Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein A-conjugated iron oxide nanoparticles for separation of Vibrio cholerae from water samples
Tran Quang
Huy
a,
Pham
Van Chung
a,
Nguyen Thanh
Thuy
a,
Cristina
Blanco-Andujar
bc and
Nguyễn Thị Kim
Thanh
*bc
aNational Institute of Hygiene and Epidemiology, 1 Yersin Street, Hai Ba Trung District, Hanoi, Vietnam
bDepartment of Physics and Astronomy, University College London, London, WC1E 6BT, UK
cUCL Healthcare Biomagnetic and Nanomaterials Laboratories, 21 Albemarle Street, London W1S 4BS, UK. E-mail: ntk.thanh@ucl.ac.uk; Tel: +44 (0) 207 4916509
First published on 5th August 2014
Abstract
Pathogen separation is of great significance for precise detection and prevention of disease outbreaks. For the first time, protein A conjugated with chitosan-coated iron oxide nanoparticles was prepared for pathogen separation at low concentrations from liquid samples. Vibrio cholerae O1 (VO1) bacteria were used for testing the effectiveness of this conjugate. Transmission electron microscopy (TEM) was used to confirm the presence of captured VO1. The results showed that, after binding with a specific antibody, the conjugate allows separation of VO1 bacteria from water samples at a concentration as low as 10 cfu mL−1. Moreover, the conjugate can be used in parallel with conventional or modern diagnostic tests for quick and accurate detection of pathogens.
Introduction
Recently, the world has witnessed an increased number of emerging and re-emerging infectious disease outbreaks caused by viruses and bacteria, such as ebola, severe acute respiratory syndrome-associated coronavirus (SARS-CoV), avian influenza (A/H5N1, A/H7N9), foot–hand–mouth diseases, measles, diarrhea and cholera. These outbreaks not only have public health consequences, but also socio-economic tolls.1 Rapid and accurate detection of pathogens is of paramount important for the control of these infectious diseases and prevention of their spread to the community.In particular, cholera outbreak caused by V. cholerae is one of the continuous risks to public health and socio-economic development in many developing countries.2V. cholerae is a Gram-negative, comma-shaped bacterium, which causes abrupt onset of watery diarrhoea, occasionally vomiting, renal failure, coma and even death. There are more than 200 known V. cholerae serogroups, but only the O1 and O139 strains are pathogenic and cause epidemics. V. cholerae bacteria can also live for up to one year in an aquatic environment due to a protective bio-film.2 It is not easy to detect V. cholerae using conventional diagnostic methods, because it is non-culturable and generally found in low levels in water.3 Several techniques have been developed for detection of V. cholerae in environmental water including immunomagnetic beads, nucleic acid-based methods (such as PCR and DNA probe hybridization) or fluorescent labeling techniques.4 Immunomagnetic bead detection offers good sensitivity, but has the possibility of false positives. Nucleic acid-based methods require biological agents such as primers, standardised bio-safe laboratories, and can only detect nucleic acids rather than live bacterial cells. The fluorescent labeling technique is preferred for the detection of V. cholerae in environmental water samples, but filtering is necessary to concentrate the bacteria from a large volume of water.
In recent years, many studies have shown the potential applications of magnetic nanoparticles in biomedicine such as imaging, targeted drug delivery and therapy,5 but only a few studies have used magnetic nanoparticles for the detection of infectious disease pathogens so far.6 The main challenges in the preparation of magnetic nanoparticles for pathogen detection are surface modification and biofunctionalisation, particularly the immobilisation of biomolecules on the surface of nanoparticles as probes.7
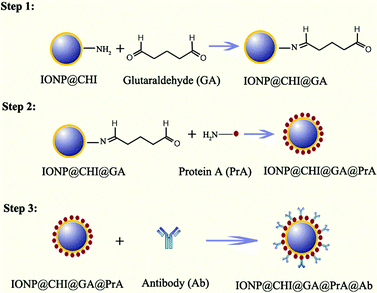
In this study, a conjugate of protein A and chitosan-coated magnetic nanoparticles was prepared for separation of V. cholerae bacteria at low concentrations from water samples. Chitosan, a polysaccharide obtained from the N-deacetylation of chitin, is one of the most abundant polysaccharides in nature. It has been used for a wide range of biological applications, thanks to its biocompatibility and biodegradability.8 Its use for the functionalisation of nanoparticles not only allows the incorporation of amine groups on the surface of the nanoparticles, but also adds a positive charge to the system, which has been shown to be preferred by some cell lines.9 On the other hand, protein A is well known as a molecule which binds with high affinity to the Fc region of IgG antibodies and orientates the binding sites of the antibody molecule outwards from the surface of nanoparticles for capturing antigens.10
Herein, chitosan-coated iron oxide nanoparticles (IONPs) were first conjugated with protein A and then the conjugate was incubated with the specific IgG antibody against V. cholerae strain just before testing. The V. cholerae bacteria could be easily magnetically separated and collected by binding to iron oxide nanoparticles.
