Metabolomic effects in HepG2 cells exposed to four TiO2 and two CeO2 nanomaterials†‡
Kirk T.
Kitchin
*a,
Eric
Grulke
b,
Brian L.
Robinette
a and
Benjamin T.
Castellon
 a
a
aIntegrated Systems Toxicology Division, National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, 109 Alexander Drive, Mail Drop B105-03, Research Triangle Park, NC 27711, USA. E-mail: kitchin.kirk@epa.gov; robinette.brian@epa.gov; ben.castellon@gmail.com; Fax: +1 919 541 0694; Tel: +1 919 541 7502
bUniversity of Kentucky, Department of Chemical & Materials Engineering, Lexington, KY 40506, USA. E-mail: eric.grulke@uky.edu
First published on 13th August 2014
Abstract
It is difficult to evaluate nanomaterials potential toxicity and to make science-based societal choices. To better assess potential hepatotoxicity issues, human liver HepG2 cells were exposed to four TiO2 and two CeO2 nanomaterials at 30 ug ml−1 for three days with dry mean primary particle sizes ranging from 8 to 142 nm. Two nanomaterials were also run at 3 ug ml−1. A metabolomics study was then performed using three mass spectroscopy dependent platforms (LC and GC). Five of the six nanomaterials strongly reduced glutathione concentration. The two strongest effects were from exposures to a TiO2 (59 nm) and a CeO2 (8 nm), both from NanoAmor. The decreases in the GSH system were observed in (a) GSH precursors (glutamate and cysteine), (b) GSH itself and (c) GSH metabolites (the gamma-glutamyl condensation products with glutamate, glutamine, alanine, valine and also 5-oxoproline and cysteine–GSH). The glutathione decreases were the largest decreases seen among the 265 biochemical metabolites determined and is consistent with nanomaterials acting via an oxidative stress mode of action. CeO2, but not TiO2, increased asymmetric dimethylarginine concentration and thus possible decreases in iNOS activity and NO concentration could result. One CeO2 (8 nm from NanoAmor) increased concentrations of many lipids, particularly fatty acids. Similar statistically significant elevations were seen in several other classes of lipids (lysolipid, monoacylglycerol, diacylglycerol and sphingolipid) but not in all classes of lipids (glycerolipid, carnitine and fatty acid dicarboxylate). None of the other 5 nanomaterials had this lipid effect. Thus, metabolomic analysis of nanomaterial treated HepG2 cells revealed several previously unknown biochemical effects.
Nano impactThis metabolomics-based study of the four TiO2 and two CeO2 nanomaterials in HepG2 cells has shown substantial evidence for the oxidative stress theory of nanomaterial toxicity in the depletion of glutathione, glutathione precursors and glutathione metabolites. Nanomaterial exposures may also perturb the nitric oxide physiology of organisms by increasing asymmetric dimethylarginine (ADMA) concentration, an iNOS inhibitor. Many lipids were increased by one CeO2 nanomaterial exposure. |
Introduction
It is difficult to evaluate nanomaterials to determine their degree and type of toxicity.1–3 For nanomaterials a major determinant of their biological action may be their surface properties, particularly their ability to donate or accept electrons4 and/or to generate free radicals and to form reactive oxygen species (ROS).5 Thus the oxidative stress mode of action has frequently been prominently mentioned among the major possible modes of action of nanomaterials.6,7After the development of the genomics and proteomics technologies, metabolomics has more recently been developed and used as an analytical tool in general biological research8–10 and toxicological studies.11 The analytical platforms most commonly used to determine cellular metabolites are LC-MS/MS, GC-MS and NMR. This study partnered with the Metabolon corporation which used three analytical platforms to determine as many cellular metabolites as possible – liquid chromatography–tandem mass spectroscopy with positive ionization (LC-MS/MS+), liquid chromatography–tandem mass spectroscopy with negative ionization (LC-MS/MS-) and gas chromatography mass spectroscopy (GC-MS). With metabolomics tools such as these, cellular biochemicals from many major different groups can be determined – amino acids, peptides, carbohydrates, energy molecules, lipids, nucleotides, cofactors and vitamins and xenobiotics. For example metabolomics methods can determine many members of the amino acid superfamily such as glycine, serine, threonine, alanine, aspartate, glutamate, histidine, lysine, phenylalanine, tyrosine, tryptophan, valine, leucine, isoleucine, cysteine, methionine, S-Adenylmethionine (SAM), taurine, urea cycle, arginine, proline, creatine, butanoate, polyamine and glutathione.
To assess potential hepatotoxicity issues from oral and/or inhalation routes, human liver HepG2 cells were used. In the structure-activity and dose–response studies presented here, four TiO2 (C from Alfa Aesar of 25 nm, A from Degussa of 31 nm, B from NanoAmor of 59 nm and I from Acros of 142 nm dry size) and two CeO2 (L from Alfa Aesar of 58 nm and M from NanoAmor of 8 nm dry size) nanomaterials (at either 3 or 30 ug ml−1 for three days) were examined for their ability to cause cellular toxicity and effects on the concentrations of cellular metabolites in HepG2 cells. Only nanomaterials M and I were run at 3 ug ml−1. A large variety of physical chemical techniques were employed on dry and wet (dynamic light scattering) nanomaterials to better characterize their inherent properties. In our study 265 cellular metabolites were found and relatively quantified. This metabolomics study included enough parameters related to reduced glutathione (GSH) and other cellular redox molecules to be able to evaluate one of the main theories of nanomaterial-induced toxicity – the oxidative stress hypothesis. The metabolomics experimental results are discussed in terms of systems biology, toxicology and the oxidative stress theory of nanomaterial toxicity.
