstract
In this paper we describe a pneumatically actuated fibre-optic spanner integrated into a microfluidic Lab-on-a-Chip device for the controlled trapping and rotation of living cells. The dynamic nature of the system allows interactive control over the rotation speed with the same optical power. The use of a multi-layer device makes it possible to rotate a cell both in the imaging plane and also in a perpendicular plane allowing tomographic imaging of the trapped living cell. The integrated device allows easy operation and by combining it with high-resolution confocal microscopy we show for the first time that the pattern of rotation can give information regarding the sub-cellular composition of a rotated cell.
Optical trapping is a powerful tool for manipulating microscopic objects1 including living cells. In addition to holding the trapped object in place, it is also possible to rotate it using the intrinsic spin,2,3 orbital angular momentum4,5 or anisotropic beam shape6 however all of these systems require complex optical equipment.
Dual-beam optical traps7 are created when two, diverging counter propagating beams overlap and form a stable trap in the centre of the two beams. The large area and diverging nature provides a less photo-damaging technique for holding and manipulating living cells.6,8,9 Recent developments have increased the range of possible manipulations which can be carried out with dual beam traps to include mechanical phenotyping of living cells,9,10 optical rotation6 and optofluidic rotation. The development of dual-beam fibre traps11 simplifies the creation and integration of such traps into microfluidic systems12–14 allowing for high throughput measurements and more complex microfluidic processing.
The fibre optic spanner15 is a form of dual-beam trap where the optical fibres are intentionally misaligned to provide a torque on the trapped object from the asymmetry of the trapping light beams, and it can be used to rotate cells around an axis perpendicular to the misalignment. In theory this allows cells to be rotated around any angle by correctly arranging the fibre positions, however accurate control of the fibre position and angle limits the usability of such a system.
Here we present a dynamically reconfigurable system for positioning the optical fibres within a dual-beam trap. The positioning is achieved using the elastic nature of PDMS devices and by pneumatically applying air pressure to specifically designed membranes.16,17 Multilayer PDMS microfabrication allows such membranes to be placed both in the plane of the fibre as well as above it, to allow adjustment of the fibre in orthogonal directions, opening the door to rotate cells around an axis perpendicular to the imaging plane of the microscope for single cell tomography.
Experimental
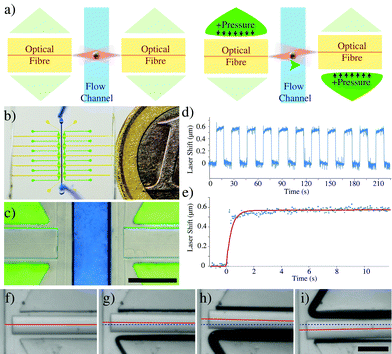
The device is made up of a PDMS chip housing two optical fibres aligned to each other across a microfluidic flow channel, and pneumatic actuators next to the fibres to dynamically adjust the positions of the fibres. The actuators move the fibres by applying a pressure to one side of a thin PDMS membrane which in turn pushes the fibre away. By moving the fibres from their aligned position, it is possible to create a reconfigurable fibre optic spanner capable of controlling the rotation speed of single cells held in the optical trap.The additional freedom of controlling the fibre misalignment allows more detailed characterisation of the rotating cell.
Device fabrication
Devices were fabricated using standard PDMS moulding techniques from a photolithography defined SU8 master. Briefly SU8-2025 (Microchem Inc) was spin coated onto 3′′ Si wafers (silicon materials) to form a 125 μm thick layer to allow the easy insertion of the optical fibres,18 and then features were created using UV photolithography from an acetate mask. Single-layer devices were produced from 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 elastomer to curing agent (Syl-Guard185), which was mixed and poured onto the SU8 master. After baking for at least 3 hours at 80 °C, the device was peeled from the SU8 and plasma bonded to a microscope coverslip using air plasma generated by a plasma oxidiser (Diener) and further baked for 1 hour at 80 °C.
1 elastomer to curing agent (Syl-Guard185), which was mixed and poured onto the SU8 master. After baking for at least 3 hours at 80 °C, the device was peeled from the SU8 and plasma bonded to a microscope coverslip using air plasma generated by a plasma oxidiser (Diener) and further baked for 1 hour at 80 °C.