Experimental procedures
Materials
Protein A (PrA, extracellular from Staphylococcus aureus, lyophilized, ≥90% protein basis), glutaraldehyde solution (GA, grade I, 25%), paraformaldehyde (reagent grade), sodium cacodylate trihydrate ((CH3)2AsO2Na·3H2O, ≥98%), and chitosan (C6H11NO4)n, Mw ∼ 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 were obtained from Sigma Aldrich, UK. V. cholerae O1 bacteria, anti-V. cholerae O1 polyclonal antibody, and Escherichia coli (E. coli) were provided by Professor Nguyen Binh Minh and Dr Tran Huy Hoang at Department of Bacteriology, National Institute of Hygiene and Epidemiology (NIHE), Hanoi. Bacterial strains were inoculated at the laboratory of enteric bacteria, certified by ISO 15189: 2012, and all proper and standard biosafety procedure were followed when handling them. All chemicals used were of analytical grade.
000 g mol−1 were obtained from Sigma Aldrich, UK. V. cholerae O1 bacteria, anti-V. cholerae O1 polyclonal antibody, and Escherichia coli (E. coli) were provided by Professor Nguyen Binh Minh and Dr Tran Huy Hoang at Department of Bacteriology, National Institute of Hygiene and Epidemiology (NIHE), Hanoi. Bacterial strains were inoculated at the laboratory of enteric bacteria, certified by ISO 15189: 2012, and all proper and standard biosafety procedure were followed when handling them. All chemicals used were of analytical grade.
Synthesis and characterization of chitosan-coated IONPs
Chitosan-coated IOPNs were synthesized by a chemical method,11 and then characterized by transmission electron microscope (TEM), dynamic light scattering (DLS), zeta-potential (ζ-potential), and attenuated total reflectance-Fourier transformed infrared spectroscopy (ATR-FTIR).Bio-conjugation of protein A to IONPs
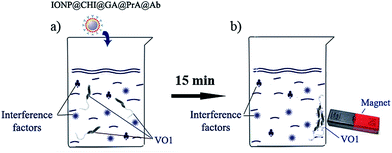
Protein A was conjugated to the surface of chitosan-coated IONPs via a cross-linkage with GA (Fig. 1). The two functional aldehyde groups of GA can easily co-bind with amine groups of the chitosan layer on the surface of IONPs and those of PrA. Firstly, the solution of chitosan-coated IONP (IONP@CHI) was diluted in distilled water to a concentration of 10 μg mL−1, and the solution pH adjusted to pH 6.5. Next, 5% GA was added, incubated for 45 min at room temperature (RT), and washed with phosphate-buffered saline, PBS (0.01 M, pH 7.4) three times to remove residual GA (Fig. 1, step 1). 2 μg mL−1 of protein A was then added to the solution of IONPs and incubated overnight at 4 °C (Fig. 1, step 2). After washing twice with PBS, the non-specific binding sites on the surfaces of the nanoparticles were blocked with 0.5% bovine serum albumin (BSA) in PBS for 20 min at RT. Finally, the conjugate was washed with PBS twice, and then re-suspended in 0.5 mL PBS and kept at 4 °C for testing. Due to the two carbonyl functional groups of GA, as well as multiple amine groups of protein A, cross-linking could occur between IONP@CHI leading to aggregation or precipitation; however, these agglomerates can easily be discarded. This phenomenon can easily be checked by naked eye or using DLS to observe the increase in size of particle aggregates in the solution. Also, round-shaped IONPs may help to minimize the effect of aggregation, because there might be some free spaces remaining on the surface of each IONP@CHI for binding to target molecules, even with several particles stuck together. In our study, the preparation of the conjugate was done in triplicate and no flocculation was detected. The conjugate can then be incubated with specific IgG antibodies against pathogens in advance before separation (Fig. 1, step 3).To separate V. cholerae bacteria from a water sample, 0.5 mL of IONP@CHI@GA@PrA was incubated in advance with polyclonal antibody against V. cholerae O1 at a concentration of 2 μg mL−1, for 45 min at RT. In this step, IgG antibody molecules would bind with high affinity to protein A–IONPs (IONP@CHI@GA@PrA). A magnet was used to collect the product of IgG antibody conjugated IONPs (IONP@CHI@GA@PrA@Ab) on the wall of the tube, washed two times with PBS to remove unbound antibodies, and then re-suspended in 0.5 mL of PBS (Fig. 2b). This procedure can be carried out just before separation and detection at RT in the laboratory or prepared in advance and then kept at 4 °C for further detection on-site.