Materials and methods
Nanomaterials, their dispersion via ultrasound and their characterization
The six nanomaterials used in this study (Table 1) were selected by a combination of perceived data needs of the US EPA and the Organization for Economic Co-operation and Development (OECD) international toxicity testing needs list. These nanomaterials are being used by multiple research laboratories at the US EPA in a coordinated research effort with many different scientific disciplines and experimental techniques.| ID | Chemical | Vendor | Cat no. | Lot number | Primary particle size (nm) | Size range (nm) | Surface area (m2 g−1) | % Purity | Crystal form | Assayer |
|---|---|---|---|---|---|---|---|---|---|---|
| A | TiO2 | Degussa | Aeroxide® TiO2 P25 | 4165012298 | 27.5 | 14–64 | 49 | 95.10 | 86% Anatase, 14% rutile | Vendor |
| 31 | 12–88 | 52.9 | 99.9 | Anatase and rutile | Univ Kentucky | |||||
| B | TiO2 | NanoAmor | 5485HT | 5485-030007 | 30–40 | 30–40 | >30 | 95.10 | Rutile | Vendor |
| 59 | 36–97 | 22.2 | 99.9 | Anatase > rutile | Univ Kentucky | |||||
| C | TiO2 | Alfa Aesar | 44690 | D22T034 | 10 | 100–130 | Anatase | Vendor | ||
| 25 | 6–60 | 118 | 98.8 | Anatase | Univ Kentucky | |||||
| I | TiO2 | Acros | 21358 | A0075656 | Ave. 200 | Anatase | Vendor | |||
| 142 | 67–322 | 6.99 | 99.9 | Anatase | Univ Kentucky | |||||
| L | CeO2 | Alfa Aesar | 44758 | J02S055 | 70–105 | 8–12 | 99.9 | Vendor | ||
| 58 | 36–99 | 10.1 | 99.9 | Cerianite | Univ Kentucky | |||||
| M | CeO2 | NanoAmor | 1406RE | 1406-111607 | 15–30 | 30–50 | 99.9 | Vendor | ||
| 8 | 5–13 | 73.2 | 99.9 | Cerianite | Univ Kentucky |
Nanomaterial physical–chemical characterization was done by a variety of techniques for primary particle size, range of particle size, surface area, % purity and crystal form by their manufacturer and by an independent party (University of Kentucky, Chemical & Engineering Department) via a US EPA contract. Table 1 presents the six nanomaterials and some of the physical characterization data on the dry powders obtained from both the vendors and the University of Kentucky. In the text of this paper the primary particle size presented is that from the University of Kentucky and not the vendors. Other physical–chemical characterization data available from the University of Kentucky studies on our study compounds includes elemental analysis by TEM/EDX, elemental analysis by ICP/MS, primary and agglomerated particle size, crystal structure by XRD, and particle shape and morphology by TEM and SEM (ESI‡ files S1–S6).
The nanomaterials were obtained from four different vendors (Alfa Aesar, Degussa, NanoAmor, and Acros). The chemical purity was high (>98.8% for all cases and as high as 99.9% for five cases), the primary dry particle sizes ranged from 8 to 142 nm. With respect to crystal form, two of the TiO2 nanomaterials contained both the anatase and rutile crystal forms. The two other TiO2 nanomaterials were all anatase. Both CeO2 nanomaterials were cerianite.
For dispersion, measured amounts of bovine serum albumin at 200 mg ml−1 and physiological buffered saline (PBS) were added to the dry nanomaterials in a glass vial. The general protein coating recipe of Dale Porter12 was followed with the mass ratio of the nanomaterial to BSA of 1/0.6. For example in preparation of CeO2 “M” for study, the recipe was 10.0 mg nanomaterial “M”, 6 mg bovine serum albumin, 3.0 ml of saline. Sonication occurred at a nanomaterial concentration of 3.33 mg ml−1 and 3.0 ml of volume. Sonication was done for two 10 minute cycles of 13 seconds on, 7 seconds off with a total typical power of about 150 watts and 175![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 joules with a S-4000 Misonix Ultrasonic Liquid Processor with a 2.5 inch cup horn (part #431-A, Farmington, NY). Excess unbound albumin was removed by pelleting (10
000 joules with a S-4000 Misonix Ultrasonic Liquid Processor with a 2.5 inch cup horn (part #431-A, Farmington, NY). Excess unbound albumin was removed by pelleting (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 minutes) the nanomaterials and resuspending them in cell culture media by 20 seconds of rapid vortexing. This process avoids any sonication of the media.
000 rpm for 5 minutes) the nanomaterials and resuspending them in cell culture media by 20 seconds of rapid vortexing. This process avoids any sonication of the media.
After nanomaterial dispersion, the degree of agglomeration was determined by dynamic light scattering at 35 ° C. Refractive index values for TiO2 were 2.488 for anatase crystal structure, 2.609 for rutile crystal structure, 2.504 for the Degussa TiO2 nanomaterial (86% anatase, 14% rutile from manufacturer's description) and 2.31 for CeO2. Size and zeta potential determinations were done both just after sonication and 3 days later at the end of cell culture (data in ESI‡ Table S1) with a Malvern Model Zen3600 Zetasizer.
Chemicals and cell culture methods
The chemicals and suppliers used in this study were: bovine serum albumin (Sigma) and fetal bovine serum (Invitrogen). Human hepatocellular carcinoma cells, designation HepG2 (ATCC cat # HB-8065, originally obtained from a male 15 year old human hepatocellular carcinoma), were obtained and expanded through passage seven using growth medium (Basal Medium Eagle) containing 2 mM GlutaMAXTM, 1 mM sodium pyruvate, and 10% Fetal Bovine Serum (this combined cell culture media is called EMEM) and then frozen in liquid nitrogen. Cells were subsequently carefully thawed and expanded before experimentation at passage 10 and 11. Cultures were maintained in a humidified incubator at 37 °C and 95%Air/5% CO2 during the study. Cells were plated at 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells cm−2 in vented T-25 flasks (Corning) for 48 hours prior to nanomaterial exposure. Working stocks of each nano material were prepared at 1.0 mg mL−1 and diluted using culture medium. Individual flasks were dosed with 200 uL per cm2 of the appropriate nano material dilution to achieve either 30 ug ml−1 (high) or 3 ug ml−1 (mid) exposure concentrations. Cultures were then incubated for 72 hours prior to harvesting. At 72 hours, the media was vacuum aspirated and the flasks rinsed with warm Dulbecco's Phosphate-Buffered Saline (DPBS). The DPBS was removed, cells were scraped free of the flask and collected in labeled 15 mL tubes using 1 mL of warm DPBS by micropipette. The cells were then centrifuged at 8000 rpm for 5 minutes. The supernatant was removed via vacuum aspiration and the cellular pellet placed on dry ice before transfer to −80 ° C freezer for storage prior to metabolomic analysis.
000 cells cm−2 in vented T-25 flasks (Corning) for 48 hours prior to nanomaterial exposure. Working stocks of each nano material were prepared at 1.0 mg mL−1 and diluted using culture medium. Individual flasks were dosed with 200 uL per cm2 of the appropriate nano material dilution to achieve either 30 ug ml−1 (high) or 3 ug ml−1 (mid) exposure concentrations. Cultures were then incubated for 72 hours prior to harvesting. At 72 hours, the media was vacuum aspirated and the flasks rinsed with warm Dulbecco's Phosphate-Buffered Saline (DPBS). The DPBS was removed, cells were scraped free of the flask and collected in labeled 15 mL tubes using 1 mL of warm DPBS by micropipette. The cells were then centrifuged at 8000 rpm for 5 minutes. The supernatant was removed via vacuum aspiration and the cellular pellet placed on dry ice before transfer to −80 ° C freezer for storage prior to metabolomic analysis.