Multi-layer devices were produced in a similar manner to Hulme.19 Briefly, the top layer containing the z-adjustment pads were produced as above, however instead of bonding to a microscope coverslip, they were plasma bonded to a thin layer of PDMS which was spin coated onto a silanised (Sigma-Aldrich) glass slide. Once the top layer had sealed to the thin layer of PDMS, both layers were peeled away from the supporting glass slide. The bottom layer was prepared by pouring 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixed PDMS : curing agent onto the SU8 master and aligning the top layer to the fibre channels under a microscope. The multi-layer device was then baked for at least 3 hours at 80 °C to cure the liquid PDMS and seal it to the top layer. Finally the multi-layer structure was cut out of the master and sealed to a 150 μm thick layer of PDMS using air plasma treatment, to form the bottom wall of the fluidic channel.
1 mixed PDMS : curing agent onto the SU8 master and aligning the top layer to the fibre channels under a microscope. The multi-layer device was then baked for at least 3 hours at 80 °C to cure the liquid PDMS and seal it to the top layer. Finally the multi-layer structure was cut out of the master and sealed to a 150 μm thick layer of PDMS using air plasma treatment, to form the bottom wall of the fluidic channel.
Elastomer was used to fill the fibre channels to provide refractive-index matching between the device walls and the optical fibre.20,21 In order to minimise the possibility of trapping air bubbles in front of the fibres, which would cause the light to be scattered, the elastomer was filled into the channels under vacuum and the optical fibres were dipped in elastomer before insertion to ensure the surface was fully wetted. Optical fibres were then manually inserted into the fibre channels and externally secured in place with the fibre aligned with the end of the fibre channel, leaving a small volume of liquid PDMS to allow the fibre to easily move. The system was then visually checked to ensure that there was no debris or air between the end of the fibre and the channel wall, which could scatter the light and disrupt the trapping.
Cell culture
HEK293 and U2OS cells (ATCC: HBT-96 and ATCC: CRL-1573 respectively) were cultivated at 37 °C, 5% CO2 in DMEM medium containing 10% fetal calf serum and 4 mM L-glutamine. Cells were regularly passaged every three days and kept in the exponential growth phase. Nuclear staining was performed utilizing DRAQ5 (abcam) according to the manufacturers manual. For transfection experiments the coding sequence of human Lamin A was fused to the coding sequence of eGFP and inserted into the multiple cloning site of the pIRES NEO3 vector system (Clonetech). Transfection was carried out utilising Lipofectamine LTX (Invitrogen); the procedure was performed according to the manufacturers manual. For the generation of stable expressing HEK293/U20S cell lines, stable expressing cells were selected using 1 mg mL−1 G418 (Sigma).Device operation
Microfluidic flow was regulated using custom, automated air-over-liquid pumps. The microfluidic channels of the devices were first flushed with a 3% BSA solution (Sigma-Aldrich) to reduce cell adhesion to the device walls,22 then with 1× PBS (Sigma-Aldrich) and finally with the cell samples.Control of the fibre position was achieved by inflating the triangular fibre pad features using pneumatic regulators (Marsh Bellofram) as can be seen in Fig. 1. Imaging of the trapped and rotated cell was performed using either a CCD camera (ProSilica 680, AVT Technologies) or a laser-scanning confocal (SP5, Leica microsystems).