Investigation of bacterial cells before testing
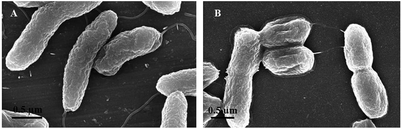
Scanning electron microscopy (SEM) (S-4800, Hitachi) was used to check the presence and morphology of bacterial cells in the samples before testing with the conjugate of IONPs and protein A. At a concentration of 106 cfu mL−1, the samples of V. cholerae and E. coli were fixed in 2% paraformaldehyde/2.5% GA for 1 h, then washed with 0.1 M cacodylate buffer before post-fixing with 2% aqueous OsO4 in 0.2 M cacodylate buffer for 2 h. After washing with 0.1 M cacodylate buffer again, samples were dehydrated with increasing ethanol solutions (from 30% to 100% with 10% increments), dried using a critical point dryer (Emitech, K850, Quorum Technologies), and coated with Pt–Pd using ion sputter before SEM imaging.Separation of V. cholerae O1 from water samples
V. cholerae O1 bacteria were dispersed in 0.01 M PBS (pH 7.4) with an initial concentration of 103 cfu mL−1. Ten-fold serial dilutions in water were then prepared in test tubes to a final volume of 5 mL from the original concentration of bacteria. A 50 μL aliquot of the complex of IONP@CHI@GA@PrA@Ab was added to the test tube (with the ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 v/v), and mixed at RT for about 15 min (Fig. 2a). The IONP-bound VO1 bacteria were collected on the wall of the tube using a bar magnet, and most of the supernatant was removed with a pipette. The conjugate was then re-suspended with the remaining water in the tube (Fig. 2b). A drop of this solution was transferred to a carbon coated copper grid to determine the presence of VO1 bacteria under a TEM (JEM1010, JEOL), operating at 80 kV.
100 v/v), and mixed at RT for about 15 min (Fig. 2a). The IONP-bound VO1 bacteria were collected on the wall of the tube using a bar magnet, and most of the supernatant was removed with a pipette. The conjugate was then re-suspended with the remaining water in the tube (Fig. 2b). A drop of this solution was transferred to a carbon coated copper grid to determine the presence of VO1 bacteria under a TEM (JEM1010, JEOL), operating at 80 kV.