Cytotoxicity assays and kits
Many common cytotoxicity assays (MTT (3-[4,5-dimethyl-2-thiazol]-2,5-diphenyl-2H-tetrazolium bromide, CAS 298-93-1)), MTS (4-[5-[3-(carboxymethoxy)phenyl]-3-(4,5-dimethyl-1,3-thiazol-2-yl)tetrazol-3-ium-2-yl]benzenesulfonate, CAS 138169-43-4), alamar blue (resazurin, CAS 62758-13-8), neutral red (3-amino-7-dimethylamino-2 methylphenazine hydrochloride, CAS 553-24-2), ATP and simple visual examination of the cells have been used by our laboratory seeking to avoid or minimize interferences from the study nanomaterials themselves. After 3 days of culture with various nanomaterials, cytotoxicity assays based on MTT (Sigma-Aldrich, St Louis, MO), MTS (Promega, Madison, WI), alamar blue (Cell Tier-Blue, Promega, Madison, WI) and ATP (Promega, Madison, WI) were performed by the enclosed kit directions. Neutral red (Sigma-Aldrich, St Louis, MO) uptake cytotoxicity assays were also performed. Cytotoxicity assays results were always checked with each other and versus visual assessment of the cells to ensure the cytotoxicity assays were working well.In nanomaterials research cytotoxicity assays are a major problem area.13 Briefly, nanomaterials may interfere with common cytotoxicity assays by scattering light, absorbing light, fluorescence and precursor dyes and/or product dyes adsorption onto the nanomaterial surface. Generally alamar blue, MTT, and MTS cytotoxicity assays were used with the TiO2 and CeO2 nanomaterials. A PerkinElmer 1420 Multilabel Counter Victor3V was used as the plate reader for all cytotoxicity assays.
Metabolite analysis and sample accessioning
Metabolomic profiling analysis was performed by Metabolon Inc. (Durham, NC) as previously described.14 Each sample received was accessioned into the Metabolon Laboratory Information Management System (LIMS) and was assigned by the LIMSQC a unique identifier that was associated with the original source identifier only. This identifier was used to track all sample handling, tasks, results etc. The samples (and all derived aliquots) were tracked by the LIMS system. All portions of any sample were automatically assigned their own unique identifiers by the LIMS when a new task is created; the relationship of these samples is also tracked. All samples were maintained at −80 °C until processed.Metabolite sample preparation
Samples were prepared using the automated MicroLab STAR® system from Hamilton Company. A recovery standard was added prior to the first step in the extraction process for QC purposes. Four volumes of 100% MeOH was added to samples to make samples containing 80% MeOH which were centrifuged to precipitate the majority of the cellular proteins while allowing maximum recovery of small molecules. The resulting extract was divided into four fractions: one for analysis by ultrahigh performance LC/MS/MS (UPLC/MS/MS) (positive mode), one for UPLC/MS/MS (negative mode), one for GC/MS, and one for backup. Samples were placed briefly on a TurboVap® (Zymark) to remove the organic solvent. Each sample was then frozen and dried under vacuum. Lastly, samples were prepared for the appropriate instrument, either UPLC/MS/MS or GC/MS.Ultrahigh performance liquid chromatography/tandem Mass Spectroscopy (UPLC/MS/MS)
UPLC/MS/MS conditions were as previously published,15 using a Thermo-Finnigan linear trap quadrupole (LTQ) mass spectrometer, which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The sample extract was dried then reconstituted in acidic or basic LC-compatible solvents, each of which contained 8 or more injection standards at fixed concentrations to ensure injection and chromatographic consistency. UPLC conditions included a flow rate of 350 uL min−1 with an acidic mobile phases of (A) 0.1% formic acid in water and (B) 0.1% formic acid in methanol. Acidic gradient elutions occurred over 0% B to 70% B in 4 min, 70–98% B in 0.5 min, 98% B for 0.9 min. Basic elutions used the same gradient with (A) 6.5 mM ammonium bicarbonate in water, pH 8, and (B) 6.5 mM ammonium bicarbonate in 95/5 methanol/water. The dedicated 2.1 mm × 100 mm Waters BEH C18 1.7 μm particle columns were heated to 40 °C. The MS interface capillary was maintained at 350 °C, with a sheath gas flow of 40 (arbitrary units) and aux gas flow of 5 (arbitrary units) for both positive and negative injections. The spray voltage for the positive ion injection was 4.5 kV, and it was 3.75 kV for the negative ion injection. The instrument scanned 99–1000 m/z and alternated between MS and MS/MS scans. Raw data files are archived and extracted as described below.Gas chromatography/Mass Spectroscopy (GC/MS)
The samples destined for GC/MS analysis were re-dried under vacuum desiccation for a minimum of 24 hours prior to being derivatized under dried nitrogen using bistrimethyl-silyl-triflouroacetamide (BSTFA). The GC column was 5% phenyl and the temperature ramp was from 40° to 300 ° C in a 16 minute period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis. The information output from the raw data files was automatically extracted as discussed below.Quality assurance/QC
For QA/QC purposes, additional samples were included with each day's analysis. These samples included extracts of a pool created from a small aliquot of the experimental samples and process blanks. QC samples were spaced evenly among the injections and all experimental samples were randomly distributed throughout the run. A selection of QC compounds was added to every sample for chromatographic alignment, including those under test. These compounds were carefully chosen so as not to interfere with the measurement of the endogenous compounds.MS data extraction and compound identification
Raw data was extracted, peak-identified and QC processed using Metabolon's hardware and software. These systems are built on a web-service platform utilizing Microsoft's.NET technologies, which run on high-performance application servers and fiber-channel storage arrays in clusters to provide active failover and load-balancing. Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. More than 2400 commercially available purified standard compounds have been acquired and registered into LIMS for distribution to both the LC and GC platforms for determination of their analytical characteristics.Study design
Two different exposure concentrations (3 or 30 ug ml−1) are used for just the nanomaterials “M” and “I”. The number of samples per treatment group is either 5 for nanomaterial treatments or 6 for controls. Two different days were used for HepG2 culturing with all the TiO2 (or CeO2) treatment groups run on the same single experimental day.Statistical analysis
Missing values (if any) are assumed to be below the level of detection. However, biochemicals that were detected in all samples from one or more groups, but not in samples from other groups, were assumed to be near the lower limit of detection in the groups in which they were not detected. In this case, the lowest detected level of these biochemicals was imputed for samples in which that biochemical was not detected. Following log transformation and imputation with minimum observed values for each compound, Welch's two-sample t-test16 was used to identify biochemicals that differed significantly between experimental groups. To account for multiple comparison testing, false-discovery rates were computed for each comparison via the q-value method. P values and false discovery q-values for all comparisons are reported in ESI‡ Table S2. If three or more P < 0.05 treatment groups were observed per 8 nanomaterial exposure groups, a higher degree of confidence was given to the finding. Pathways were assigned for each metabolite, allowing examination of overrepresented pathways. The degree of statistical significance presented in this study is both the common P < 0.05 level and the more lenient P < 0.10 (but more than 0.05) (ESI‡ Table S2). The text of the paper uses only the P < 0.05 level of significance. Some data at the P < 0.10 (but more than 0.05) is presented and tabularized in ESI‡ Table S2 because this is less likely to miss some true biological effects.Results
Nanomaterials, their dispersion via ultrasound and their characterization
The hydrodynamic diameter was often as much as 10 to 100 times larger than the dry primary particle size. In the two buffer systems used, the nanomaterials differed in hydrodynamic diameter based on dynamic light scattering (ESI‡ Table S1) (271 to 1500 nm in PBS and 260 to 822 nm in EMEM with 10% fetal bovine serum (FBS)). Neither the zeta potential nor the sizes changed greatly (more than 10-fold for both observed peaks) during the 3 days of incubation at 37 °C.ESI‡ files of physical chemical characterization data on the dry six nanomaterials are available in the electronic version of this paper (ESI‡ file S1: physical chemical characterization of TiO2 “A”, ESI‡ file S2: physical chemical characterization of TiO2 “B”, ESI‡ file 3: physical chemical characterization of TiO2 “C”, ESI‡ file S4: physical chemical characterization of TiO2 “I”, ESI‡ file S5: physical chemical characterization of TiO2 “L” and ESI‡ file S6: physical chemical characterization of TiO2 “M”).