Data analysis
Videos of the cells were processed using bespoke analysis written in MatLab. Images were aligned using the imregister function, thresholded and the position of contrast features were tracked over time. The average rotation rate was calculated by linear fitting of the angle vs. time curve. Instantaneous rotation rate was calculated for each frame by taking the numerical differentiation of the curve. The resulting data was then low-passed.Device characterisation
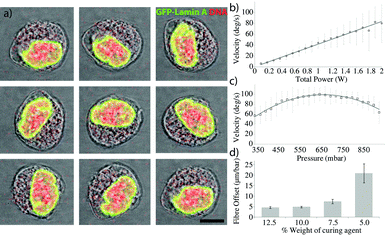
Applying pressure to one side of the membrane can provide enough force to displace the fibre (Fig. 1f–i) even at moderate pressures (~700 mbar), well below the breaking pressure of the device at ~4 bar. The pneumatic displacement of the fibre is reversible and repeatable (Fig. 1d) and can be actuated many times with little change in the resulting position. The movement of the fibre is fast (Fig. 1e) with the fibre moving to its new position in less than 100 ms. The elastic properties of the PDMS determine the size of the displacement for a given pressure. By decreasing the curing agent to elastomer ratio of the PDMS, softer devices can be produced23 which are capable of larger displacements (Fig. 2d), however the breaking pressure of the device is lower (see ESI,† Fig. S1) and the fabrication is made more difficult with the softer material. The curing temperature has also been shown to affect the material properties of the resulting devices, however we saw very little difference in the fibre displacements between devices baked at different temperatures between 45 °C and 100 °C (see ESI,† Fig. S2). Also changes in the refractive index of PDMS have been reported when the preparation method was varied. This effect is especially important, when PDMS devices are used as optical tools, for example as optofluidic lenses.17 In our case, a different refractive index will also result in changes to the rotation for a given offset. Analysis of the trap position by ray optics shows a negligible shift in the trap displacement over the range of PDMS refractive indices used.In-plane rotation
Such displacements are enough to be able to rotate a cell, as can be seen in Fig. 2. The fibres were first aligned with no additional pressure to form a stable non-rotating trap where the long axis of the cell aligns along the optical trap, before pressure was applied. Image analysis of the rotating cell provides details of the angle and rotation speed as shown in Fig. 2b.Control of the rotation rate can be achieved by modulating either the optical power or the offset between the fibres. Fig. 2 demonstrates the control over the rotation rate obtainable using the device. Fig. 2b shows the linear dependence of the rotation rate on the optical power in the trap. As expected, increasing the power linearly increases the rotation rate. Fig. 2c shows the rotation rate against fibre offset, showing the increase in rotation rate until a maximum is reached, beyond which the offset is too large and the rotation rate decreases again.
Sub-cellular dependencies
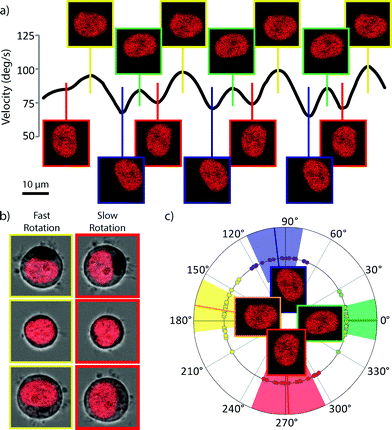
Detailed analysis of the rotation reveals that the rotation in such a system is not constant, but has periodic features. Fig. 3a is a typical rotation rate curve showing the periodic features present in optical spanner rotation. Further investigation shows that it is the orientation of the sub-cellular organelles within the optical trap which determines this substructure in the rotation rate. The optical forces acting on the trapped cell originate from differences in refractive index and so can be separated into those acting on the surface of the spheroidal cell and those acting on the organelles with a refractive index contrast to the cytoplasm. Because the distribution of refractive index inside the cell is not isotropic, as the cell rotates, the forces generated by interaction with the light beam will change, giving rise to this periodic structure. The nucleus, the largest organelle, with the large, dense packing of DNA provides most of this anisotropic force as can be seen in Fig. 3a and c where the orientation of the nuclear long axis lies parallel to the optical axis when the cell is rotating at its fastest and perpendicularly to the trap axis when the cell is rotating at its slowest. Analysis of these periodic structures in