Description of urolithin production capacity from ellagic acid of two human intestinal Gordonibacter species
María V.
Selma
*,
David
Beltrán
,
Rocío
García-Villalba
,
Juan C.
Espín
and
Francisco A.
Tomás-Barberán
Research Group on Quality, Safety and Bioactivity of Plant Foods, Department of Food Science and Technology, CEBAS-CSIC, 30100. Campus de Espinardo, Murcia, Spain. E-mail: mvselma@cebas.csic.es; Fax: +34 968396213; Tel: +34 968396102
First published on 8th May 2014
Abstract
Ellagitannin and ellagic acid metabolism to urolithins in the gut shows a large human interindividual variability and this has been associated with differences in the colon microbiota. In the present study we describe the isolation of one urolithin-producing strain from the human faeces of a healthy volunteer and the ellagic acid transformation to different urolithin metabolites by two species of intestinal bacteria. The isolate belongs to a new species described as Gordonibacter urolithinfaciens, sp. nov. The type strain of the Gordonibacter genus, Gordonibacter pamelaeae DSM 19378T, was also demonstrated to produce urolithins. Both human intestinal bacteria grew similarly in the presence and absence of ellagic acid at 30 μM concentration. Ellagic acid catabolism and urolithin formation occurred during the stationary phase of the growth of the bacteria under anaerobic conditions. The HPLC-MS analyses showed the sequential production of pentahydroxy-urolithin (urolithin M-5), tetrahydroxy-urolithin (urolithin M-6) and trihydroxy-urolithin (urolithin C), while dihydroxy-urolithins (urolithin A and isourolithin A), and monohydroxy-urolithin (urolithin B) were not produced in pure cultures. Consequently, either other bacteria from the gut or the physiological conditions found in vivo are necessary for completing metabolism until the final urolithins (dihydroxy and monohydroxy urolithins) are produced. This is the first time that the urolithin production capacity of pure strains has been demonstrated. The identification of the urolithin-producing bacteria is a relevant outcome as urolithin implication in health (cardiovascular protection, anti-inflammatory and anticarcinogenic properties) has been supported by different bioassays and urolithins can be used in the development of functional foods and nutraceuticals. This study represents an initial work that opens interesting possibilities of describing enzymatic activities involved in urolithin production that can help in understanding both the human interindividual differences in polyphenol metabolism, the microbial pathways involved, and the role of polyphenols in human health. The presence of urolithin producing bacteria can indirectly affect the health benefits of ellagitannin consumption.
1 Introduction
Dietary polyphenols have been suggested to be responsible for some health benefits associated with the consumption of fruits, vegetables and plant-derived foods.1 Their bioavailability is however, rather limited and it is now well established that most of these compounds reach the colon where they are metabolized by the gut microbiota to produce bioactive molecules that are more readily absorbed than the original polyphenols.2 The characterization of the metabolites produced from polyphenols by gut microbiota is a current trend in polyphenol research and in human nutrition due to the implication in health as a result of their antioxidant, anti-inflammatory, antiestrogenic, cardioprotective, cancer chemopreventive and neuroprotective properties.3 Examples of these are hop (beer) isoxanthohumol which is transformed to 8-prenylnaringenin, soy isoflavones that are transformed to equol, cereal lignans that are transformed to mammalian-lignans (enterolactone, enterodiol), resveratrol which is transformed to dihydroresveratrol and ellagitannins that are transformed to urolithins.2The identification of the bacteria responsible for the polyphenol and other organic compound transformation is also a relevant objective due to the potential development of functional foods and ingredients with health benefits on individuals with low production of some of these bioactive metabolites.3 Several new bacterial species that are able to produce equol from isoflavones have been identified and these include Adlercreutzia equolifaciens,4Eggerthella strain Julong 732,5Paraeggerthella hongkongensis,6Slackia equolifaciens,7 and Slackia isoflavoniconvertens.8 The transformation of the flavanone isoxanthohumol to prenylnaringenin by Eubacterium limosum as well as the deglycosylation of flavones and isoflavones by Eubacterium cellulosolvens has been described.9,10 Among dietary non-flavonoid polyphenols, the lignane secoisolariciresinol can be transformed to phytoestrogenic metabolites such as enterodiol and enterolactone by Eggerthella lenta and Peptostreptococcus productus.11 More recently, the transformations of resveratrol into dihydroresveratrol by S. equolifaciens and A. equolifaciens as well as the hydrolysis of chlorogenic acid by Bifidobacterium animalis subsp. lactis have also been reported.12,13 Alternatively, human gut bacteria such as Enterococcus casseliflavus CP1 and Escherichia coli VL8 can metabolize glucosinolates to produce isothiocyanates as chemopreventive agents.14 Apart from these examples, human gut bacteria involved in most dietary polyphenol transformations remain unknown. This is the “gap in knowledge” intended to be filled in part by the research.