Elemental composition of the six nanomaterials for 35 elements by ICP-MS has been published previously.17 ESI‡ Table S5 presents a subset of this ICP-MS information for just 8 of the more interesting and important elementary contaminants (Fe, Cu, Co, Cr, Mo, Mn, Se and V) of the TiO2 and CeO2 nanomaterials.
Cytotoxicity
The order of cytotoxicity potency for these nanomaterials was C > B > I > A for TiO2 and M > L for CeO2. No signs of cytotoxicity were observed at 3 ug ml−1 for any of the nanomaterials. At 30 ug ml−1 L and A showed no changes in cytotoxicity parameters while B, C, I and M responded to a limited degree. In the exposure range of 100 to 1000 ug ml−1 all compounds showed some degree of cytotoxic response. For example at 100, 300 and 1000 ug ml−1 exposure, the MTT values were 95, 87 and 74% control for M and 90, 82 and 81% control for C.Results overview
The majority of the data this paper is based on is presented as a large Excel data file as ESI‡ Table S2. The four major parts of this Excel file are (a) the complete data summary, (b) data summary with 3 or more changes at the P < 0.10 level, (c) glutathione and gamma-glutamyl pathway related effects and (d) data pertaining to asymmetric dimethylarginine, arginine and iNOS which is located in the ea372 location in the spreadsheet.The two treatments that changed the largest number of cellular metabolites were “M”, a CeO2 and “B” a TiO2 (ESI‡ Table S3). In ESI‡ Table S2 results are shown for the commonly used P < 0.05 statistical criteria as well as for results for the less strict standard of P values > 0.05 but still < 0.10. For CeO2 there were many more increases than decreases noted, while the opposite was true for TiO2 (decreases were more common effects). At the P < 0.05 level, all nanomaterial exposures produced effects on cellular metabolite profiles with 27 effects (“I”) being the minimum and 89 (“M”) the maximum number of effects. The large number of increases in metabolites with CeO2 nanomaterials is dominated by lipid metabolites.
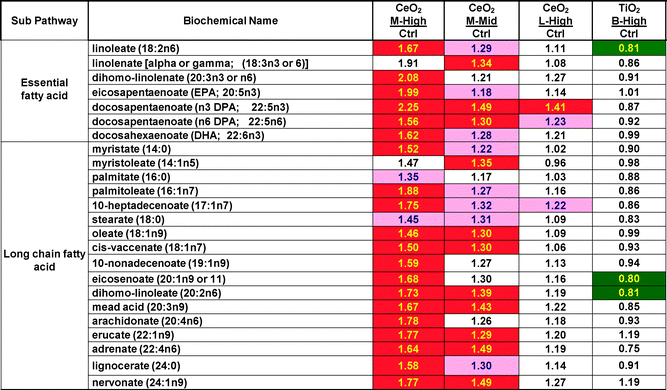
Heat maps of the overall data set show a large effect of CeO2 nanomaterial “M” (in both the high and mid concentrations) elevating many lipid related metabolite concentrations but essentially no elevations by any TiO2 nanomaterial. Table 2 shows the results from just the essential and long chain fatty acids for M, L and for B as a representative TiO2. In Tables 2 to 4, data is expressed and the mean of the treated samples divided by the mean of the control samples. Similar statistically significant (P < 0.05) elevations were seen in several other classes of lipids (lysolipid, monoacylglycerol, diacylglycerol and sphingolipid) but not in all classes of lipids (glycerolipid, carnitine and fatty acid dicarboxylate) (ESI‡ data Table S2). By the criteria of number of changed metabolite concentrations, lipid effects were the most frequent nanomaterial-induced effect of the entire data set. What is most curious about this is that the lipid majority of the increases were due to only one of the nanomaterials (“M”) and did not occur with the 2nd CeO2 tested or with any of the other 4 tested TiO2 nanomaterials. Exposure of HepG2 cells to CeO2 “M” caused significant (P < 0.05) increases in 20 of 24 fatty acid concentrations at the 30 ug ml−1 concentration (q-value false discovery rates between 0.030 and 0.056) and 11 of 24 fatty acid concentrations at 3 ug ml−1 (q-values between 0.144 and 0.182), respectively. Not a single decrease in fatty acid concentrations was observed with any of the three nanomaterial CeO2 exposures (high “M”, mid “M” or high “L”). For comparative purposes lipid results from a TiO2 exposure is included in Table 2. The TiO2 nanomaterial “B” produced no increases and only 3 significant decreases in fatty acid concentration.
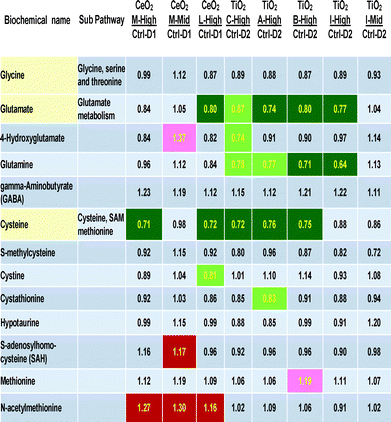
Table 3 includes the three direct precursors of glutathione (cysteine, glutamate and glycine) and other cellular metabolites related to cysteine, methionine, SAM, taurine and glutamate metabolism. A large number of statistically significant decreases were seen in three rows of the 13 biochemicals related to glutathione biosynthesis (Table 3). The three most effected metabolites are glutamine, glutamate and cysteine. Generally the size of the metabolite concentration decrease was 20–30% below control levels. Interestingly, glycine, the third amino acid precursor of glutathione was not significantly decreased by any nanomaterial treatment. The mid concentration of “M” and the “l” did not show decreases, showing a dose-related nature of this biological effect.