Results and discussion
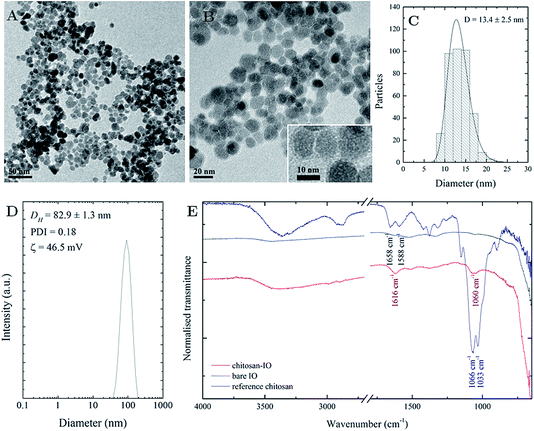
The synthesised nanoparticles are spheroidal, as observed by TEM (Fig. 3A and B), with an average particle size of 13.4 ± 2.5 nm, as calculated from the measurement of 400 particles (Fig. 3C). The hydrodynamic size of the chitosan-IONPs, as observed by DLS, is 82.9 ± 13 nm. The measured ζ-potential of 45 mV indicated the presence of chitosan around the IONPs (Fig. 3D). From ATR-FTIR spectra (Fig. 3E), we could observe the NH2 bending bands appearing at 1658 and 1588 cm−1 for free chitosan, merging to the 1616 cm−1 band after coating. The peak at 1378 cm−1 for the νCO vibration of the hydroxyl group, the C–N stretching at 1153 cm−1 and the stretching of the aliphatic CH2 group around 2900 cm−1 were found in the coated samples. The coordination of chitosan to the surface of the nanoparticles occurs via charge affinity as the reaction occurs at acidic pH where chitosan presents a high positive charge density and thus high affinity for the negatively charged nanoparticle surface. Nonetheless, not all the amine groups are bound to the nanoparticles, as surface charge was 46.5 mV, due to the presence of positively charged groups. As a result, the obtained system is highly stable due to steric repulsion of the nanoparticles provided by the polymer backbone and the surface charge obtained from the amine groups.Before testing with the complex of IONP@CHI@GA@PrA@Ab, the solution containing bacterial strains was checked by SEM. SEM images revealed the presence of V. cholerae and E. coli bacterial cells in the samples. V. cholerae bacteria were found to be comma-shaped, measuring 0.3 μm in diameter and 1.3 μm in length and have a flagellum at one cell pole (Fig. 4A). E. coli bacterial cells are rod-shape, measuring 0.5 μm in diameter and 1.2 μm in length (Fig. 4B).
To test the effectiveness of the conjugate, IONP@CHI@GA@PrA was incubated with anti-V. cholerae O1 polyclonal antibody before being dropped into the tubes containing V. cholerae bacteria.
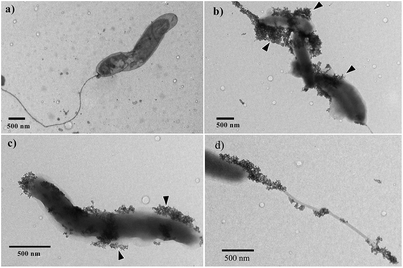
The TEM images show that before the test with the complex of IONP@CHI@GA@PrA@Ab, V. cholerae bacterial cell could be clearly seen without the presence of IONPs (Fig. 5a). After using IONP@CHI@GA@PrA@Ab to separate the bacterial cells from water, V. cholerae cells were found to aggregate on the grid and were bound by IONPs (Fig. 5b). In this study, we used polyclonal IgG antibodies specific to V. cholerae O1 bacteria, so IONPs would specifically bind around the bacterial cell wall (Fig. 5c) as well as the bacterial flagellum (Fig. 5d). All V. cholerae O1 bacterial cells found on the grids were covered with IONPs, which demonstrates that the capture efficiency of IONP@CHI@GA@PrA@Ab to V. cholerae O1 bacterial cells is up to 100%. This may be the result of the diffusion capacity of IONPs in solution, as well as the specificity of the IONP@CHI@GA@PrA@Ab complex to pathogens. By serial dilution of bacterial cultures, the results also showed that VO1 bacteria could be detected at a level as low as 10 cfu mL−1, as indicated by the IONPs presence (repeated three times). Interestingly, most of IONPs were found sticking around bacterial cells, with only a few small clusters of IONPs found on the TEM grids. After separation, the supernatant was also checked by TEM for the presence of bacterial cells remaining there, but none were found on the grids. This confirmed that most of the bacterial cells adhered to IONPs and were attracted by a magnet towards the wall of the tube.
For positive controls, the solutions containing IONP@CHI, and IONP@CHI@GA@PrA were incubated with V. cholerae bacteria diluted in water from an initial concentration of 103 cfu mL−1. The same procedures used for testing the IONP@CHI@GA@PrA@Ab complex were followed. A significant number of nanoparticles were found clustered on the grid, but no V. cholerae bacterial cells were found (data not shown).
To demonstrate the specificity of the complex to the pathogens, as negative controls E. coli bacteria were also prepared in 5 mL of PBS with an initial concentration of 103 cfu mL−1. Ten-fold serial dilutions in water were also prepared in 5 mL for testing. The complex of IONP@CHI@GA@PrA@Ab (using anti-V. cholerae O1 polyclonal IgG antibody) was then added to the tubes and mixed for 15 min at RT. The same procedure used for VO1 was followed; however, TEM images did not show the presence of E. coli bacterial cells on the grids.