Ellagitannins are present in the berries of the genera Rubus (raspberry, blackberry, cloudberry, arctic bramble) and Fragaria (strawberry), and in pomegranates, tea, walnuts and some other nuts, oak-aged wines and muscadine grapes.2,15 Upon hydrolysis in the gut, ellagitannins undergo lactonization to produce ellagic acid (EA). It is generally accepted that the intestinal absorption of ellagitannins and EA is very low and that they reach the colon where they are catabolized into urolithins by the gut microbiota, these metabolites being much more absorbed.16–18 Urolithins are bioavailable metabolites that can reach up to micromolar concentrations in the plasma of humans.17 These metabolites exert anti-inflammatory19–24 and cancer chemopreventive effects against colon cancer.24–27 In addition, urolithins can also reach different target tissues as previously reported in both animal models (rats and pigs;24,28 and human prostate biopsies).29 The main urolithin metabolite detected in the human prostate was urolithin A (range between 0.5 and 2 ng g−1 of tissue). In the case of animal models, up to 25 urolithin metabolites have been identified in the plasma, intestine, and bile from pigs.28 In rats, urolithins and their Phase-II conjugates can also reach organs including, the liver, prostate, brain, uterus, kidney, small intestine and colon.24 The highest urolithin concentrations occur (in both pigs and rats) in the colon content (ranging from 6 to 168 μM of urolithin A, the most abundant urolithin) whereas the amount detected in the kidney and liver was much lower (0.12–0.20 μg g−1, mainly urolithin A glucuronide). Urolithins undergo an active enterohepatic circulation and do not accumulate in organs.28 In this context, chronic consumption of ellagitannin or EA containing foods can provide relevant concentrations of urolithins at both digestive (colon) and systemic levels. The poor bioavailability of ellagitannins or EA and the biological effects of urolithins in different in vitro assays suggest that urolithins can be the bioactive molecules in vivo and those responsible for the health effects observed after ellagitannin or EA intake.30
Urolithin production in the gut shows a large human interindividual variability and this has been associated with differences in the colon microbiota.31 However, in the scientific literature, nothing is found about the bacterial species responsible for urolithin production from ellagitannins, or other EA derived compounds. In the present study, we describe the metabolism of two species of human intestinal bacteria which can convert EA into different urolithins.
2 Materials and methods
2.1 Isolation of urolithin producing bacteria from human faeces
Faecal samples of a healthy woman (age 31) whose intestinal microbiota was capable of converting EA to urolithins were collected in stool cups at five moments of the year. The study was conformed to ethical guidelines outlined in the Declaration of Helsinki and its amendments. The protocol (reference 03/2011) was approved by the Clinical Ethics Committee at Reina Sofia University Hospital (Murcia, Spain) and by the Spanish National Research Council's Bioethics Committee (Madrid, Spain). The volunteer provided written informed consent. Urolithins were analyzed and detected in the urine, plasma and faeces of this volunteer after the intake of walnuts as described in one of our previous studies.31 The faeces were processed for isolation of bacteria within 2 h. The samples were placed in filter bags, diluted 1/10 (w/v) in nutrient broth (NB, Oxoid, Basingstoke, Hampshire, UK) supplemented with 0.05% L-cysteine hydrochloride (Panreac química, Barcelona, Spain) and homogenized with a stomach homogenizer. The filtered suspension was diluted further in the same broth medium containing EA (Sigma-Aldrich, St. Louis, MO, USA) at 30 μM in order to determine the metabolic activity first. A portion of the culture, having metabolic activity, was seeded on differential reinforced clostridial medium (DRCM; Merck, Darmstadt, Germany) agar to improve the growth of anaerobic bacteria. Approximately 200 colonies were picked up, inoculated into 2 mL of a broth used to grow anaerobes (anaerobe basal broth, ABB; Oxoid) and after incubation, their conversion capacity of EA to urolithins was assayed. Urolithin-producing colonies were sub-cultured, until urolithin-producing strains were isolated. The isolation procedure and incubation of plates were carried out under anaerobic conditions in an anaerobic chamber (Don Whitley Scientific Limited, Shipley, UK) with an atmosphere consisting of N2/H2/CO2 (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) at 37 °C.
10) at 37 °C.
2.2 Conversion testing of EA to urolithins
Colonies (200) isolated from human faeces described in the previous section were used in in vitro experiments to investigate their capacity to grow and produce urolithins in the presence of EA. Gordonibacter pamelaeae DSM 19378T, obtained from DSMZ culture collection, was reconstituted, maintained under the conditions specified in the DSMZ online catalogue (http://www.dsmz.de) and its urolithin production capacity was also tested because it is the only species belonging to the same genus as that of the urolithin-producing isolates of the present study. Firstly, aliquots of the intestinal bacterial strains were inoculated into 2 mL of ABB. After incubation, subcultures were inoculated into fresh ABB containing EA (Sigma-Aldrich, St. Louis, MO, USA) at 30 μM. Duplicate cultures were prepared in parallel from each bacterial strain suspension. In addition, controls were used, some without bacteria and others without EA. The test culture was incubated for 3 days at 37 °C under anaerobic conditions at 37 °C. Samples (5 mL) were collected and prepared for LC analyses of urolithins as described below.2.3 Growth curve and time-course production of urolithins by Gordonibacter urolithinfaciens and Gordonibacter pamelaeae
The stocks of G. pamelaeae DSM 19378T and the isolated strain able to produce urolithins (G. urolithinfaciens strain CEBAS 1/15P), preserved frozen, were thawed and incubated on an ABB agar plate for 4 days. A single colony was then transferred to a 5 mL ABB tube. One milliliter of diluted inoculum was transferred to ABB (50 mL) in 100 mL flasks to give an initial concentration of about 100 cfu mL−1. EA was added to the 50 mL culture to achieve a final concentration of 9–10 μM. Flasks inoculated with G. pamelaeae and G. urolithinfaciens were incubated for 8 days at 37 °C under anaerobic conditions at 37 °C. Samples (5 mL) were collected at appropriate time intervals during 8 day incubation at 37 °C, and clean-up for LC analyses of urolithins as described below. Similarly, 1 mL were collected every 2 h during 2 day incubation at 37 °C. Dilutions were made if necessary in NB and counts in ABB agar plates were performed. Triplicate growth curves were obtained.2.4 LC-UV/Vis and LC-MS/MS analyses
Samples obtained in the fermentation experiments with colonies isolated from human faeces and with G. pamelaeae were extracted and analyzed by LC-UV/Vis and LC-MS as described previously.31 Briefly, 5 mL of fermented medium was extracted with 5 mL of ethyl acetate (Labscan, Dublin, Ireland), acidified with 1.5% of formic acid (Panreac), vortexed for 2 minutes and centrifuged at 3500g for 10 min. The organic phase was separated and evaporated and the dry samples were then re-dissolved in 250 μl of methanol (Romil, Barcelona, Spain). The analyses were performed using an HPLC system (1200 Series, Agilent Technologies, Madrid, Spain) equipped with a photodiode-array detector and a single quadrupole mass spectrometer detector in series (6120 Quadrupole, Agilent Technologies, Madrid, Spain) as described previously.28 Calibration curves were obtained for EA (Sigma-Aldrich, St. Louis, MO, USA), urolithin A (chemically synthesized by Villapharma SL (Parque tecnológico de Fuente Álamo, Murcia, Spain)) and urolithin C (Dalton Pharma Services (Toronto, Canada)) with good linearity (R2 > 0.998). Urolithins A and C were quantified at 305 nm with their own standards; urolithins M-5 and M-6 with an EA calibration curve at 360 nm and EA was quantified at 360 nm with its own standard.2.5 Data modelling
Growth curves were fitted using the function of Baranyi et al.32 to estimate the main growth parameters (maximum specific growth rate, lag time of microorganisms before the onset of growth and estimated correlation coefficient, which indicates the goodness of fit of the parameters derived from experimental data). Only growth curves with at least 10 data points were used for modeling, as suggested by the authors.3 Results
3.1 Identification of urolithin producing bacteria
One bacterial strain isolated from the faecal sample, named G. urolithinfaciens strain CEBAS 1/15P, obtained from a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 106 dilution plated on DRCM agar showed the capacity to convert EA to urolithins under anaerobic conditions. We have proposed the strain as a novel species belonging to the genus Gordonibacter based on several different physiological/biochemical, chemotaxonomic and phylogenetic characteristics from more closely related species.33 The 16S rRNA gene sequence of G. urolithinfaciens has been deposited in the GenBank nucleotide sequence database under the accession number HG000667. G. urolithinfaciens has been deposited in two public culture collections with the accession number (=DSM 27213T = CCUG 64261T). We also investigated the capacity of the type strain and only species of Gordonibacter genus (G. pamelaeae DSM 19378T) to convert EA to urolithins under anaerobic conditions and also produced urolithins.
106 dilution plated on DRCM agar showed the capacity to convert EA to urolithins under anaerobic conditions. We have proposed the strain as a novel species belonging to the genus Gordonibacter based on several different physiological/biochemical, chemotaxonomic and phylogenetic characteristics from more closely related species.33 The 16S rRNA gene sequence of G. urolithinfaciens has been deposited in the GenBank nucleotide sequence database under the accession number HG000667. G. urolithinfaciens has been deposited in two public culture collections with the accession number (=DSM 27213T = CCUG 64261T). We also investigated the capacity of the type strain and only species of Gordonibacter genus (G. pamelaeae DSM 19378T) to convert EA to urolithins under anaerobic conditions and also produced urolithins.
3.2 Analysis of urolithins produced by Gordonibacter species
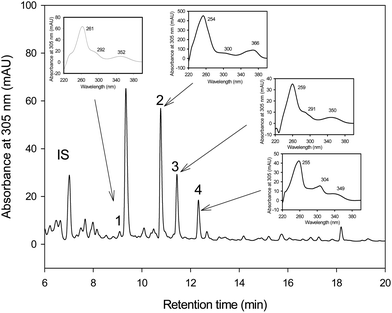