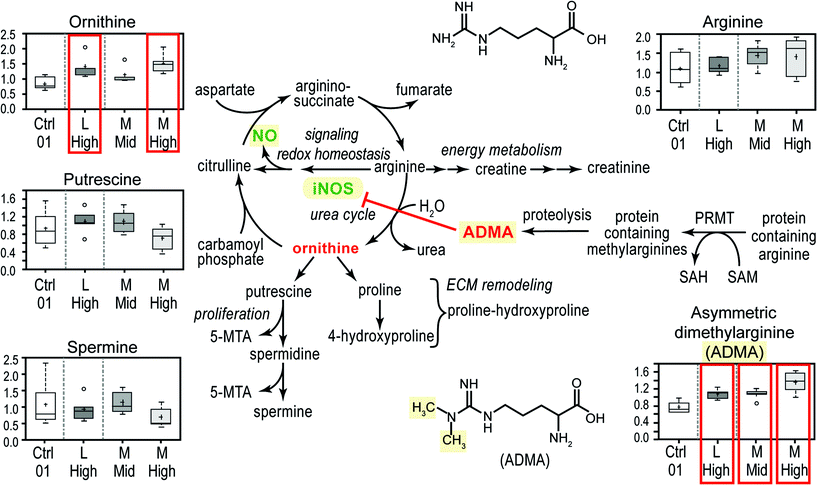
ESI‡ Fig. S1 presents both a graphical scheme of nitric oxide production and arginine metabolism as well as data on the concentrations of 5 metabolites in the arginine metabolism pathway. Box plots display mean (+), median (−), extreme data points, upper and lower quartile (box) and the maximum and minimum of the distribution (whiskers). The chief interesting observation is that all three CeO2 nanomaterial exposures significantly (P < 0.05, false discovery q-values of 0.0299, 0.105 and 0.065 for M high, M mid and L high, respectively) increased the concentration of asymmetric dimethylarginine (ADMA) (ESI‡ Fig. S1). ADMA is a known competitive inhibitor of inducible nitric oxide synthase (iNOS). Thus the imbalance of ADMA to arginine suggests that CeO2 nanomaterial exposure might decrease cellular NO concentrations (Fig. 3). Interestingly exposures to TiO2 may exhibit the opposite biology, sometimes increasing arginine concentrations and possibly increasing cellular NO concentration (ESI‡ Table S2). Cellular NO is an important signaling molecule in vasodilation and immunology.18
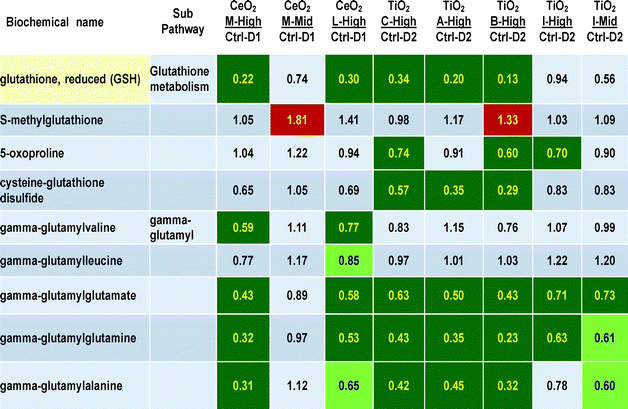
The effects of nanomaterial treatments on glutathione and gamma-glutamyl metabolites is shown in Table 4. A large number of P < 0.05 statistically significant decreases are observed in metabolites related to glutathione, particularly in respect to glutathione itself (with the largest decreases seen in this 265 metabolite data set, reaching 13 to 34% of control values, for M high, L high, C high, A high and B high the q values were 0.0299, 0.0799, 0.2756, 0.0750 and 0.0154, respectively). The gamma-glutamyl compounds with glutamate, glutamine and alanine as acceptors were significantly decreased to 31 to 73% of control values. Lesser decreases were seen in the two gamma-glutamyl acceptors with leucine and valine. At nanomaterials concentrations of 30 ug ml−1, decreases were seen in between 3 to 6 of these 9 glutathione related metabolites with all six different nanomaterials. Two other glutathione metabolites, 5-oxoproline and cysteine–glutathione disulfide, showed 3 significant decreases particularly after exposures to TiO2 nanomaterials “C” and “B”. These decreases seem dose-related to exposure concentration as the mid doses of nanomaterials “M” and “I” showed lesser or no effects than the higher doses (Table 4).
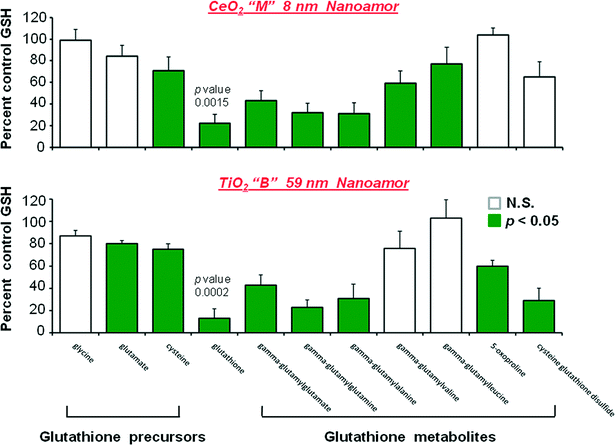
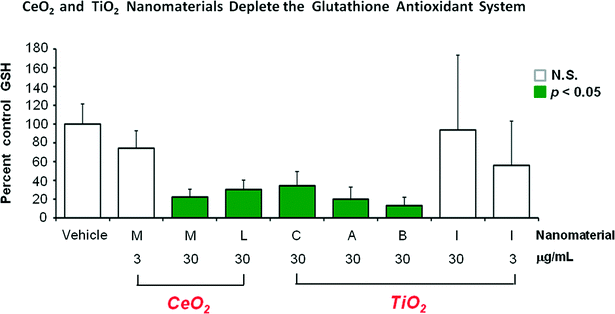
The magnitude of the glutathione related decreases following CeO2 “M” and TiO2“B” is shown in Fig. 1. With the data graphically presented in this way it is clearer that the largest perturbation from homeostatsis is in GSH concentration itself, followed by the gamma-glutamyl acceptors and finally the two glutathione precursors of glutamate and cysteine showing the smallest degree of decrease.