After incubation of the protein A-conjugated IONPs with anti-V. cholerae O1 polyclonal IgG antibody, the separation ability of VO1 bacteria from water samples could be found at concentrations as low as 10 cfu mL−1, as confirmed by TEM. More importantly, the detection level was found to be better than with conventional diagnostic techniques, where bacterial pathogens at concentrations lower than 102 cfu mL−1 cannot be detected without inoculation, especially non-culturable bacteria that present their own bio-films such as V. cholerae.12 Following isolation and culture, a direct fluorescent antibody (DFA) test is known to be an effective method for direct detection of V. cholerae. Hasan et al.13 applied DFA to detect V. cholerae O139 in water samples, and the sensitivity of V. cholerae O139 detection reached 150 cfu mL−1, which was 15-fold higher than the detection levels reported here. In this work, the complex serves as a convenient and versatile agent for separation of different pathogens on-site or in the laboratory. It can be used for selective enrichment of pathogens for early diagnostic tests and preliminary pathogenic screening for a large number of patients who were infected in a short period of time during outbreaks. Noticeably, before testing, the samples and the complex should be adjusted to pH 7.0–7.4 to ensure their bioactivity, but all procedures for separation and detection can be performed at RT.
There are several reports on the preparation of magnetic nanoparticles for pathogen detection. Gu et al.14 showed that biofunctionalised magnetic nanoparticles could capture vancomycin-resistant Enterococci and other Gram-positive bacteria at concentrations of ∼10 cfu mL−1 within 1 h. In this study, the authors proposed the use of either optical microscopy or SEM to confirm the presence of “magnetized” bacterial cells. However, to the best of our knowledge, it is very difficult to use optical microscopy to identify the differences between bacterial strains, due to the limitation of optical resolution. On the other hand, SEM is time-consuming, as it requires complex sample preparation by dehydration and critical point drying steps before imaging. Gao et al.15 also prepared vancomycin-functionalised FePt nanoparticles, but in combination with a vancomycin-conjugated fluorescent probe to detect bacteria in human blood with a response time of 2 h. Cheng et al.16 combined biofunctional magnetic nanoparticles and adenosine triphosphate (ATP) bioluminescence to detect E. coli inoculated into pasteurized milk with a detection limit of 20 cfu mL−1 and a detection time of about 1 h. Recently, the development of a magneto-DNA platform based on magnetic nanoparticles that could detect a panel of 13 bacterial species within 2 h was reported.17 The reported literature demonstrates the use of biofunctionalised magnetic nanoparticles for detection of different pathogens with high sensitivity; however, they all present some intrinsic limitations. Firstly, biofunctionalised nanoparticles need to be prepared in advance against specific pathogens. Moreover, modern complex facilities and equipment are needed, and response time is still longer than 1 h. It is important to consider that for magnetic nanoparticles conjugated directly to antibodies, bio-activity may be reduced due to manipulation or with time.
In this work, the conjugate of protein A and IONPs overcomes some of the aforementioned limitations. Firstly, it serves as a convenient platform for enrichment and separation of various pathogens in different liquid samples, after just a prior incubation with their specific IgG antibodies. Moreover, this conjugate can be used as a smart material to selectively concentrate pathogens from a large volume of water, which may indirectly improve the sensitivity of other diagnostic techniques. Finally, it can also be applied for purifying water infected by pathogens.
Concluding remarks
Our preliminary study showed that the conjugate of protein A and IONPs was successfully prepared and tested for separation of V. cholerae bacteria at low concentrations from water samples in the laboratory. The results showed by TEM observation that the conjugate could easily separate V. cholerae bacteria from water samples at concentrations as low as 10 cfu mL−1. The selective separation of specific pathogens from liquid samples is achieved by incubation of the conjugate with a specific monoclonal or polyclonal antibody. The pathogens separated from a large volume of liquid samples can then be detected simply by conventional diagnostic methods or immunochromatographic strip tests on-site. More importantly, it has the potential to be used for the separation and detection of even unknown pathogens in clinical samples collected from emerging outbreaks. This conjugate would be incubated in advance with serum collected from patients presenting typical symptoms of the outbreak to perform the immunolabelling reaction. This procedure may allow the preliminary recognition of pathogens by studying their morphology using SEM or TEM. This approach may help other advanced techniques to confirm the causing agent of the outbreak. Further investigation is planned to optimize the bio-activity and stability of the conjugate over time.Acknowledgements
Nguyễn T. K. Thanh thanks the Royal Society for her Royal Society University Research Fellowship and Foreign Common Wealth Office for a travel grant.References
- E. K. Jones, G. N. Patel, A. M. Levy, A. Storeygard, D. Balk, L. J. Gittleman and P. Daszak, Nature, 2008, 451, 990 CrossRef CAS PubMed; J. R. Koker, M. B. Hunter, W. J. Rudge, M. Liverani and P. Hanvoravongchai, Lancet, 2011, 377, 599 CrossRef; A. S. Fauci and D. M. Morens, N. Engl. J. Med., 2012, 366, 454 CrossRef PubMed.