The HPLC-MS analyses showed that pentahydroxy-urolithin (urolithin M-5), tetrahydroxy-urolithin (urolithin M-6) and urolithin C were produced from EA by both strains G. urolithinfaciens and G. pamelaeae cultured separately (Fig. 1). Identification of all metabolites was carried out by direct comparison (UV spectra and MS) with pure standards and confirmed by their spectral properties and molecular mass as reported previously.313.3 In vitro catabolism of EA by Gordonibacter species
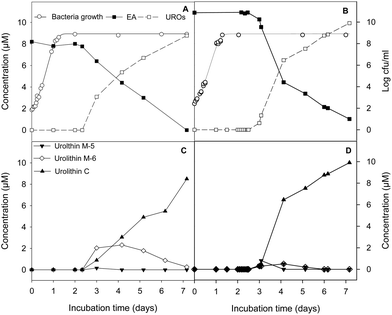
G. urolithinfaciens and G. pamelaeae separately cultured grew similarly with and without EA at 30 μM, the lag phase being 4.87 ± 0.81 h and 3.96 ± 0.56 h while the growth rate being 0.27 ± 0.03 and 0.30 ± 0.01 h−1, in the presence and absence of EA, respectively. EA catabolism and urolithin formation occurs during the stationary phase of the growth of the G. urolithinfaciens and G. pamelaeae species (Fig. 2A and B). Disappearance of EA from the medium was observed, at the same time urolithins appeared. Urolithin M-5 was only observed in the sample obtained on day 3, suggesting that this is the first metabolite produced. Urolithin M-6 reached a maximum on day four, and then decreased steadily while urolithin C is produced reaching the complete transformation of EA into urolithin C on day seven (Fig. 2C and D). Longer incubation periods did not produce further hydroxyl removals from the urolithin nucleus.4 Discussion
G. urolithinfaciens DSM 27213T, which is able to produce urolithins, was isolated from the human intestine of a healthy volunteer where it was found at high concentrations (≥107 cfu g−1 faeces). On the basis of 16S rRNA gene sequence identity, we tested the closest relatives of G. urolithinfaciens from the Coriobacteriaceae family including, P. hongkongensis HKU10T (94.3%), E. sinensis HKU14T (94.3%) and E. lenta DSM 2243T (93.0%) and they were not able to produce urolithins.33 In contrast, G. pamelaeae DSM 19378T (97.0% identity) was able to transform EA and the time-course production of urolithins is shown in the present study. G. pamelaeae DSM 19378T was previously isolated by Würdemann et al.34 from a patient with crohn's disease. However, strain ARC-1 whose 16S rRNA gene sequence (EF413639) has 100% identity with that of G. pamelaeae (AM886059) was isolated by Jin et al. from a healthy volunteer.35 This suggests that G. pamelaeae could also be found in healthy human guts as occurs with G. urolithinfaciens.Several studies have related species from the Coriobacteriaceae family with the transformation of different polyphenols to bioactive molecules such as equol, dihydroresveratrol, prenylnaringenin enterodiol and enterolactone,4–12 but not with urolithin production. In the present study, the sequential production of urolithins M-5, M-6 and C by G. urolithinfaciens and G. pamelaeae was observed when the production of these metabolites was represented against time. Urolithin M-5 is transformed into urolithin M-6 by the removal of the hydroxyl at the 9-position, which means that this dehydroxylase enzyme is more active than the one responsible for the opening of the lactone ring and sequential decarboxylation (Fig. 3). Urolithin C seems to be an end product for the catabolism of EA by G. urolithinfaciens and G. pamelaeae (Fig. 3). The studies carried out so far with human biological fluids (urine and plasma) allowed the identification of urolithin A, urolithin B, urolithin C and isourolithin A.17,31 Other metabolites such as urolithins M-5, M-6 and M-7 had been reported in rat faecal samples and other animal materials.36–38 More recently, we reported the in vitro production of urolithins M-5, M-6 and M-7, E, C and A by human gut microbiota.31 However, it is in the present study where two particular bacterial species from human gut microbiota are identified as producers of urolithins such as urolithins M-5, M-6 and C. Consequently, other bacteria from the gut are necessary for completing metabolism to urolithin A, urolithin B, and isourolithin A. Further studies should be carried out to find out if the absence of Gordonibacter species capable of transforming EA to urolithin C is the limiting factor in the production of urolithins in vivo.
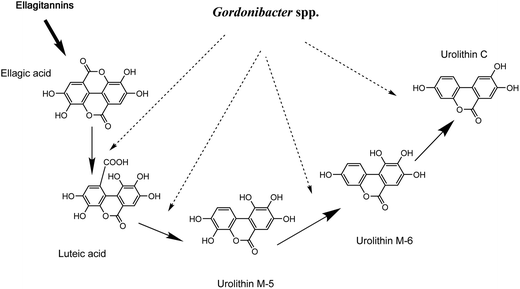 | ||
| Fig. 3 Proposed pathway for ellagitannins and ellagic acid metabolism by Gordonibacter urolithinfaciens and Gordonibacter pamelaeae leading to urolithin formation. | ||
No antimicrobial effect of EA at a concentration of 30 μM was observed during in vitro incubations of G. urolithinfaciens and G. pamelaeae as both bacteria grew similarly in the presence and absence of this substrate. First urolithins were obtained on day three of incubation and seven days were necessary for the complete transformation of EA. This indicates that urolithins are microbial secondary metabolites that do not play a role in growth because they are produced by Gordonibacter species during the stationary phase of growth. Other polyphenol transformations also occur in the stationary phase. This is the case of transformation of polyphenol lignans (dihydroxyenterodiol and dihydroxyenterolactone) to mammalian lignans (enterodiol and enterolactone) by strain ARC-1.35 Equol production from daidzein by Lactococcus garvieae also occurs after the stationary phase of growth has been achieved.39 The production of urolithins in vivo is also quite slow, and it is possible to detect urolithin metabolites in urine after the intake of ellagitannins for three or even more days.17–19 This could be associated with the slow catabolism of ellagitannins and EA by the human gut microbiota, and by additional enterohepatic recirculation. Therefore, the results found in the present work suggest that the slow metabolism by the bacteria can be a relevant factor for the persistency of urolithins in urine after EA and ellagitannin intake.
Microbial secondary metabolites including antibiotics, antitumor agents, cholesterol-lowering drugs, and others are now increasingly being used against diseases previously treated only by synthetic drugs. Production of the metabolites directly by microbial fermentation is often more convenient than using synthetic chemistry. The present challenges in microbial pharmaceutical development are the discovery of novel secondary metabolites with significant biological activities.40 The identification of bacteria responsible for the urolithin production is a relevant outcome due to the urolithin implication in health (cardiovascular protection, anti-inflammatory and anticarcinogenic properties). Urolithin producing bacteria could potentially be applied either as probiotics or in the industrial production of urolithins to develop food ingredients, drinks, dietary complements, pharmaceuticals and/or functional foods enriched in urolithins produced in a similar form as in the intestine (bacterial metabolism from dietary polyphenols). However, further studies to demonstrate the probiotical potential of these bacteria in functional foods should be carried out.
Acknowledgements
This work has been supported by the Projects CSD2007-00063 (Fun-C-Food; Consolider Ingenio 2010), Fundación Séneca de la Región de Murcia (grupo de excelencia GERM 06, 04486) and CICYT AGL2011-22447. R.G.V. holds a JAE-DOC grant from CSIC (Spain) co-financed by European Social Fund (ESF).References
- L. H. Yao, Y. M. Jiang, J. Shi, F. A. Tomás-Barberán, N. Datta, R. Singanusong and S. S. Chen, Plant Foods Hum. Nutr., 2004, 59, 113–122 CrossRef CAS PubMed.
- M. V. Selma, J. C. Espín and F. A. Tomás-Barberán, J. Agric. Food Chem., 2009, 57, 6485–6501 CrossRef CAS PubMed.
- F. A. Tomás-Barberán and C. Andrés-Lacueva, J. Agric. Food Chem., 2012, 60, 8773–8775 CrossRef PubMed.
- T. Maruo, M. Sakamoto, C. Ito, T. Toda and Y. Benno, Int. J. Syst. Evol. Microbiol., 2008, 58, 1221–1227 CrossRef CAS PubMed.
- M. Kim, S. Kim, J. Han, X. Wang, D. Song and S. Kim, Appl. Environ. Microbiol., 2009, 75, 3062–3068 CrossRef CAS PubMed.