Fig. 2 shows the glutathione-depleting abilities of the 6 different nanomaterials. Generally nanomaterials “M”, “L”, “C”, “A” and “B” were fairly similar showing about 20% of control glutathione levels. The mid (3 ug ml−1) concentration of “M” showed a good dose-related lessening of depletion effect. However, at 30 ug ml−1 the TiO2 nanomaterial “I” was much less active than the other 5 nanomaterials, failing to reach a statistically significant level of glutathione depletion. With “I” a greater degree of glutathione depletion was observed with a smaller exposure concentration (3 ug ml−1), suggesting that the single time point of 72 hours after the start of nanomaterial exposure may not tell the entire time related story of what is happening. The “I” high group showed a large standard error of the mean. The five individual glutathione values in group “I” high were 32, 59, 80, 101 and 201% of control values. Thus, at 30 ug ml−1 treatment group “I” did have two samples that showed a substantial degree of glutathione depletion.
Discussion
Dispersion and agglomeration of nanomaterials (size and zeta potential)
These nanomaterial samples displayed a fairly large hydrodynamic diameter by dynamic light scattering in both this study with water based cell culture media EMEM with 10% fetal bovine serum (ESI‡ Table S1) and in a prior study using only 100 mM NaCl, 50 mM phosphate at pH 7.4 (210 to 797 nm).17Cytotoxicity results
The exposure concentrations used in this study (3 and 30 ug ml−1) were below concentrations which produced a full degree of cytotoxicity in HepG2 cells via common colorimetric and fluorimetric assays.Non oxidative stress effects
iNOS & NO
The finding that CeO2 exposures lead to increases in the iNOS-inhibiting metabolite ADMA is a major novel finding of this study. Nanomaterial exposures have caused increases in mRNA for iNOS and/or nitric oxide concentration in several studies. ICR mouse bronchoalveolar lavage fluid showed an increase in iNOS protein by Western blot analysis 1, 7, 14 and 28 days after intratracheal instillation of semi-single-walled carbon nanotubes.20 mRNA for iNOS was increased in cultured peritoneal macrophages harvested from nano-sized silica.21 Similarly, both nitric oxide concentration and mRNA for iNOS were elevated in peritoneal macrophages of mice treated with 50 mg kg−1 (ip) of nano-sized silica.21 After ultrafine TiO2 administration to rat lung NR8383 alveolar macrophages, the cells showed increased iNOS mRNA levels.22Thus at this time, little understanding of the overall effects of common nanomaterials on the NO and iNOS systems is available. Thus, nitric oxide, iNOS, arginine and ADMA should be included in future nanomaterial toxicity studies because this important biochemical pathway merits more research attention. The true overall impact of nanomaterial exposures on NO concentrations will depend on the multiple factors in the iNOS and NO system (enzyme induction and ADMA inhibition of enzyme activity).23
Potency of nanomaterials & dose–response relationship
Generally the mid concentration of 3 ug ml−1 demonstrated fewer effects than the high nanomaterial exposure concentration of 30 ug ml−1 (Tables 2–4, ESI‡ Table S2). From the limited dose–response information available in this study, the dose–response relationships generally are monotonic for the nanomaterials studied, although there are exceptions to this such as more glutathione depletion happening with 3 ug ml−1 than 30 ug ml−1 of “I”. In other related genomic studies from our group, both monotonic and non-monotonic dose response relationships have been observed.24Glutathione depletion and oxidative stress effects
The two most common theories of nanomaterial mode of action are probably the oxidative stress and inflammation theories. This study does not have any experimental parameters germane to inflammation but does contain many parameters pertaining to oxidative stress, mainly glutathione-related parameters. Overall the evidence in this metabolomics study of nanomaterial effects in HepG2 cells shows a great deal of positive evidence for the oxidative stress theory of nanomaterial-mediated toxicity. Indeed the glutathione concentration decreases observed, often in the range of only 20% of control values, were the largest decreases observed in this study. There is a consistency of findings for both glutathione precursors (Fig. 1) and glutathione metabolites of the gamma-glutamyl pathway (Table 4) that argue for a massive depletion of cellular glutathione and subsequent cellular attempts to restore homeostasis. From the information that is available it is not decidable if this is happening via ROS, reactive nitrogen species (RNS) or a combination of the two. At least ROS is expected to be a major contributor.25–27 It would be of interest to confirm the finding of oxidative stress observed in this study which is based on glutathione-related evidence with other oxidative stress parameters such as isoprostanes, aldehydes and 8-hyroxydeoxyguanosine.It may be that cellular reduced glutathione concentrations are under severe attack by ROS and/or RNS free radicals generated by the surfaces of the TiO2 and CeO2 nanomaterials and that this is the reason for the large decreases in glutathione concentration. Secondarily to that, the concentrations of gamma-glutamyl acceptors is also driven down because of the shortage of glutathione and also shortages of glutamate and glutamine. The glutathione shortage is so severe that the precursor pools of cysteine and glutamate are also depleted by the rapid consumption of glutathione in the HepG2 cells. Interestingly, the lipid soluble antioxidant molecule alpha-tocopherol was not generally decreased in this metabolomics study (ESI‡ Table S2), showing a major difference with the glutathione depletion results (Fig. 1 and 2).
Treatment group “I” high concentration did not show substantial glutathione depletion at the single time point tested although some individual samples did show glutathione depletion. It may be that any “I”-induced glutathione depletion reached its maximal degree of depletion either before or after the 72 hour time point used in this study. In contrast to common organic compounds, cells have few effective ways of getting rid of ingested nanomaterials. But cells can enclose nanomaterials in endosomes or alter the surface of nanomaterials by coating the redox active surfaces with cellular constituents and lessen the redox activity of the coated surface.
In rats given ultrafine TiO2 intratracheally at 2 mg per rat, depletion of GSH was seen in alveolar macrophages along with changes consistent with oxidative stress in GSH peroxidase, GSH reductase, glucose 6 phosphate dehydrogenase, GSH-transferase and ascorbic acid.28 Other investigators have demonstrated that nanomaterial exposure has produced depletion of cellular glutathione in lung29–31 and in liver cells.28,32,33 In human lung A549 cells exposed to nanomaterials of TiO2,29 CeO230 and CuO,31 decreased cellular GSH concentrations as well as increased concentrations of cellular ROS as determined by a fluorescent probe30,31 have been observed. In HepG2 human liver cells, silica of 14 nm size decreased hepatic GSH, increased malondialdehyde (MDA) and cellular ROS and showed apoptotic cellular indications.32 Nanosized ZnO depleted human hepatocyte L02 cellular GSH concentrations and gave increased MDA and comet assay results.33 However, in none of these studies were the amino acid precursors of GSH or the metabolites of the gamma-glutamyl transpeptidase system assessed along with GSH. Thus, our study confirms and extends the oxidative stress theory of nanomaterial toxicity via our data on the depletion of overall glutathione system. Many of the glutathione depletion studies mentioned above and some others related to iNOS due to either nanomaterial or fine particle exposures are summarized in ESI‡ Table S4. Glutathione depletion has been demonstrated following at least nano or fine particulate exposures to single walled carbon nanotubes, metals, metal oxides, carbon black and silica.