- S. Chaiyanan, A. Huq, T. Maugel and R. R. Colwell, Syst. Appl. Microbiol., 2001, 24, 331 CrossRef CAS PubMed; P. R. Mason, J. Infect. Dev. Ctries., 2009, 3, 148 Search PubMed; N. Sithivong, T. Morita-Ishihara, A. Vongdouangchanh, T. Phouthavane and K. Chomlasak, et al. , Emerging Infect. Dis., 2011, 17, 2060 Search PubMed; D. D. Anh, A. L. Lopez, V. D. Thiem, S. L. Grahek and T. N. Duong, et al. , PLoS Neglected Trop. Dis., 2011, 5, e1006 Search PubMed; A. M. Devault, G. B. Golding, N. Waglechner, J. M. Enk and M. Kuch, N. Engl. J. Med., 2014, 370, 334 CrossRef PubMed.
- S. Banoo, D. Bell, P. Bossuyt, A. Herring and D. Mabey, et al. , Nat. Rev. Microbiol., 2010, 8(suppl. 12), S17 Search PubMed; F. S. Nolte, Clin. Infect. Dis., 2008, 47(suppl. 3), S123 CrossRef CAS PubMed; P. Yager, G. J. Domingo and J. Gerdes, Annu. Rev. Biomed. Eng., 2008, 10, 107 CrossRef PubMed; A. van Belkum, N. H. Renders, S. Smith, S. E. Overbeek and H. A. Verbrugh, FEMS Immunol. Med. Microbiol., 2000, 27, 51 CrossRef.
- K. H. Seo, R. E. Brackett and J. F. Frank, Int. J. Food Microbiol., 1998, 44, 115 CrossRef CAS; S. Chakraborty, J. Khanam, Y. Takeda and G. B. Nair, Trans. R. Soc. Trop. Med. Hyg., 1999, 93, 527 CrossRef; E. K. Lipp, I. N. Rivera, A. I. Gil, E. M. Espeland, N. Choopun, V. R. Louis, E. Russek-Cohen, A. Huq and R. R. Colwell, Appl. Environ. Microbiol., 2003, 69, 3676 CrossRef; D. Wang, X. Xu, X. Deng, C. Cheng and B. Li, et al. , Appl. Environ. Microbiol., 2010, 76, 5520 CrossRef PubMed.
- Q. A. Pankhurst, N. T. K. Thanh, S. K. Jones and J. Dobson, J. Phys. D: Appl. Phys., 2009, 42, 224001 CrossRef CAS; I. Robinson, L. D. Tung, S. Maenosono, C. Walti and N. T. K. Thanh, Nanoscale, 2010, 2, 2624 RSC; O. Veiseh, J. W. Gunn and M. Zhang, Adv. Drug Delivery Rev., 2010, 62, 284 CrossRef PubMed; X. Meng, H. Seton, L. T. Lu, I. Prior, N. T. K. Thanh and B. Song, Nanoscale, 2011, 3, 977 RSC; Y. Wang, C. Blanco-Andujar, Z. Zhi, P. W. So, N. T. K. Thanh and J. C. Pickup, Chem. Commun., 2013, 49, 7255 RSC; R. Hachani, M. Lowdell, M. Birchall and N. T. K. Thanh, Nanoscale, 2013, 5, 11362 RSC; A. Hervault and N. T. K. Thanh, Nanoscale, 2014 10.1039/c4nr03482a.
- N. Tran and T. J. Webster, J. Mater. Chem., 2010, 20, 8760 RSC; P. Tallury, A. Malhotra, L. M. Byrne and S. Santra, Adv. Drug Delivery Rev., 2010, 62, 424 CrossRef CAS PubMed; T. S. Hauck, S. Giri, Y. Gao and W. C. W. Chan, Adv. Drug Delivery Rev., 2010, 62, 438 CrossRef PubMed.