- X. Wang, H. Hur, J. H. Lee, K. T. Kim and S. Kim, Appl. Environ. Microbiol., 2005, 71, 214–219 CrossRef CAS PubMed.
- J. S. Jin, M. Kitahara, M. Sakamoto, M. Hattori and Y. Benno, Int. J. Syst. Evol. Microbiol., 2010, 60, 1721–1724 CrossRef CAS PubMed.
- A. Matthies, M. Blaut and A. Braune, Appl. Environ. Microbiol., 2009, 75, 1740–1744 CrossRef CAS PubMed.
- S. Possemiers, S. Rabot, J. C. Espín, A. Bruneau, C. Philippe, A. González-Sarrías, A. Heyerick, F. A. Tomás-Barberán, D. D. Keukeleire and W. Verstraete, J. Nutr., 2008, 138, 1310–1316 CAS.
- A. Braune and M. Blaut, Intestinal bacterium, Appl. Environ. Microbiol., 2012, 78, 8151–8153 CrossRef CAS PubMed.
- T. Clavel, G. Henderson, C. A. Alpert, C. Philippe, L. Rigottier-Gois, J. Dore and M. Blaut, Appl. Environ. Microbiol., 2005, 71, 6077–6085 CrossRef CAS PubMed.
- L. M. Bode, D. Bunzel, M. Huch, G. S. Cho, D. Ruhland, M. Bunzel, A. Bub, C. M. Franz and S. E. Kulling, Am. J. Clin. Nutr., 2013, 97, 295–309 CrossRef CAS PubMed.
- F. A. Tomás-Barberán, R. García-Villalba, A. Quartieri, S. Raimondi, A. Amaretti, A. Leonardi and M. Rossi, Mol. Nutr. Food Res., 2013, 00, 1–10 Search PubMed.
- L. I. Vijitra, A. Narbad, C. Nueno-Palop, R. Mithen, M. Bennett and J. T. Rossiter, Mol. Nutr. Food Res., 2013, 00, 1–9 Search PubMed.
- D. A. Marshall, S. J. Stringer and J. D. Spiers, Pharm. Crops, 2012, 3, 69–77 CrossRef.
- B. Cerdá, R. Llorach, J. J. Cerón, J. C. Espín and F. A. Tomás-Barberán, Eur. J. Nutr., 2003, 42, 18–28 CrossRef PubMed.
- B. Cerdá, J. C. Espín, S. Parra, P. Martínez and F. A. Tomás-Barberán, Eur. J. Nutr., 2004, 43, 205–220 CrossRef PubMed.
- B. Cerdá, F. A. Tomás-Barberán and J. C. Espín, J. Agric. Food Chem., 2005, 53, 227–235 CrossRef PubMed.
- M. Dell'Agli, G. V. Galli, M. Bulgari, M. Basilico, S. Romeo, D. Bhattacharya, D. Taramelli and E. Bosisio, Malar. J., 2010, 9, 208 CrossRef PubMed.
- J. A. Giménez-Bastida, A. González-Sarrías, M. Larrosa, F. A. Tomás-Barberán, J. C. Espín and M. T. García-Conesa, Mol. Nutr. Food Res., 2012, 56, 784–796 Search PubMed.
- J. A. Giménez-Bastida, M. Larrosa, A. González-Sarrías, F. A. Tomás-Barberán, J. C. Espín and M. T. García-Conesa, J. Agric. Food Chem., 2012, 60, 8866–8876 CrossRef PubMed.
- A. González-Sarrías, M. Larrosa, F. A. Tomás-Barberán, P. Dolara and J. C. Espín, Br. J. Nutr., 2010, 104, 503–512 CrossRef PubMed.
- A. K. Kiss, S. Granica, M. Stolarczyk and M. F. Melzig, Food Chem., 2012, 131, 1015–1020 CrossRef CAS PubMed.
- M. Larrosa, A. González-Sarrías, M. J. Yáñez-Gascón, M. V. Selma, M. Azorín-Ortuño, S. Toti, F. A. Tomás-Barberán, P. Dolara and J. C. Espín, J. Nutr. Biochem., 2010, 21, 717–725 CrossRef CAS PubMed.
- G. Cozza, A. Gianoncelli, P. Bonvini, E. Zorzi, R. Pasquale, A. Rosolen, L. A. Pinna, F. Meggio, G. Zagotto and S. Moro, ChemMedChem, 2011, 6, 2273–2286 CrossRef CAS PubMed.
- V. Furlanetto, G. Zagotto, R. Pasquale, S. Moro and B. Gatto, J. Agric. Food Chem., 2012, 60, 9162–9170 CrossRef CAS PubMed.
- S. G. Kasimsetty, D. Bialonska, M. K. Reddy, C. Thornton, K. L. Willett and D. Ferreira, J. Agric. Food Chem., 2009, 57, 10636–10644 CrossRef CAS PubMed.
- J. C. Espín, R. González-Barrio, B. Cerdá, C. Lopez-Bote, A. I. Rey and F. A. Tomás-Barberán, J. Agric. Food Chem., 2007, 55, 10476–10485 CrossRef PubMed.
- A. González-Sarrías, J. A. Giménez-Bastida, M. T. García-Conesa, M. B. Gomez-Sanchez, N. V. Garcia-Talavera, A. Gil-Izquierdo, C. Sanchez-Alvarez, L. Fontana-Compiano, J. P. Morga-Egea, F. A. Pastor-Quirante, F. Martinez-Diaz, F. A. Tomas-Barberan and J. C. Espín, Mol. Nutr. Food Res., 2010, 54, 311–322 Search PubMed.
- J. C. Espín, M. Larrosa, M. T. García-Conesa and F. A. Tomás-Barberán, J. Evidence-Based Complementary Altern. Med., 2013, 00, 1–15 CrossRef PubMed.
- R. García-Villalba, D. Beltrán, J. C. Espín, M. V. Selma and F. A. Tomás-Barberán, J. Agric. Food Chem., 2013, 61, 8797–8806 CrossRef PubMed.
- J. Baranyi, T. A. Roberts and P. McClure, Food Microbiol., 1993, 10, 43–59 CrossRef.
- M. V. Selma, F. A. Tomás-Barberán, D. Beltrán, R. García-Villalba and J. C. Espín, Int. J. Syst. Evol. Microbiol., 2014, 64 DOI:10.1099/ijs.0.055095-0.
- D. Würdemann, B. J. Tindall, R. Pukall, H. Lünsdorf, C. Strömpl, T. Namuth, H. Nahrstedt, M. Wos-Oxley, S. Ott, S. Schreiber, K. N. Timmis and A. P. A. Oxley, Int. J. Syst. Evol. Microbiol., 2009, 59, 1405–1415 CrossRef PubMed.
- J. S. Jin, Y. F. Zhao, N. Nakamura, T. Akao, N. Kakiuchi, B. S. Min and M. Hattori, Biol. Pharm. Bull., 2007, 30, 2113–2119 CAS.
- R. González-Barrio, P. Truchado, H. Ito, J. C. Espín and F. A. Tomás-Barberán, J. Agric. Food Chem., 2011, 59, 1152–1162 CrossRef PubMed.
- R. González-Barrio, P. Truchado, R. García-Villalba, G. Hervás, P. Frutos, J. C. Espín and F. A. and Tomás-Barberán, J. Agric. Food Chem., 2012, 60, 3068–3077 CrossRef PubMed.
- H. Ito, A. Iguchi and T. Hatano, J. Agric. Food Chem., 2008, 56, 393–400 CrossRef CAS PubMed.
- S. Uchiyama, T. Ueno and T. Suzuki, J. Intestinal Microbiol., 2007, 21, 217–220 Search PubMed.
- L. Z. Wu and B. Hong, Yaoxue Xuebao, 2013, 48, 155–160 CAS.
| This journal is © The Royal Society of Chemistry 2014 |