Our studies also suggest the use of several of GSH-related molecules as possible biomarkers of nanomaterial toxicity, namely the amino acids glutamate, glutamine and the gamma-glutamyl acceptors with three amino acids alanine, glutamate and glutamine (Tables 3 and 4). No previous studies showing nanomaterial induced decreases in gamma-glutamyl acceptors were found in a PubMed search, so it is likely this is a novel metabolomics finding. It is curious and unexplained why our study did not show a decrease in hepatic alpha-tocopherol concentrations while other published nanotoxicology studies have shown decreases in this antioxidant in A540 human lung cells30 and also in ascorbic acid concentration in alveolar macrophages.28
Comparison with prior immuno-spin trapping results
In a prior published study from this group, nanomaterials “L” and “C” showed the highest degree of free radical formation as determined by immuno-spin trapping in vitro.17 In cell culture experiments nanomaterials “M” and “B” were the most active in the number of changes they produced. In glutathione depleting ability nanomaterials “M” and “B” were again the most powerful followed by “L”, “C” and “A”. There is general but not exact agreement between these two very different techniques (immuno-spin trapping versus this cellular metabolomics study) to measure the formation and biochemical effects of free radicals. The TiO2 nanomaterial “I” was surprisingly weak in its ability to deplete glutathione. The time at which the cells were harvested (3 days) may play a major role in determining the degree of glutathione depletion effect that is measured.Comparison with other published metabolomics studies of nanomaterials
At least four metabolic studies have been published although two of these nano TiO2 studies are written in Chinese (Zhao et al., Dulixue Zazhi, 2009, 23(3), 201–204 (a rat kidney study); Wang, Zhonghua Yu Fang Yi Xue Za Zhi, 2009, 43(5), 399–403 (a serum study)). One other nano TiO2 study was an exposure to Eisenia fetida earthworms which concluded oxidative stress occurred.34 The second metabolomics study was of rat serum following intratracheally instilled nano TiO2. This rat serum study observed increased ketone bodies, low density lipoprotein (LDL), blood urea nitrogen and creatinine, lactate dehydrogenase, aspartate aminotransferase and alkaline phosphatase suggesting slight rat liver and kidney injury.35Conclusion
This metabolomics-based study of the four TiO2 and two CeO2 nanomaterials on HepG2 cells has shown substantial evidence for the oxidative stress theory of nanomaterial toxicity in the depletion of glutathione, glutathione precursors and glutathione metabolites. Nanomaterial exposures may perturb the nitric oxide physiology of organisms by increasing asymmetric dimethylarginine (ADMA) concentration, an iNOS inhibitor. Many lipids, particularly free fatty acids, were increased in concentration by one CeO2 nanomaterial (M) but not by any other nanomaterial treatments. All but the glutathione depletion were novel experimental findings showing the value of metabolomics explorations of toxicology modes of action and adverse outcome pathways.Funding
U.S. Environmental Protection Agency.Disclaimer
The information in this document has been funded wholly by the U. S. Environmental Protection Agency. It has been subjected to review by the National Health and Environmental Effects Research Laboratory and approved for publication. Approval does not signify that the contents necessarily reflect the views of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.Acknowledgements
We are grateful for the participation of many individuals of the Metabolon Inc. group who performed the analysis of the HepaG2 cells, the statistical data reduction and some of the figures and tables. In particular Ryan Michalek assisted us in interpreting metabolite effects and preparing figures. We thank Drs. Urmila Kodavanti and Michael F. Hughes for reviewing this manuscript as part of EPA clearance procedures. Dr. William Boyes is the overall coordinator of US EPA nanomaterial research. Dr. Kevin Dreher coordinated the characterization of the 8 nanomaterials used in this study via a contract for analytical chemistry and physical chemistry parameters with the University of Kentucky which was administered by Dr. Eric Grulke in the Engineering School. Kevin Dreher also compiled the physical characterization data presented in Table I. Kathleen Wallace set up many of the cytotoxicity assays and cell culture procedures and forms used in this study.References
- D. B. Warheit, et al., Testing strategies to establish the safety of nanomaterials: conclusions of an ECETOC workshop, Inhalation Toxicol., 2007, 19(8), 631–643 CrossRef CAS PubMed.
- N. J. Walker and J. R. Bucher, A 21st century paradigm for evaluating the health hazards of nanoscale materials?, Toxicol. Sci., 2009, 110(2), 251–254 CrossRef CAS PubMed.
- M. P. Holsapple, et al., Research strategies for safety evaluation of nanomaterials, part II: toxicological and safety evaluation of nanomaterials, current challenges and data needs, Toxicol. Sci., 2005, 88(1), 12–17 CrossRef CAS PubMed.
- T. L. Thompson and J. T. Yates Jr., Surface science studies of the photoactivation of TiO2--new photochemical processes, Chem. Rev., 2006, 106(10), 4428–4453 CrossRef CAS PubMed.
- P. Møller, J. K. Folkmann, P. H. Danielsen, K. Jantzen and S. Loft, Oxidative stress generated damage to DNA by gastrointestinal exposure to insoluble particles, Curr. Mol. Med., 2012, 12(6), 732–745 CrossRef.
- P. Kovacic and R. Somanathan, Nanoparticles: toxicity, radicals, electron transfer, and antioxidants, Methods Mol. Biol., 2013, 1028, 15–35, DOI:10.1007/978-1-62703-475-3_2.
- A. Nel, et al., Toxic potential of materials at the nanolevel, Science, 2006, 311(5761), 622–627 CrossRef CAS PubMed.
- S. Jeong, W. Cho, J. Kim and M. Cho, Systems Toxicological Approach to the Risk Assessment of Nanomaterials, in General, Applied, and Systems Toxicology, ed. D. A. Casciano and S. C. Sahu, published: John Wiley & Sons Ltd., Chichester, UK, 2011, vol. 2, pp. 871–890, DOI:10.1002/9780470744307.gat247.
- A. Roux, et al., Applications of liquid chromatography coupled to mass spectrometry-based metabolomics in clinical chemistry and toxicology: A review, Clin. Biochem., 2011, 44(1), 119–135 CrossRef CAS PubMed.
- D. G. Robertson, P. B. Watkins and M. D. Reily, Metabolomics in toxicology: preclinical and clinical applications, Toxicol. Sci., 2011, 120(Suppl 1), S146–170 CrossRef CAS PubMed.
- S. Nesnow, W. T. Padgett and T. Moore, Propiconazole induces alterations in the hepatic metabolome of mice: relevance to propiconazole-induced hepatocarcinogenesis, Toxicol. Sci., 2011, 120(2), 297–309 CrossRef CAS PubMed.
- D. Porter, et al., A biocompatible medium for nanoparticle dispersion, Nanotoxicology, 2008, 2(3), 144–154 CrossRef CAS.
- N. A. Monteiro-Riviere, A. O. Inman and L. W. Zhang, Limitations and relative utility of screening assays to assess engineered nanoparticle toxicity in a human cell line, Toxicol. Appl. Pharmacol., 2009, 234(2), 222–235 CrossRef CAS PubMed.
- Z. J. Reitman, et al., Profiling the effects of isocitrate dehydrogenase 1 and 2 mutations on the cellular metabolome, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(8), 3270–3275 CrossRef CAS PubMed.
- A. M. Evans, C. D. DeHaven, T. Barrett, M. Mitchell and E. Milgram, Integrated, Nontargeted Ultrahigh Performance Liquid Chromatography/Electrospray Ionization Tandem Mass Spectrometry Platform for the Identification and Relative Quantification of the Small-Molecule Complement of Biological Systems, Anal. Chem., 2009, 81, 6656–6667 CrossRef CAS PubMed.
- B. L. Welch, The generalisation of student's problems when several different population variances are involved, Biometrika, 1947, 34(1–2), 28–35 CAS.
- K. T. Kitchin, R. Y. Prasad and K. A. Wallace, Oxidative stress studies of six TiO(2) and two CeO(2) nanomaterials: immuno-spin trapping results with DNA, Nanotoxicology, 2011, 5(4), 546–556 CrossRef CAS PubMed.
- Y. Gao, The multiple actions of NO, Pfluegers Arch., 2010, 459(6), 829–839 CrossRef CAS PubMed.
- I. Celardo, E. Traversa and L. Ghibelli, Cerium oxide nanoparticles: a promise for applications in therapy, J. Exp. Ther. Oncol., 2011, 9(1), 47–51 CAS.
- E. J. Park, et al., Biological toxicity and inflammatory response of semi-single-walled carbon nanotubes, PLoS One, 2011, 6(10), e25892 CAS.
- E. J. Park and K. Park, Oxidative stress and pro-inflammatory responses induced by silica nanoparticles in vivo and in vitro, Toxicol. Lett., 2009, 184(1), 18–25 CrossRef CAS PubMed.
- A. M. Scherbart, et al., Contrasting macrophage activation by fine and ultrafine titanium dioxide particles is associated with different uptake mechanisms, Part. Fibre Toxicol., 2011, 8, 31–50 CrossRef CAS PubMed.
- S. M. Hirst, et al., Anti-inflammatory properties of cerium oxide nanoparticles, Small, 2009, 5(24), 2848–2856 CrossRef CAS PubMed.
- S. F. Thai, K. A. Wallace, C. P. Jones, H. Ren, B. T. Castellon, J. Crooks and K. T. Kitchin, Differential genomic effects on signaling pathways by two different CeO2 nanoparticles in HepG2 cells, Manuscript submitted.
- A. E. Nel, et al., Understanding biophysicochemical interactions at the nano-bio interface, Nat. Mater., 2009, 8(7), 543–557 CrossRef CAS PubMed.
- A. A. Shvedova, et al., Mechanisms of carbon nanotube-induced toxicity: focus on oxidative stress, Toxicol. Appl. Pharmacol., 2012, 261(2), 121–133 CrossRef CAS PubMed.
- E. Fireman, Ultrafine and nanoparticles-induced oxidative stress: the role of heme oxygenase-1 and carbon monoxide as anti-inflammatory pathways, J. Asthma, 2012, 49(1), 8–9 CrossRef CAS PubMed.
- F. Afaq, et al., Cytotoxicity, pro-oxidant effects and antioxidant depletion in rat lung alveolar macrophages exposed to ultrafine titanium dioxide, J. Appl. Toxicol., 1998, 18(5), 307–312 CrossRef CAS.
- C. Monteiller, et al., The pro-inflammatory effects of low-toxicity low-solubility particles, nanoparticles and fine particles, on epithelial cells in vitro: the role of surface area, Occup. Environ. Med., 2007, 64(9), 609–615 CrossRef CAS PubMed.
- W. Lin, et al., Toxicity of cerium oxide nanoparticles in human lung cancer cells, Int. J. Toxicol., 2006, 25(6), 451–457 CrossRef CAS PubMed.
- M. Ahamed, et al., Genotoxic potential of copper oxide nanoparticles in human lung epithelial cells, Biochem. Biophys. Res. Commun., 2010, 396(2), 578–583 CrossRef CAS PubMed.
- J. Ahmad, et al., Apoptosis induction by silica nanoparticles mediated through reactive oxygen species in human liver cell line HepG2, Toxicol. Appl. Pharmacol., 2012, 259(2), 160–168 CrossRef CAS PubMed.
- R. Guan, et al., Cytotoxicity, oxidative stress, and genotoxicity in human hepatocyte and embryonic kidney cells exposed to ZnO nanoparticles, Nanoscale Res. Lett., 2012, 7(1), 602 CrossRef PubMed.
- M. L. Whitfield-Åslund, H. McShane, M. J. Simpson, A. J. Simpson, J. K. Whalen, W. H. Hendershot and G. I. Sunahara, Earthworm sublethal responses to titanium dioxide nanomaterial in soil detected by 1H NMR metabolomics, Environ. Sci. Technol., 2012, 46(2), 1111–1118, DOI:10.1021/es202327k. Epub 2011 Dec 22.
- M. Tang, T. Zhang, Y. Xue, S. Wang, M. Huang, Y. Yang, M. Lu, H. Lei, L. Kong, Y. Wang and Y. Pu, Metabonomic studies of biochemical changes in the serum of rats by intratracheally instilled TiO2 nanoparticles, J. Nanosci. Nanotechnol., 2011, 11(4), 3065–3074 CrossRef CAS PubMed.
Footnotes |
| † The authors declare that they have no conflicts of interest. |
| ‡ Electronic supplementary information (ESI) available: Supplementary data (Tables, Figures and Files) are available online with the electronic version of this article. See DOI: 10.1039/c4en00096j |
| This journal is © The Royal Society of Chemistry 2014 |