the rotation rate could provide novel insight into the size, shape and refractive index of nuclei, in a non-contact, label-free technique and may lead to rapid diagnostic tools for diseases which affect nuclear morphology such as Emery–Dreifuss dystrophy,24 Hutchinson–Gilford progeria syndrome25 as well as a wide range of other laminopathies.26,27 As well as diseases related to the nuclear proteins, it may also be possible to diagnose and characterise other serious diseases. Changes in nuclear morphology are one of the primary markers for the diagnosis of cancer28,29 and the ability to rapidly measure, at the single-cell level, properties of the nuclear morphology may provide a powerful diagnostic tool.Tomographic rotation
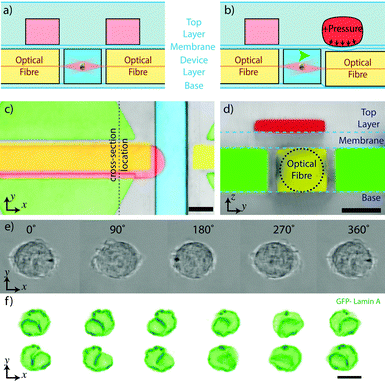
By applying pressure from above the fibre in a multi-layer device, it is possible to push the fibre downwards into the PDMS layer below the device and thus create a vertical offset between the fibres. In order to test this possibility we created a multi-layer version of the device which included additional pressure pads above one of the optical fibres, separated by a flexible, thin PDMS membrane. Such a displacement induces a rotation around the fluidic flow axis, as shown in Fig. 4e and f. In contrast to the in-plane rotation, such tomographic rotation visualises the trapped cell from different directions. Although rotation properties in this direction are harder to measure, it offers the possibility to perform tomography on the trapped cell by imaging it from a series of different angles. It can be assumed that the tomographic rotation would experience the same periodic features as seen in the in-plane rotation, resulting in an uncertainty regarding the orientation of the cell during image acquisition. However this can be compensated for in post-processing using algorithms to register the unknown angles during tomographic reconstruction.30,31Conclusions
The integration of pneumatic valves into a dual-beam optical trap allows for the reconfigurable rotation of living cells around both the optical axis and a perpendicular axis. Rotation around the optical axis allows for accurate observation of the rotation rate, which is dependent on the size and composition of the particular cell. The system allows further details of the rotation rate properties of living cells held in the trap and may offer new insights and diagnostic possibilities for rapid cell characterisation. The reconfigurable nature of the device also allows the rotator to be optimised for the particular cell type, or even the particular cell, to give the optimum rotation rate for the available optical power and account for differences in cell size between samples. The rotation rate observed in the system is sensitive to the distribution of refractive index within the cell and it may be possible to gain further insight into the current cell state by observing the details of the rotation curve. In particular, changes to the morphology and localization of the nucleus may give rise to observable changes in the substructure of the rotation pattern, giving rise to a contactless, non-destructive, marker-free analysis of the sub-cellular conditions.The extension of the system into multi-layer devices allows the optical fibres to be manipulated in two orthogonal directions and creates the possibility to rotate a living cell around an axis orthogonal to the optical axis. This allows the cell to be visualised from different directions and opens the door to using the system for single-cell tomography.
Notes and references
- A. Ashkin, J. M. Dziedzic, J. E. Bjorkholm and S. Chu, Opt. Lett., 1986, 11, 288 CrossRef CAS.
- M. Friese and T. Nieminen, Nature, 1998, 350, 348–350 CrossRef PubMed.
- S. L. Neale, M. P. MacDonald, K. Dholakia and T. F. Krauss, Nat. Mater., 2005, 4, 530–3 CrossRef PubMed.
- N. B. Simpson, K. Dholakia, L. Allen and M. J. Padgett, Opt. Lett., 1997, 22, 52 CrossRef.
- A. O'Neil, I. MacVicar, L. Allen and M. Padgett, Phys. Rev. Lett., 2002, 88, 053601 CrossRef.
- M. K. M. Kreysing, A. Fritsch, C. Dietrich, T. Kiessling, J. R. Guck and J. A. Kaes, Opt. Express, 2008, 16, 16984–16992 CrossRef CAS.
- A. Ashkin, Phys. Rev. Lett., 1970, 24, 156–159 CrossRef.
- J. Guck, R. Ananthakrishnan, T. J. Moon, C. C. Cunningham and J. Käs, Phys. Rev. Lett., 2000, 84, 5451–5454 CrossRef.
- A. E. Ekpenyong, G. Whyte, K. Chalut, S. Pagliara, F. Lautenschläger, C. Fiddler, S. Paschke, U. F. Keyser, E. R. Chilvers and J. Guck, PLoS One, 2012, 7, e45237 Search PubMed.
- J. Guck, S. Schinkinger, B. Lincoln, F. Wottawah, S. Ebert, M. Romeyke, D. Lenz, H. M. Erickson, R. Ananthakrishnan, D. Mitchell, J. Käs, S. Ulvick and C. Bilby, Biophys. J., 2005, 88, 3689–3698 CrossRef PubMed.
- A. Constable, J. Kim and J. Mervis, Opt. Lett., 1993, 18, 1867–1869 CrossRef.
- B. Lincoln, S. Schinkinger, K. Travis, F. Wottawah, S. Ebert, F. Sauer and J. Guck, Biomed. Microdevices, 2007, 9, 703–710 CrossRef PubMed.
- N. Bellini, F. Bragheri, I. Cristiani, J. Guck, R. Osellame and G. Whyte, Biomed. Opt. Express, 2012, 3, 2658–2668 CrossRef PubMed.
- J. T. Blakely, R. Gordon and D. Sinton, Lab Chip, 2008, 8, 1350–1356 RSC.
- B. J. Black and S. K. Mohanty, Opt. Lett., 2012, 37, 5030–5032 CrossRef CAS PubMed.
- A. R. Abate and D. A. Weitz, Appl. Phys. Lett., 2008, 92, 243509 CrossRef PubMed.
- W. Song, A. E. Vasdekis and D. Psaltis, Lab Chip, 2012, 12, 3590–3597 RSC.
- M. J. Kennedy, S. J. Stelick, L. G. Sayam, A. Yen, D. Erickson and C. A. Batt, Lab Chip, 2011, 11, 1138–1143 RSC.
- S. Hulme, S. Shevkoplyas and G. Whitesides, Lab Chip, 2009, 9, 79–86 RSC.
- L. M. Fidalgo, G. Whyte, B. T. Ruotolo, J. L. P. Benesch, F. Stengel, C. Abell, C. V. Robinson and W. T. S. Huck, Angew. Chem., Int. Ed., 2009, 48, 3665–3668 CrossRef CAS PubMed.
- C. L. Bliss, J. N. McMullin and C. J. Backhouse, Lab Chip, 2007, 7, 1280–1287 RSC.
- Z. Long, E. Nugent, A. Javer, P. Cicuta, B. Sclavi, M. Cosentino Lagomarsino and K. D. Dorfman, Lab Chip, 2013, 13, 947–954 RSC.
- X. Q. Brown, K. Ookawa and J. Y. Wong, Biomaterials, 2005, 26, 3123–3129 CrossRef CAS PubMed.
- A. Fidziańska and I. Hausmanowa-Petrusewicz, J. Neurol. Sci., 2003, 210, 47–51 CrossRef.
- K. N. Dahl, P. Scaffidi, M. F. Islam, A. G. Yodh, K. L. Wilson and T. Misteli, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10271–10276 CrossRef CAS PubMed.
- M. Zwerger, C. Y. Ho and J. Lammerding, Nuclear mechanics in disease, 2011, vol. 13 Search PubMed.
- H. J. Worman, L. G. Fong, A. Muchir and S. G. Young, J. Clin. Invest., 2009, 119, 1825–36 CrossRef CAS PubMed.
- D. Zink, A. H. Fischer and J. A. Nickerson, Nat. Rev. Cancer, 2004, 4, 677–87 CrossRef CAS PubMed.
- P. Dey, Diagn. Cytopathol., 2010, 38, 382–390 Search PubMed.
- O. Renaud, J. Viña, Y. Yu, C. Machu, A. Trouvé, H. Van der Voort, B. Chalmond and S. L. Shorte, Biotechnol. J., 2008, 3, 53–62 CrossRef CAS PubMed.
- B. Le Saux, B. Chalmond, Y. Yu, A. Trouvé, O. Renaud and S. L. Shorte, J. Microsc., 2009, 233, 404–416 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3lc51277k |
| This journal is © The Royal Society of Chemistry 2014 |