- N. T. K. Thanh and L. A. W. Green, Nano Today, 2010, 5, 213 CrossRef CAS PubMed; T. D. Schladt, K. Schneider, H. Schild and W. Tremel, Dalton Trans., 2011, 40, 6315 RSC; R. A. Sperling and W. J. Parak, Philos. Trans. R. Soc. London, Ser. A, 2010, 368, 1333 CrossRef PubMed; H. Xu, Z. P. Aguilar, L. Yang, M. Kuang, H. Duan, Y. Xiong, H. Wei and A. Wang, Biomaterials, 2011, 32, 9758 CrossRef PubMed; A. K. Gupta and M. Gupta, Biomaterials, 2005, 26, 3995 CrossRef PubMed.
- M. N. V. Ravi Kumar, R. A. A. Muzzarelli, C. Muzzarelli, H. Sashiwa and A. J. Domb, Chem. Rev., 2004, 104, 6017 CrossRef PubMed; Y. Yi, L. Q. Wu, W. E. Bentley, R. Ghodssi, G. W. Rubloff, J. N. Culver and G. F. Payne, Biomacromolecules, 2005, 6, 2881 CrossRef PubMed.
- E. Frohlich, Int. J. Nanomed., 2012, 7, 5577 CrossRef CAS PubMed; Y. Ge, Y. Zhang, J. Xia, M. Ma, S. He, F. Nie and N. Gu, Colloids Surf., B, 2009, 73, 294 CrossRef PubMed; Z. G. Yue, W. Wei, P. P. Lv, H. Yue, L. Y. Wang, Z. G. Su and G. H. Ma, Biomacromolecules, 2011, 12, 2440 CrossRef PubMed.
- I. Takeshi, H. Yumehiro, N. Ken-ichi, I. Yoshiaki, T. Keigo, A. Satoka, H. Ryuichi and K. Akio, Anal. Biochem., 2009, 385, 132 CrossRef CAS PubMed; A. K. Minkstimiene, A. Ramanaviciene, J. Kirlyte and A. Ramanavicius, Anal. Chem., 2010, 82, 6401 CrossRef PubMed; G. Shen, C. Cai, K. Wang and J. Lu, Anal. Biochem., 2011, 409, 22 CrossRef PubMed; T. Q. Huy, N. T. H. Hanh, P. V. Chung, D. D. Anh, P. T. Nga and M. A. Tuan, Appl. Surf. Sci., 2011, 257, 7090 CrossRef PubMed.
- C. Blanco-Andujar, Sodium carbonate mediated synthesis of iron oxide nanoparticles to improve magnetic hyperthermia efficiency and induce apoptosis, PhD thesis, University College London, London, UK, 2014.
- M. Alam, M. Sultana, G. B. Nair, A. K. Siddique, N. A. Hasan, R. B. Sack, D. A. Sack, K. U. Ahmed, A. Sadique, H. Watanabe, C. J. Grim, A. Huq and R. R. Colwell, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17801 CrossRef CAS PubMed; A. Safa, G. B. Nair and R. Y. C. Kong, Trends Microbiol., 2010, 18, 46 CrossRef PubMed; K. Bharati and S. K. Bhattacharya, Curr. Top. Microbiol. Immunol., 2014, 379, 87–116 Search PubMed.
- J. A. Hasan, A. Huq, G. B. Nair, S. Garg, A. K. Mukhopadhyay, L. Loomis, D. Bernstei and R. R. Colwell, J. Clin. Microbiol., 1995, 33, 2935 CAS.
- H. W. Gu, P. L. Ho, K. W. T. Tsang, L. Wang and B. Xu, J. Am. Chem. Soc., 2003, 125, 15702 CrossRef CAS PubMed.
- J. Gao, L. Li, P.-L. Ho, G. C. Mak, H. Gu and B. Xu, Adv. Mater., 2006, 18, 3145 CrossRef CAS.
- Y. Cheng, Y. Liu, J. Huang, K. Li, W. Zhang, Y. Xian and L. Jin, Talanta, 2009, 77, 1332 CrossRef CAS PubMed.
- H. J. Chung, C. M. Castro, H. Im, H. Lee and R. Weissleder, Nat. Nanotechnol., 2013, 8, 369 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |