Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Tri- and tetra-nuclear polypyridyl ruthenium(II) complexes as antimicrobial agents
Anil K.
Gorle
a,
Marshall
Feterl
bc,
Jeffrey M.
Warner
bc,
Lynne
Wallace
a,
F. Richard
Keene
*cde and
J. Grant
Collins
*a
aSchool of Physical, Environmental and Mathematical Sciences, University of New South Wales, Australian Defence Force Academy, Canberra, ACT 2600, Australia. E-mail: g.collins@adfa.edu.au
bSchool of Veterinary and Biomedical Sciences, James Cook University, Townsville, QLD 4811, Australia
cCentre for Biodiscovery and Molecular Development of Therapeutics, James Cook University, Townsville, QLD 4811, Australia
dSchool of Pharmacy and Molecular Sciences, James Cook University, Townsville, QLD 4811, Australia
eSchool of Chemistry and Physics, University of Adelaide, Adelaide, SA 5005, Australia. E-mail: richard.keene@adelaide.edu.au
First published on 1st October 2014
Abstract
A series of inert tri- and tetra-nuclear polypyridylruthenium(II) complexes that are linked by the bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane ligand (“bbn” for n = 10, 12 and 16) have been synthesised and their potential as antimicrobial agents examined. Due to the modular nature of the synthesis of the oligonuclear complexes, it was possible to make both linear and non-linear tetranuclear ruthenium species. The minimum inhibitory concentrations (MIC) of the ruthenium(II) complexes were determined against four strains of bacteria − Gram positive Staphylococcus aureus (S. aureus) and methicillin-resistant S. aureus (MRSA), and Gram negative Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa). In order to gain an understanding of the relative antimicrobial activities, the cellular uptake and water–octanol partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were determined for a selection of the ruthenium complexes. Although the trinuclear complexes were the most lipophilic based upon log
P) were determined for a selection of the ruthenium complexes. Although the trinuclear complexes were the most lipophilic based upon log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values and showed the greatest cellular uptake, the linear tetranuclear complexes were generally more active, with MIC values <1 μM against the Gram positive bacteria. Similarly, although the non-linear tetranuclear complexes were slightly more lipophilic and were taken up to a greater extent by the bacteria, they were consistently less active than their linear counterparts. Of particular note, the cellular accumulation of the oligonuclear ruthenium complexes was greater in the Gram negative strains compared to that in the Gram positive S. aureus and MRSA. The results demonstrate that the lower antimicrobial activity of polypyridylruthenium(II) complexes towards Gram negative bacteria, particularly P. aeruginosa, is not strongly correlated to the cellular accumulation but rather to a lower intrinsic ability to kill the Gram negative cells.
P values and showed the greatest cellular uptake, the linear tetranuclear complexes were generally more active, with MIC values <1 μM against the Gram positive bacteria. Similarly, although the non-linear tetranuclear complexes were slightly more lipophilic and were taken up to a greater extent by the bacteria, they were consistently less active than their linear counterparts. Of particular note, the cellular accumulation of the oligonuclear ruthenium complexes was greater in the Gram negative strains compared to that in the Gram positive S. aureus and MRSA. The results demonstrate that the lower antimicrobial activity of polypyridylruthenium(II) complexes towards Gram negative bacteria, particularly P. aeruginosa, is not strongly correlated to the cellular accumulation but rather to a lower intrinsic ability to kill the Gram negative cells.
Introduction
Infectious diseases remain a leading cause of death worldwide, and as there is also an increasing emergence of drug-resistant bacteria1 it is clear that there is a need for new antimicrobial agents. Based upon the versatile nature of transition metal complexes and their recent success as anticancer agents,2–4 there has been growing interest in their use as antimicrobial agents, and in particular ruthenium complexes have been widely examined.5–10 Dwyer and his co-workers first reported the antimicrobial potential of mononuclear iron and ruthenium complexes containing polypyridyl ligands.11,12 However, while these complexes exhibited excellent activity against drug-sensitive strains, they were significantly less active against current drug-resistant strains.13 In an attempt to increase the activity of inert polypyridylruthenium complexes against drug-resistant bacteria, we have examined the antimicrobial properties of inert and chlorido-containing dinuclear ruthenium and iridium metal analogues.13–15 The inert dinuclear polypyridylruthenium(II) complexes [{Ru(phen)2}2{μ-bbn}]4+ {“Rubbn”; where phen = 1,10-phenanthroline; bbn = bis[4(4′-methyl-2,2′-bipyridyl)]-1,n-alkane for n = 5, 7, 10, 12 and 16} showed excellent activity, with minimum inhibitory concentrations (MIC) of 1 and 2 μg mL−1 for the Rubb12 and Rubb16 complexes against a range of Gram positive and Gram negative bacterial strains, and they maintained the activity against drug-resistant strains such as methicillin-resistant Staphylococcus aureus (denoted as MRSA).13 Furthermore, preliminary toxicity assays against human red blood cells and a human monocytic leukemia cell line (THP-1) indicated that the Rubbn complexes were not significantly toxic to human cells.13The inert dinuclear Rubbn complexes with an overall charge of 4+ can interact reversibly with various intra-cellular receptors such as proteins and nucleic acids to stop bacterial cell replication. Rubb16 was shown to condense ribosomes when they existed as polysomes, and it was postulated that the condensation of polysomes would halt protein production and thereby inhibit bacterial growth.16 Although it would be expected that complexes which have a higher positive charge would condense polysomes more efficiently, cellular uptake experiments with mononuclear and dinuclear polypyridylirdium(III) complexes (3+ and 6+ respectively) demonstrated that they could not easily cross the cellular membrane, and hence they showed no antimicrobial activity.15,17 An alternative approach of increasing the charge of the dinuclear ruthenium complexes is to synthesise tri- and tetra-nuclear species. While the tri- and tetra-nuclear ruthenium complexes will be more positively charged – 6+ and 8+ respectively – they will also be more lipophilic than the dinuclear counterparts due to the additional non-polar linking ligands. Preliminary experiments with the tri- and tetra-nuclear complexes of Rubb7 demonstrated the potential of this approach.13 The Rubb7-tri and Rubb7-tetra were 2–4 times more active against a range of bacteria than the corresponding dinuclear Rubb7.
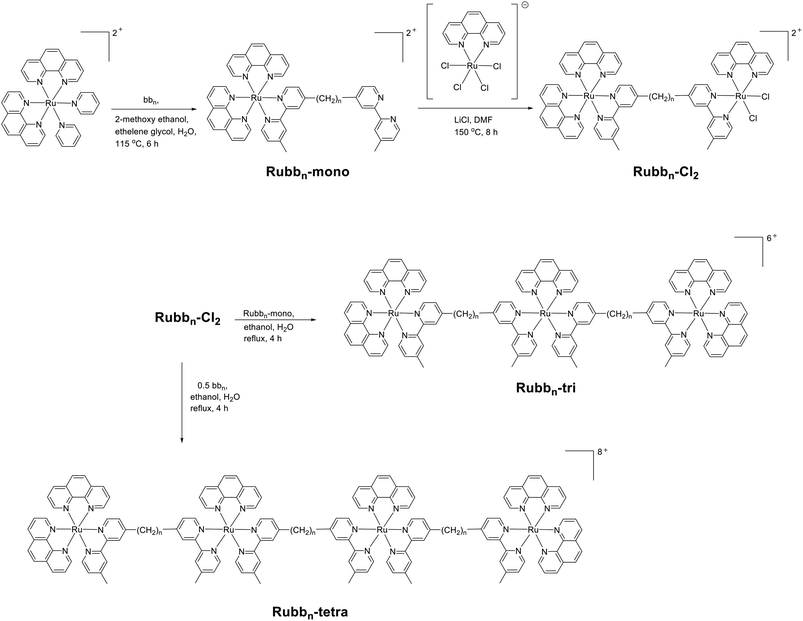
Over the last decade there has been considerable interest in developing inert dinuclear ruthenium(II) complexes as nucleic acid binding probes, anticancer agents and cellular imaging agents.18–27 More recently, there has been increasing interest in using higher nuclearity ruthenium complexes as anticancer agents.28–30 Predominantly, research has focused on ruthenium clusters or cages, as these bulky complexes may preferentially accumulate in tumours due to the enhanced permeability and retention effect.28–30 Alternatively, while several tri- and tetra-nuclear copper(II) complexes with modest antimicrobial activities have been reported,31,32 there have been very few studies on the potential of tri- or tetra-nuclear ruthenium complexes as antimicrobial agents. In the present study, we aimed to synthesise the tri- and tetra-nuclear analogues of the most active dinuclear complexes, Rubb12 and Rubb16, and examine their antimicrobial activities, log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values and cellular uptake. Additionally, due to the modular nature of the synthesis of these complexes, it was possible to synthesise both linear and non-linear tetranuclear complexes. The structures of the multinuclear complexes and the important precursor complex Rubbn-Cl2, which was also examined for antimicrobial activity, are shown in Fig. 1. The results of this study indicate that inert tri- and tetra-nuclear ruthenium polypyridyl complexes can be highly active antimicrobial agents.
P values and cellular uptake. Additionally, due to the modular nature of the synthesis of these complexes, it was possible to synthesise both linear and non-linear tetranuclear complexes. The structures of the multinuclear complexes and the important precursor complex Rubbn-Cl2, which was also examined for antimicrobial activity, are shown in Fig. 1. The results of this study indicate that inert tri- and tetra-nuclear ruthenium polypyridyl complexes can be highly active antimicrobial agents.
 | ||
| Fig. 1 Chlorido-containing dinuclear complexes (Rubbn-Cl2); and the inert trinuclear (Rubbn-tri) and tetranuclear (linear Rubbn-tetra, and non-linear Rubbn-tetra-nl) ruthenium(II) complexes. | ||
Results
Synthesis
The synthesis of flexibly-linked dinuclear (Rubbn-Cl2), trinuclear (Rubbn-tri), tetranuclear (Rubbn-tetra) and non-linear tetranuclear (Rubbn-tetra-nl) ruthenium complexes incorporating bis[4′-(4-methyl-2,2′-bipyridyl)]-1,n-alkane bridging ligands (bbn) has been achieved in good yield, as shown in Schemes 1 and 2. The chlorido-containing dinuclear species (Rubbn-Cl2) were synthesised by reacting the mononuclear complex, Rubbn-mono, with (phenH+)[Ru(phen)Cl4] in DMF at reflux temperature. The characterisation of the chlorido complexes was carried out by NMR spectroscopy and they were used as precursors for the synthesis of the tri- and tetra-nuclear complexes.The trinuclear and tetranuclear (both linear and non-linear) complexes were characterised by microanalysis, NMR (1H and 13C) and high resolution electrospray ionisation mass spectroscopy methods. Consistent with the observations previously reported for Rubb7-tetra,33 satisfactory ESI-MS could not be obtained for the linear tetranuclear complexes when they were dissolved in acetonitrile. However, good mass spectra could be obtained using acetone as the solvent. The synthesis of non-linear complexes was achieved by the reaction between the mononuclear complex, Rubbn-mono, and cis-[Ru(DMSO)4Cl2]. The reaction was carried out in ethanol–water at reflux temperatures for 5–6 hours, whereupon all the DMSO and chlorido ligands were replaced by the free ‘2,2′-bpy’ entities of Rubbn-mono complexes. All the inert complexes (tri, tetra, tetra-nl species) were purified by cation exchange on an SP Sephadex C-25 column, whereas the chlorido-containing complexes (Rubbn-Cl2) were purified by size exclusion on an Sephadex LH-20 column.
It is noted that geometric isomers will exist for the oligonuclear complexes in this study. In the bridging ligands bbn, the 2,2′-bipyridine coordinating moieties are unsymmetrically substituted – one pyridine entity with a methyl group in the 4-position and the other with the bridging methylene chain. Accordingly, for cases where there are two bbn ligands attached to one metal centre – as is the case for the central ruthenium in the trinuclear complexes and the two central ruthenium centres in the linear tetranuclear species – the chain-bearing pyridine entities may bear either relative ‘trans’ or one of two possible ‘cis’ orientations (one symmetrical in the sense that the centre will have C2 point group symmetry – denoted s-cis – and the other has C1 point group symmetry – denoted u-cis). For the trinuclear case (Rubbn-tri) there are three isomers possible based on the central metal centre (trans, s-cis and u-cis), and six isomers based on the two internal metal centres (trans,trans; s-cis,s-cis; u-cis,u-cis; trans,s-cis; trans,u-cis; s-cis,u-cis) for the linear tetranuclear case (Rubbn-tetra). For the dinuclear Rubbn-Cl2 complexes the two chlorido ligands may bear either a cis,cis or a cis,trans relationship to the chain-bearing pyridine entity, so that there are two isomers. Finally, in the case of the non-linear tetranuclear complexes, Rubbn-tetra-nl, the central Ru centre has three bbn ligands attached so that the chain-bearing pyridine entities may bear either a facial or meridional orientation to one another – giving rise to two isomers (fac and mer). In the case of fac/mer geometric isomerism, the mer isomer is thermodynamically preferred for statistical reasons (3/1) but the mer/fac ratio is often considerably greater than that because of steric congestion in the fac form.34 In the present case for the tetra-nl species, 2D NMR studies (COSY and ROESY – not reported) indicated the existence of both isomers with the fac isomer comprising about 5% (mer/fac = 19/1). The separation of possible geometrical isomers was not attempted for any of the complex systems in this study at this stage, and would represent a significant challenge in a number of these cases.
In agreement with previous studies, the Rubbn-Cl2 complexes hydrolysed when they were dissolved in water.35,36 The aromatic region of the 1H NMR spectrum of a Rubbn-Cl2 complex dissolved in water was extremely complicated suggesting a mixture of [Rubbn(OH2)Cl]+ and [Rubbn(OH2)2]2+ species, as has been previously observed for the hydrolysis of cis-[Ru(bpy)2Cl2].36 Consistent with this proposal, the addition of AgNO3 to a Rubbn-Cl2 complex that had been in an aqueous solution for two days did induce some changes in the NMR spectrum. However, even after the addition of AgNO3, the aromatic region in the NMR spectrum was still very complex. This could indicate that both the cis- and trans-diaqua species were formed, as has been noted in the acid hydrolysis of [Ru(CO3)(bpy)2].37
Electrochemistry
Cyclic voltammetry was used to determine the redox potentials for the multinuclear complexes with n = 12 (Table 1). All complexes showed a single, reversible Ru(II/III) oxidation peak at ≈+1.27 V vs. Ag/AgCl, and a series of ligand-based reductions, consistent with the well-established behaviour of tris(bidentate) polypyridylruthenium complexes.38,39 It was thought that differences might be discernible in the potentials, particularly the reduction patterns, given the subtle structural variations in the different metal centres in the multinuclear complexes. For example it has been established that in the homoleptic complexes [Ru(phen)3]2+ and [Ru(Me2bpy)3]2+, the Me2bpy ligand is slightly more difficult to reduce than phen due to the electron-donating effect of the methyl groups, and this also translates to an effect on the Ru(II) centre, which undergoes oxidation more readily.39,40 Either terminal of the bb12 ligand could be expected to mimic a Me2bpy ligand, as has been observed for the complex [{(bpy)2Ru}2(bb2)]4+, which shows very similar redox potentials to the mononuclear complex [Ru(bpy)2(Me2bpy)]2+.41 In the present case, no significant differences were observed in the oxidation potentials of the complexes, even for the non-linear tetranuclear complex, in which the central ruthenium differs somewhat more from the others. The reduction patterns were also quite similar, with each complex showing two closely-spaced reductions. A third clear reduction peak was observed for Rubb12, though this was distorted by adsorption. This three-reduction set is consistent with the pattern reported for both mononuclear [Ru(L)2(Me2bpy)]2+ (L = bpy, phen) and dinuclear [{Ru(bpy)2}2}(bbn)]4+ (n = 2,3).41–43 For the tri- and tetra-nuclear complexes only a small peak was evident at potentials where a third reduction might be expected, and a desorption peak was generally observed in the re-oxidation scan. Adsorption of reduction products has previously been noted for other multinuclear ruthenium bipyridyl complexes.44,45 The results indicate that redox differences between the complexes are negligible and unlikely to underpin any differences in biological activity.Antimicrobial activity
The minimum inhibitory concentrations (MIC) for the di-, tri- and tetra-nuclear ruthenium complexes against four bacterial strains {Staphylococcus aureus (S. aureus), methicillin-resistant S. aureus (MRSA), Escherichia coli (E. coli) and Pseudomonas aeruginosa (P. aeruginosa)} have been determined and the results are summarised in Table 2. The results demonstrate that some of the ruthenium(II) complexes in the present study have significant antimicrobial activity against both classes of bacteria, and most of the complexes are more active against Gram positive bacteria than the Gram negative strains. Of particular note, the inert tri- and tetra-nuclear ruthenium complexes showed better antimicrobial activity than the corresponding dinuclear ruthenium(II) complexes (Rubbn), particularly when the MIC values are given on a molar basis. Interestingly, the non-linear tetranuclear ruthenium(II) complexes (Rubbn-tetra-nl) were two-fold less active than the dinuclear Rubbn complexes. Of all the complexes, Rubb12-tri, Rubb16-tri, Rubb12-tetra and Rubb16-tetra are the most active compounds, and are up to four-times more active than the dinuclear counterparts against Gram positive and slightly more active against the Gram negative strains. Furthermore, Rubb12-tri, Rubb16-tri, Rubb12-tetra and Rubb16-tetra are 4–8 times more active than the previously reported Rubb7-tri and Rubb7-tetra complexes.13 Even though the overall charge of the linear and non-linear version of the tetranuclear complexes is the same (8+), the non-linear tetranuclear ruthenium complexes (Rubbn-tetra-nl) are less active when compared to the corresponding linear species, suggesting that the linearity could play an important role in inhibiting bacterial growth. The dinuclear ruthenium(II) complexes with two chlorido ligands (Rubbn-Cl2) also showed good activity, but are fractionally less active than the tri- and tetra-nuclear complexes. In general, complexes with the bb12 linking ligand were the most active against S. aureus, MRSA, and E. coli, with the complexes having the shortest (bb10) and longest (bb16) linking chain being the least active. However, against P. aeruginosa, complexes with the bb16 linking ligand showed better activity.| Compounds | Gram positive | Gram negative | ||
|---|---|---|---|---|
| S. aureus | MRSA | E. coli | P. aeruginosa | |
| Rubb12 | 0.6 | 0.6 | 2.5 | 20.1 |
| Rubb16 | 1.2 | 1.2 | 2.4 | 9.8 |
| Rubb10-tri | 0.8 | 1.7 | 1.7 | 13.5 |
| Rubb12-tri | 0.4 | 0.8 | 1.6 | 13.1 |
| Rubb16-tri | 0.4 | 0.8 | 3.1 | 12.6 |
| Rubb10-tetra | 1.2 | 2.5 | 2.5 | 19.9 |
| Rubb12-tetra | 0.3 | 0.6 | 1.2 | 9.7 |
| Rubb16-tetra | 0.3 | 0.6 | 2.3 | 9.2 |
| Rubb10-tetra-nl | 2.5 | 2.5 | 5.0 | 19.9 |
| Rubb12-tetra-nl | 1.2 | 1.2 | 4.9 | 19.4 |
| Rubb16-tetra-nl | 1.1 | 2.3 | 4.6 | 18.5 |
| Rubb10-Cl2 | 5.9 | 5.9 | 5.9 | 46.9 |
| Rubb12-Cl2 | 1.4 | 1.4 | 2.9 | 23.0 |
| Rubb16-Cl2 | 1.4 | 2.8 | 5.5 | 44.2 |
| Ampicillin | <0.7 | 183.0 | 11.4 | > 366.0 |
| Gentamicin | <0.5 | 66.9 | 1.0 | 0.5 |
Due to their greater lipophilicity (see below), it was considered that the antimicrobial activity of the tri- and tetra-nuclear complexes could decrease to a greater extent with longer incubation times than would the dinuclear complexes. Table 3 summarises the MIC values for the ruthenium complexes when the assays were carried out over a 20–22 hour timeframe. The MIC values for the tri- and tetra-nuclear complexes increased by as much as four-fold for some of the complexes when compared to the 16–18 hour values. Alternatively, the dinuclear complexes Rubb12 and Rubb16 only exhibited a maximum two-fold decrease in activity across the four bacteria. These observations suggest that the tri- and tetra-nuclear complexes rapidly accumulate within the bacteria, thereby decreasing the concentration in the incubation broth. However, even at the longer incubation time Rubb12-tetra and Rubb16-tetra are the most active of all the ruthenium complexes tested.
| Compound | Gram positive | Gram negative | ||
|---|---|---|---|---|
| S. aureus | MRSA | E. coli | P. aeruginosa | |
| Rubb12 | 1.3 | 2.5 | 5.0 | 40.3 |
| Rubb16 | 1.2 | 2.4 | 4.9 | 19.6 |
| Rubb10-tri | 1.7 | 3.4 | 3.4 | 27.0 |
| Rubb12-tri | 1.6 | 1.6 | 3.3 | 26.3 |
| Rubb16-tri | 1.6 | 1.6 | 6.3 | 25.1 |
| Rubb10-tetra | 2.5 | 5.0 | 5.0 | 19.9 |
| Rubb12-tetra | 0.6 | 1.2 | 2.4 | 9.7 |
| Rubb16-tetra | 1.1 | 1.1 | 2.3 | 9.2 |
| Rubb10-tetra-nl | 5.0 | 5.0 | 9.9 | 19.9 |
| Rubb12-tetra-nl | 1.2 | 2.4 | 4.9 | 19.4 |
| Rubb16-tetra-nl | 2.3 | 2.3 | 4.6 | 18.5 |
| Rubb10-Cl2 | 5.9 | 11.7 | 5.9 | 46.9 |
| Rubb12-Cl2 | 2.9 | 1.4 | 5.7 | 23.0 |
| Rubb16-Cl2 | 2.8 | 2.8 | 11.0 | 44.2 |
| Ampicillin | <0.7 | 183.0 | 11.4 | >366.0 |
| Gentamicin | <0.5 | 66.9 | 1.0 | 0.5 |
The minimum bactericidal concentrations (MBC) of a selection of the ruthenium complexes were determined after the MIC values were obtained for the 20–22 hour incubation experiment. The results are summarised in Table 4. As the MBC values are generally ≤2 × MIC for the 20–22 hour incubation, it can be concluded that the tri- and tetra-nuclear complexes are bactericidal rather than bacteriostatic.
| Compound | Gram positive | Gram negative | ||
|---|---|---|---|---|
| S. aureus | MRSA | E. coli | P. aeruginosa | |
| Rubb12 | 2.6 | 2.5 | 10.1 | 40.3 |
| Rubb12-tri | 3.3 | 3.3 | 6.6 | 52.6 |
| Rubb16-tri | 1.6 | 6.3 | 12.6 | 50.3 |
| Rubb12-tetra | 1.2 | 1.2 | 2.4 | 19.4 |
| Rubb16-tetra | 2.3 | 2.3 | 4.6 | 37.0 |
Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P
P
Lipophilicity is a significant factor that affects the biological activity of any metal complex, as it is generally correlated to the capacity of the drug to penetrate through the cell membrane. The standard octanol-water partition coefficient (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) was determined for the mononuclear species [Ru(phen)2(Me2bpy)]2+ and Rubbn (as control experiments), Rubbn-tri, Rubbn-tetra and Rubbn-tetra-nl complexes, and the results are summarised in Table 5. From the results, the trinuclear ruthenium complexes are more lipophilic than the tetranuclear ruthenium complexes. For the dinuclear Rubbn complexes, the antimicrobial activity was directly related to the log
P) was determined for the mononuclear species [Ru(phen)2(Me2bpy)]2+ and Rubbn (as control experiments), Rubbn-tri, Rubbn-tetra and Rubbn-tetra-nl complexes, and the results are summarised in Table 5. From the results, the trinuclear ruthenium complexes are more lipophilic than the tetranuclear ruthenium complexes. For the dinuclear Rubbn complexes, the antimicrobial activity was directly related to the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, with activity increasing with increasing lipophilicity. However, although the trinuclear complexes were more lipophilic than their corresponding linear tetranuclear complexes, they were less active. This suggests lipophilicity is an important determinant of activity, but only to the level that allows the ruthenium complex to easily diffuse across the cellular membrane.
P, with activity increasing with increasing lipophilicity. However, although the trinuclear complexes were more lipophilic than their corresponding linear tetranuclear complexes, they were less active. This suggests lipophilicity is an important determinant of activity, but only to the level that allows the ruthenium complex to easily diffuse across the cellular membrane.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) for the ruthenium complexes
P) for the ruthenium complexes
| Metal complex | Charge | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
|---|---|---|
| [Ru(phen)2(Me2bpy)]2+ | +2 | −2.9 |
| Rubb12 | +4 | −2.9 |
| Rubb16 | +4 | −1.9 |
| Rubb10-tri | +6 | −1.3 |
| Rubb12-tri | +6 | −1.0 |
| Rubb16-tri | +6 | −0.8 |
| Rubb10-tetra | +8 | −1.7 |
| Rubb12-tetra | +8 | −1.6 |
| Rubb16-tetra | +8 | −0.95 |
| Rubb10-tetra-nl | +8 | −1.9 |
| Rubb12-tetra-nl | +8 | −1.4 |
| Rubb16-tetra-nl | +8 | −1.1 |
Cellular accumulation
The cellular accumulations of the tri- and tetra-nuclear ruthenium complexes in S. aureus, MRSA, E. coli, and P. aeruginosa were determined by measuring the concentration of the complex remaining in the culture supernatant after removing the bacteria by centrifugation. The concentration of the ruthenium complex in the supernatant was calculated from an absorbance calibration curve obtained by adding known concentrations of the ruthenium complex to a blank supernatant. As the absorbance of the ruthenium complexes varied with the different broths and supernatants for each bacterial strain, a calibration curve was determined for each complex in the supernatant of each bacterial strain. The uptake of complexes into bacterial strains was measured at various incubation time points, however the uptake did not significantly change with incubation time after 30 minutes. Fig. 2 shows the uptake of the ruthenium complexes into the bacteria after 30 minutes. Surprisingly, the uptake of the complexes is slightly higher for the Gram negative bacteria than the Gram positive. The accumulation of Rubb16-tri was the highest in both Gram positive and Gram negative bacteria. For all the complexes lower levels of accumulation were observed for S. aureus, compared to the other bacteria. For both the Gram positive and Gram negative bacteria, the cellular accumulation of the tetranuclear metal complexes was slightly lower than with the trinuclear counterparts. Surprisingly, the uptake of Rubb12-tetra-nl is greater than Rubb12-tetra and Rubb12-tri.Discussion
We have previously shown that dinuclear ruthenium(II) complexes (Rubbn) exhibit excellent antimicrobial properties in terms of MIC values, cellular uptake, time-kill curves and show low toxicity towards human cells.13 In order to further improve the antimicrobial properties, we have synthesised the tri- and tetra-nuclear ruthenium(II) analogues of the most active dinuclear complexes, Rubb12 and Rubb16, and examined their in vitro susceptibility, lipophilicity and cellular accumulation. The results of the MIC assays indicate that the linear tetranuclear complexes, Rubbn-tetra, are consistently more active across the four bacteria used in this study than Rubb12 and Rubb16. Alternatively, the trinuclear analogues of Rubb12 and Rubb16 are slightly more active against some bacteria than the corresponding dinuclear complexes, but slightly less active in others. In a similar manner to Rubb12 and Rubb16, the tri- and tetra-nuclear linear complexes maintain their activity against MRSA compared to S. aureus, and are bactericidal. The non-linear tetranuclear complexes, Rubbn-tetra-nl, are consistently less active than their linear counterparts and Rubb12 and Rubb16. Significantly, whereas Rubb10-tri was generally more active than Rubb10-tetra, the activity of Rubbn-tetra was greater than Rubbn-tri for n = 12 and 16 over the four bacterial strains used in the study.The lipophilicity of the ruthenium complexes, as determined by log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, increased in the order Rubbn < Rubbn-tetra < Rubbn-tri, with all tri- and tetra-nuclear complexes being significantly more lipophilic than Rubb16. As the ruthenium complexes enter bacterial cells by passive diffusion,46 it is not surprising that the cellular accumulation experiments demonstrated that the tri- and tetra-nuclear complexes rapidly accumulate to high concentrations within the bacteria. However, the extent of the cellular accumulation of the tri- and tetra-nuclear complexes in the Gram negative bacteria, particularly P. aeruginosa, is surprising. The results of this study demonstrate that the lower toxicity of the dinuclear complexes towards P. aeruginosa is probably not strongly correlated to the cellular accumulation – as we previously concluded – but rather to a lower intrinsic ability to kill P. aeruginosa cells. Preliminary pharmacokinetic experiments with mice have indicated that Rubb12 and Rubb16 are rapidly cleared from the blood;47 consequently, the greater lipophilicity and greater cellular uptake may be advantageous for in vivo antimicrobial studies.
P, increased in the order Rubbn < Rubbn-tetra < Rubbn-tri, with all tri- and tetra-nuclear complexes being significantly more lipophilic than Rubb16. As the ruthenium complexes enter bacterial cells by passive diffusion,46 it is not surprising that the cellular accumulation experiments demonstrated that the tri- and tetra-nuclear complexes rapidly accumulate to high concentrations within the bacteria. However, the extent of the cellular accumulation of the tri- and tetra-nuclear complexes in the Gram negative bacteria, particularly P. aeruginosa, is surprising. The results of this study demonstrate that the lower toxicity of the dinuclear complexes towards P. aeruginosa is probably not strongly correlated to the cellular accumulation – as we previously concluded – but rather to a lower intrinsic ability to kill P. aeruginosa cells. Preliminary pharmacokinetic experiments with mice have indicated that Rubb12 and Rubb16 are rapidly cleared from the blood;47 consequently, the greater lipophilicity and greater cellular uptake may be advantageous for in vivo antimicrobial studies.
Interestingly, the one non-linear tetranuclear complex examined in cellular accumulation experiments, Rubb12-tetra-nl, showed greater accumulation than the corresponding linear complex Rubb12-tetra. Despite Rubb12-tetra-nl being slightly more lipophilic than Rubb12-tetra, it would be expected that the linear complex would cross a cell membrane more easily than a non-linear complex. However, as the ruthenium complexes enter bacterial cells by passive diffusion,47 the level of the cellular accumulation is a function of the binding of the ruthenium complex to intra-cellular receptors, such as nucleic acids and proteins. Despite the relatively greater accumulation of Rubb12-tetra-nl compared to Rubb12-tetra, the linear complex exhibited greater activity. Previous studies have shown that because of the flexibility of the alkane chain in the bbn ligand, both ruthenium metal centres in the Rubbn complexes can closely associate with the DNA minor groove.48 Similarly, the linear tetranuclear complexes could also follow the curvature of the DNA groove allowing close association of the four ruthenium centres with the DNA backbone. Alternatively, due to the three-dimensional shape of the non-linear tetranuclear complexes, the interactions with DNA would be substantially different and a different biological response would be expected.
There has been increasing interest in trinuclear and higher nuclearity ruthenium complexes as anticancer agents.28–30 However, there have been very few studies on the potential of tri- or tetra-nuclear ruthenium complexes as antimicrobial agents. The present study indicates that these multinuclear complexes are highly active antimicrobial agents. Consequently, using the Rubbn scaffold as a starting point, oligonuclear ruthenium complexes can be synthesised which vary by almost two orders of magnitude in lipophilicity, but contain a higher charge and remain water-soluble. Furthermore, the higher nuclearity complexes would be expected to display different nucleic acid binding or condensation potential and exhibit different pharmacokinetic profiles. Given the rapid uptake and high level of accumulation of the tetranuclear complexes in bacteria, it is unlikely that pentanuclear or higher nuclearity complexes would be more effective antimicrobial agents.
Experimental
Physical measurements
1H NMR and 13C NMR spectra were recorded on a Varian Advance 400 MHz spectrometer at room temperature in D2O {99.9%, Cambridge Isotope Laboratories (CIL)}, CDCl3 (99.8%, CIL), or CD3CN (>99.8%, Aldrich), or CD3OD (>99.8%, Aldrich). UV absorbance was measured on a Jenway 6300 spectrophotometer. Microanalyses were performed by the Microanalytical Unit, Research School of Chemistry, Australian National University, Canberra. High-resolution mass spectral measurements were made using a Waters LCT mass spectrometer (Research School of Chemistry, Australian National University).Materials and methods
Tetraethylammonium chloride, 2-methoxyethanol, lithium chloride, potassium hexafluorophosphate (KPF6) and ammonium hexafluorophosphate (NH4PF6) were purchased from Aldrich and used as supplied; Amberlite® IRA-402 (chloride form) anion-exchange resin, SP-Sephadex C-25 cation exchanger and Sephadex® LH-20 were obtained from GE Health Care Bioscience. Cation-adjusted Mueller-Hinton broth (CAMHB) was purchased from Fluka, Gillingham, UK; the control antibiotics gentamicin and ampicillin were purchased from Oxoid, Australia. The syntheses of ligands bbn (n = 10, 12 and 16) and [Ru(phen)2(py)2]Cl2, (phenH+)[Ru(phen)Cl4], cis-[Ru(DMSO)4Cl2] were performed according to reported literature methods.33,49–51Bacterial strains
All bacterial strains are classified as C2 risk group and must be handled within a PC2 laboratory. Two Staphylococcus aureus (Gram positive) isolates, a methicillin-susceptible S. aureus strain (ATCC 25923), a clinical multidrug-resistant MRSA strain (a wild clinical strain from the JCU culture collection) and two Gram negative isolates Escherichia coli (ATCC 25922) and Pseudomonas aeruginosa (ATCC 27853) were used for in vitro antimicrobial studies.MIC and MBC determination
The MIC tests were conducted by the broth micro-dilution method in duplicate as outlined in the CLSI guidelines.52 The MBC tests were performed in duplicate according to the standard microbiological techniques protocol.53 The bacteria were grown on Mueller-Hinton agar and suspended in growth medium CAMHB. Bacterial inocula were adjusted to a turbidity equivalent to that of a 0.5 McFarland standard and diluted to a final concentration of 4–8 × 105 cfu mL−1. Compounds tested were dissolved and serially diluted in cation-adjusted Mueller–Hinton broth (CAMHB; Fluka, Gillingham, UK) in sterile 96-well flat-bottom plates to a final volume of 100 μL in each well. An equal volume of inocula was added to each well, making a final concentration range of the compounds tested, including the control antibiotics gentamicin and ampicillin (Oxoid, Australia), of between 0.25 and 128 mg L−1. MICs were recorded after 16–18 h and 20–22 h of incubation at 37 °C. Colony counts of the inocula were performed for determination of the MBC. After MIC results were noted, the incubation was continued for another 4 h, the wells with no visible growth were taken into colony counting and the concentration of compounds that produced a 99.9% kill relative to the starting inoculum was recorded as the MBC.Cellular uptake
The cellular uptake of the ruthenium complexes was measured by monitoring the UV absorbance of the complexes remaining in the supernatant of the cultures after incubation for various periods of time. Bacterial inocula in log phase were adjusted to a cell concentration from 1–5 × 107 cfu mL−1. Aliquots (2 mL) of the adjusted inocula were placed in glass culture tubes and 50 μL of stock solution (330 mg L−1) of the ruthenium complex was added to give a final concentration of 8 mg L−1. Control flasks containing 50 mL of each bacterial suspension were set up as blank samples to obtain UV calibration curves for each complex. Culture tubes and control flasks were incubated with agitation at 150 rpm at 37 °C for 0.5, 1, 1.5 or 2 h. At the end of incubation, the culture tubes were centrifuged (S. aureus and MRSA at 6000g; E. coli and P. aeruginosa at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g) at 4 °C for 10 min. Supernatants (1.6 mL) were carefully transferred to 2 mL tubes and the UV absorbance of the remaining ruthenium complex was measured at λ = 488 nm. Volumes (10, 30, 40, 50 and 65 μL) of a stock solution (330 mg L−1) of each complex were added to 2 mL aliquots of the supernatant from each control bacterial suspension (untreated with drug) to acquire a UV-concentration linear correlation chart for calibration. The uptake of the complexes was calculated by using the calibration curve obtained from control bacterial aliquots.
000g) at 4 °C for 10 min. Supernatants (1.6 mL) were carefully transferred to 2 mL tubes and the UV absorbance of the remaining ruthenium complex was measured at λ = 488 nm. Volumes (10, 30, 40, 50 and 65 μL) of a stock solution (330 mg L−1) of each complex were added to 2 mL aliquots of the supernatant from each control bacterial suspension (untreated with drug) to acquire a UV-concentration linear correlation chart for calibration. The uptake of the complexes was calculated by using the calibration curve obtained from control bacterial aliquots.
Lipophilicity (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/h3_char_2009.gif) P) determination
P) determination
The partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were measured using the shake-flask technique: each ruthenium complex (at 0.1 mM) was dissolved in the water phase and an equal volume of n-octanol was added. The two phases were mutually saturated by shaking overnight at ambient temperature and then were allowed to separate on standing. The concentration of the metal complex in each phase was determined spectrophotometrically at λ = 450 nm.
P) were measured using the shake-flask technique: each ruthenium complex (at 0.1 mM) was dissolved in the water phase and an equal volume of n-octanol was added. The two phases were mutually saturated by shaking overnight at ambient temperature and then were allowed to separate on standing. The concentration of the metal complex in each phase was determined spectrophotometrically at λ = 450 nm.
Electrochemistry
Cyclic voltammetry was carried out using an eDAQ EA161 potentiostat operated via an eDAQ ED401 e-corder. A glassy carbon working electrode, platinum wire counter electrode and Ag/AgCl reference electrode were used. Ferrocene was used as an internal reference check.54 HPLC grade acetonitrile was used as solvent and the supporting electrolyte was 0.1 M tetra-n-butylammonium tetrafluoroborate.Synthesis of metal complexes
[{Ru(phen)2}(μ-bb16){Ru(phen)Cl2}](PF6)2 and [{Ru(phen)2}(μ-bb10){Ru(phen)Cl2}](PF6)2 complexes were synthesised as reported above for [{Ru(phen)2}(μ-bb12){Ru(phen)Cl2}](PF6)2. [{Ru(phen)2}(μ-bb16){Ru(phen)Cl2}](PF6)2·3CH2Cl2·C3H6O. Anal. Calcd for C80H86N10Cl8F12OP2Ru2: C, 48.5%; H, 4.38%; N, 7.1%. Found: C, 48.2%; H, 4.66%; N, 6.7%. 1H NMR (400 MHz, CD3CN): δ 8.64 (d, J = 8.1 Hz, 4H); 8.53 (d, J = 7.9 Hz, 4H); 8.42–8.34 (m, 2H); 8.27–8.22 (m, 4H); 8.21–8.16 (m, 4H); 7.89–7.85 (m, 4H); 7.79 (dd, J = 7.9 Hz, 5.0 Hz, 2H); 7.55 (dd, J = 8.0 Hz, 5.1 Hz, 4H); 7.48 (t, J = 5.2 Hz, 4H); 7.13–7.08 (m, 4H); 2.79–2.73 (m, 4H); 2.51 (s, 6H); 1.69–1.60 (m, 4H); 1.39–1.19(m, 24H). [{Ru(phen)2}(μ-bb10){Ru(phen)Cl2}](PF6)2·NH4PF6·2CH2Cl2. Anal. Calcd for C70H70N11Cl6F18P3Ru2: C, 43.9%; H, 3.68%; N, 8.1%. Found: C, 43.8%; H, 3.54%; N, 7.9%. 1H NMR (400 MHz, CD3CN): δ 8.65 (d, J = 7.9 Hz, 4H); 8.54 (d, J = 7.7 Hz, 4H); 8.42–8.34 (m, 2H); 8.28–8.16 (m, 8H); 7.90–7.84 (m, 4H); 7.82–7.75 (m, 2H); 7.55 (dd, J = 8.3 Hz, 5.3 Hz, 4H); 7.48 (t, J = 5.1 Hz, 4H); 7.14–7.07 (m, 4H); 2.80–2.72 (m, 4H); 2.51 (s, 6H); 1.72–1.58 (m, 4H); 1.42–1.21 (m, 12H).
The PF6− salts were converted to chloride salts with Amberlite IRA-402 (chloride form) anion-exchange resin. The PF6− salt of the complex was taken up in methanol (25 mL) and resin added and the mixture stirred at room temperature for 1–2 h until the solution was clear, the resin was then filtered and methanol was evaporated and the resultant solid dried in an oven at 70 °C for 16 h to obtain a dark brownish-orange solid of [{Ru(phen)2}(μ-bbn){Ru(phen)Cl2}]Cl2 (Rubbn-Cl2), typical yields were 60–65%, based on the synthetic starting material [Ru(phen)2(bbn)](PF6)2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 60 mL) and the mixture refluxed at 80 °C in the dark under a nitrogen atmosphere for 4 h. The colour of the reaction mixture slowly turned from dark brown to dark red during the course of the reaction. The reaction mixture was cooled to room temperature and the solvent was evaporated under reduced pressure to obtain a dark orange solid, the resulting solid was converted to chloride salt by stirring it in methanol using Amberlite IRA-402 (chloride form) anion-exchange resin for 1–2 h, after filtration of the resin, methanol was evaporated and the resultant chloride salt was dissolved in water (10 mL) and loaded onto an SP Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), the column was washed with water and eluted with 0.6 M and then 0.8 M NaCl solutions to remove mono- and di-nuclear impurities. The desired trinuclear complex was eluted with 1 M NaCl solution containing 20% acetone. After removing the acetone, solid KPF6 was added to the eluate and the complex was extracted into dichloromethane (2 × 30 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain PF6− salt of the complex. The complex was further purified on Sephadex LH-20 (2 cm diam. × 30 cm) using acetone as the eluent. The major first orange band was collected and the acetone removed and the product crystallised using acetonitrile–toluene to obtain a bright red-orange solid of [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)6. [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)6. Anal. Calcd for C136H140N18F36P6Ru3: C, 51.0%; H, 4.41%; N, 7.9%. Found: C, 51.2%; H, 4.29%; N, 7.8%. 1H NMR (400 MHz, CD3CN): δ 8.67–8.60 (m, 5H); 8.57–8.49 (m, 5H); 8.43–8.34 (m, 8H); 8.29–8.17 (m, 18H); 7.88 (m, 5H); 7.82–7.76 (m, 5H); 7.59–7.51 (m, 8H); 7.48 (m, 5H); 7.14–7.07 (m, 5H); 2.82–2.72 (m, 8H); 2.57 (s, 3H); 2.51 (s, 6H); 2.47 (s, 3H); 1.72–1.59 (m, 8H); 1.39–1.17 (m, 48H). 13C NMR (CD3CN): δ 157.82, 157.76, 157.5, 155.7, 155.4, 153.57, 153.53, 153.48, 152.3, 152.1, 151.2, 148.8, 148.6, 137.55, 137.42, 131.9, 129.0, 128.9, 128.2, 126.86, 126.76, 125.7, 124.9, 35.6, 30.8, 30.4, 30.3, 30.2, 30.0, 29.9, 28.7, 21.4 and 21.0. TOF MS (ESI+): most abundant ion found for [M − 4PF6]4+, m/z 654.94. Calc. for Ru3[C136H140N18](PF6)24+, m/z 654.97; most abundant ion found for [M − 3PF6]3+, m/z 921.59. Calc. for Ru3[C136H140N18](PF6)33+, m/z 921.61; most abundant ion found for [M − 2PF6]2+, m/z 1454.87. Calc. for Ru3[C136H140N18](PF6)42+, m/z 1454.90.
1, 60 mL) and the mixture refluxed at 80 °C in the dark under a nitrogen atmosphere for 4 h. The colour of the reaction mixture slowly turned from dark brown to dark red during the course of the reaction. The reaction mixture was cooled to room temperature and the solvent was evaporated under reduced pressure to obtain a dark orange solid, the resulting solid was converted to chloride salt by stirring it in methanol using Amberlite IRA-402 (chloride form) anion-exchange resin for 1–2 h, after filtration of the resin, methanol was evaporated and the resultant chloride salt was dissolved in water (10 mL) and loaded onto an SP Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), the column was washed with water and eluted with 0.6 M and then 0.8 M NaCl solutions to remove mono- and di-nuclear impurities. The desired trinuclear complex was eluted with 1 M NaCl solution containing 20% acetone. After removing the acetone, solid KPF6 was added to the eluate and the complex was extracted into dichloromethane (2 × 30 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain PF6− salt of the complex. The complex was further purified on Sephadex LH-20 (2 cm diam. × 30 cm) using acetone as the eluent. The major first orange band was collected and the acetone removed and the product crystallised using acetonitrile–toluene to obtain a bright red-orange solid of [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)6. [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)6. Anal. Calcd for C136H140N18F36P6Ru3: C, 51.0%; H, 4.41%; N, 7.9%. Found: C, 51.2%; H, 4.29%; N, 7.8%. 1H NMR (400 MHz, CD3CN): δ 8.67–8.60 (m, 5H); 8.57–8.49 (m, 5H); 8.43–8.34 (m, 8H); 8.29–8.17 (m, 18H); 7.88 (m, 5H); 7.82–7.76 (m, 5H); 7.59–7.51 (m, 8H); 7.48 (m, 5H); 7.14–7.07 (m, 5H); 2.82–2.72 (m, 8H); 2.57 (s, 3H); 2.51 (s, 6H); 2.47 (s, 3H); 1.72–1.59 (m, 8H); 1.39–1.17 (m, 48H). 13C NMR (CD3CN): δ 157.82, 157.76, 157.5, 155.7, 155.4, 153.57, 153.53, 153.48, 152.3, 152.1, 151.2, 148.8, 148.6, 137.55, 137.42, 131.9, 129.0, 128.9, 128.2, 126.86, 126.76, 125.7, 124.9, 35.6, 30.8, 30.4, 30.3, 30.2, 30.0, 29.9, 28.7, 21.4 and 21.0. TOF MS (ESI+): most abundant ion found for [M − 4PF6]4+, m/z 654.94. Calc. for Ru3[C136H140N18](PF6)24+, m/z 654.97; most abundant ion found for [M − 3PF6]3+, m/z 921.59. Calc. for Ru3[C136H140N18](PF6)33+, m/z 921.61; most abundant ion found for [M − 2PF6]2+, m/z 1454.87. Calc. for Ru3[C136H140N18](PF6)42+, m/z 1454.90.
The chloride salts were obtained by stirring the PF6− salts in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin for 1–2 h until the solution was clear. The resin was removed by filtration, and the orange-red solution was evaporated and the solid dried in an oven at 70 °C for 16 h to obtain dark red solid of [{Ru(phen)2}(μ-bbn){Ru(phen)}(μ-bbn){Ru(phen)2}]Cl6 (Rubbn-tri), typical yields were 25–30%, based on the synthetic starting material [Ru(phen)2(bbn)](PF6)2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 50 mL) and bb16 ligand (32 mg, 0.057 mmol) was added and the reaction mixture heated at reflux for 4 h in the dark under a nitrogen atmosphere. The colour of the reaction slowly turned from dark brown to dark red during the course of the reaction. The reaction mixture was cooled to room temperature and the solvent was evaporated under reduced pressure to obtain a dark orange-red solid, which was then converted to the chloride salt by stirring in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. After filtration of the resin, the methanol was evaporated to obtain the chloride salt which was dissolved in water (10 mL) and loaded onto an SP-Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), the column washed with water and eluted with 0.6 M and then 0.8 M NaCl solutions to remove the impurities. The desired tetranuclear complex was eluted with a 1 M NaCl solution containing 30% acetone. After removing the acetone, solid KPF6 was added to the eluate followed by extraction into DCM (2 × 20 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain the PF6− salt of the complex. The complex was further purified using Sephadex LH-20 with acetone as the eluent. The major first orange band was collected, the acetone evaporated and the resultant solid crystallised from acetonitrile–toluene to yield a bright red-orange precipitate of [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)8. [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)8. Anal. Calcd for C186H198N24F48P8Ru4: C, 51.5%; H, 4.61%; N, 7.8%. Found: C, 51.4%; H, 4.42%; N, 7.8%. 1H NMR (400 MHz, CD3CN): δ 8.66–8.52 (m, 16H); 8.42–8.34 (m, 10H); 8.30–8.18 (m, 18H); 7.91–7.86 (m, 8H); 7.83–7.77 (m, 8H); 7.55 (m, 12H); 7.51–7.46 (m, 6H); 7.11 (m, 6H); 2.76 (m, 12H); 2.57 (s, 3H); 2.51 (s, 12H); 2.47 (s, 3H); 1.74–1.58 (m, 12H); 1.43–1.14 (m, 72H). 13C NMR (CD3CN): δ 157.8, 157.75, 156.9, 155.6, 153.55, 153.50, 153.46, 152.2, 152.1, 151.2, 148.8, 148.5, 137.5, 137.4, 131.85, 131.82, 128.9, 128.8, 128.7, 128.2, 126.8, 126.7, 125.6, 124.9, 35.6, 30.8, 30.7, 30.4, 30.3, 30.24, 30.19, 30.05, 30.01, 29.8, 29.7, 21.1 and 20.6. TOF MS (ESI+): most abundant ion found for [M − 6PF6]6+, m/z 577.36. Calc. for Ru4[C186H198N24] (PF6)26+, m/z 577.33; most abundant ion found for [M − 4PF6]4+, m/z 938.55. Calc. for Ru4[C186H198N24](PF6)44+, m/z 938.48; most abundant ion found for [M − 3PF6]3+, m/z 1299.40. Calc. for Ru4[C186H198N24](PF6)53+, m/z 1299.63. [{Ru(phen)2}(μ-bb12){Ru(phen)}(μ-bb12){Ru(phen)}(μ-bb12){Ru(phen)2}](PF6)8. Anal. Calcd for C174H174N24F48P8Ru4: C, 50.2%; H, 4.21%; N, 8.1%. Found: C, 50.2%; H, 4.35%; N, 8.0%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.50 (m, 16H); 8.43–8.33 (m, 10H); 8.29–8.22 (m, 10H); 8.21–8.16 (m, 8H); 7.88 (m, 8H); 7.82–7.75 (m, 8H); 7.59–7.52 (m, 12H); 7.48 (m, 6H); 7.10 (m, 6H); 2.75 (m, 12H); 2.59 (s, 3H); 2.51 (s, 12H); 2.46 (s, 3H); 1.71–1.57 (m, 12H); 1.43–1.21 (m, 48H). 13C NMR (CD3CN): δ 157.79, 157.73, 155.6, 153.55, 153.52, 153.48, 152.29, 152.17, 151.2, 148.8, 148.5, 137.52, 137.41, 131.85, 131.81, 128.95, 128.91, 128.86, 128.1, 126.8, 126.7, 125.7, 124.9, 35.6, 30.8, 30.6, 30.35, 30.27, 30.23, 30.16, 30.04, 29.9, 29.8, 29.7, 21.1 and 20.5. TOF MS (ESI+): most abundant ion found for [M − 6PF6]6+, m/z 549.47. Calc. for Ru4[C174H174N24](PF6)26+, m/z 549.28; most abundant ion found for [M − 5PF6]5+, m/z 688.18. Calc. for Ru4[C174H174N24](PF6)35+, m/z 688.13; most abundant ion found for [M − 4PF6]4+, m/z 896.73. Calc. for Ru4[C174H174N24](PF6)44+, m/z 896.40. [{Ru(phen)2}(μ-bb10){Ru(phen)}(μ-bb10){Ru(phen)}(μ-bb10){Ru(phen)2}](PF6)8. Anal. Calcd for C168H162N24F48P8Ru4: C, 49.4%; H, 4.00%; N, 8.2%. Found: C, 49.4%; H, 3.92%; N, 8.0%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.51 (m, 16H); 8.41–8.33 (m, 10H); 8.28–8.22 (m, 10H); 8.21–8.15 (m, 8H); 7.87 (m, 8H); 7.81–7.76 (m, 8H); 7.54 (m, 12H); 7.50–7.46 (m, 6H); 7.12 (m, 6H); 2.78 (m, 12H); 2.56 (s, 3H); 2.51 (s, 12H); 2.45 (s, 3H); 1.73–1.56 (m, 12H); 1.45–1.22 (m, 36H). 13C NMR (CD3CN): δ 157.8, 157.7, 155.6, 153.56, 153.51, 153.46, 152.3, 152.2, 151.2, 148.8, 148.5, 137.5, 137.4, 131.8, 131.6, 128.94, 128.89, 128.82, 128.2, 126.8, 126.7, 125.6, 124.9, 35.6, 30.9, 30.6, 30.3, 30.2, 30.16, 30.08, 30.01, 29.9, 29.8, 29.7, 21.1 and 20.8. TOF MS (ESI+): most abundant ion found for [M − 5PF6]5+, m/z 671.40. Calc. for Ru4[C168H162N24](PF6)35+, m/z 671.29; most abundant ion found for [M − 4PF6]4+, m/z 875.77. Calc. for Ru4[C168H162N24](PF6)44+, m/z 875.36; most abundant ion found for [M − 3PF6]3+, m/z 1215.32. Calc. for Ru4[C168H162N24](PF6)53+, m/z 1215.47.
1, 50 mL) and bb16 ligand (32 mg, 0.057 mmol) was added and the reaction mixture heated at reflux for 4 h in the dark under a nitrogen atmosphere. The colour of the reaction slowly turned from dark brown to dark red during the course of the reaction. The reaction mixture was cooled to room temperature and the solvent was evaporated under reduced pressure to obtain a dark orange-red solid, which was then converted to the chloride salt by stirring in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. After filtration of the resin, the methanol was evaporated to obtain the chloride salt which was dissolved in water (10 mL) and loaded onto an SP-Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), the column washed with water and eluted with 0.6 M and then 0.8 M NaCl solutions to remove the impurities. The desired tetranuclear complex was eluted with a 1 M NaCl solution containing 30% acetone. After removing the acetone, solid KPF6 was added to the eluate followed by extraction into DCM (2 × 20 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain the PF6− salt of the complex. The complex was further purified using Sephadex LH-20 with acetone as the eluent. The major first orange band was collected, the acetone evaporated and the resultant solid crystallised from acetonitrile–toluene to yield a bright red-orange precipitate of [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)8. [{Ru(phen)2}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)}(μ-bb16){Ru(phen)2}](PF6)8. Anal. Calcd for C186H198N24F48P8Ru4: C, 51.5%; H, 4.61%; N, 7.8%. Found: C, 51.4%; H, 4.42%; N, 7.8%. 1H NMR (400 MHz, CD3CN): δ 8.66–8.52 (m, 16H); 8.42–8.34 (m, 10H); 8.30–8.18 (m, 18H); 7.91–7.86 (m, 8H); 7.83–7.77 (m, 8H); 7.55 (m, 12H); 7.51–7.46 (m, 6H); 7.11 (m, 6H); 2.76 (m, 12H); 2.57 (s, 3H); 2.51 (s, 12H); 2.47 (s, 3H); 1.74–1.58 (m, 12H); 1.43–1.14 (m, 72H). 13C NMR (CD3CN): δ 157.8, 157.75, 156.9, 155.6, 153.55, 153.50, 153.46, 152.2, 152.1, 151.2, 148.8, 148.5, 137.5, 137.4, 131.85, 131.82, 128.9, 128.8, 128.7, 128.2, 126.8, 126.7, 125.6, 124.9, 35.6, 30.8, 30.7, 30.4, 30.3, 30.24, 30.19, 30.05, 30.01, 29.8, 29.7, 21.1 and 20.6. TOF MS (ESI+): most abundant ion found for [M − 6PF6]6+, m/z 577.36. Calc. for Ru4[C186H198N24] (PF6)26+, m/z 577.33; most abundant ion found for [M − 4PF6]4+, m/z 938.55. Calc. for Ru4[C186H198N24](PF6)44+, m/z 938.48; most abundant ion found for [M − 3PF6]3+, m/z 1299.40. Calc. for Ru4[C186H198N24](PF6)53+, m/z 1299.63. [{Ru(phen)2}(μ-bb12){Ru(phen)}(μ-bb12){Ru(phen)}(μ-bb12){Ru(phen)2}](PF6)8. Anal. Calcd for C174H174N24F48P8Ru4: C, 50.2%; H, 4.21%; N, 8.1%. Found: C, 50.2%; H, 4.35%; N, 8.0%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.50 (m, 16H); 8.43–8.33 (m, 10H); 8.29–8.22 (m, 10H); 8.21–8.16 (m, 8H); 7.88 (m, 8H); 7.82–7.75 (m, 8H); 7.59–7.52 (m, 12H); 7.48 (m, 6H); 7.10 (m, 6H); 2.75 (m, 12H); 2.59 (s, 3H); 2.51 (s, 12H); 2.46 (s, 3H); 1.71–1.57 (m, 12H); 1.43–1.21 (m, 48H). 13C NMR (CD3CN): δ 157.79, 157.73, 155.6, 153.55, 153.52, 153.48, 152.29, 152.17, 151.2, 148.8, 148.5, 137.52, 137.41, 131.85, 131.81, 128.95, 128.91, 128.86, 128.1, 126.8, 126.7, 125.7, 124.9, 35.6, 30.8, 30.6, 30.35, 30.27, 30.23, 30.16, 30.04, 29.9, 29.8, 29.7, 21.1 and 20.5. TOF MS (ESI+): most abundant ion found for [M − 6PF6]6+, m/z 549.47. Calc. for Ru4[C174H174N24](PF6)26+, m/z 549.28; most abundant ion found for [M − 5PF6]5+, m/z 688.18. Calc. for Ru4[C174H174N24](PF6)35+, m/z 688.13; most abundant ion found for [M − 4PF6]4+, m/z 896.73. Calc. for Ru4[C174H174N24](PF6)44+, m/z 896.40. [{Ru(phen)2}(μ-bb10){Ru(phen)}(μ-bb10){Ru(phen)}(μ-bb10){Ru(phen)2}](PF6)8. Anal. Calcd for C168H162N24F48P8Ru4: C, 49.4%; H, 4.00%; N, 8.2%. Found: C, 49.4%; H, 3.92%; N, 8.0%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.51 (m, 16H); 8.41–8.33 (m, 10H); 8.28–8.22 (m, 10H); 8.21–8.15 (m, 8H); 7.87 (m, 8H); 7.81–7.76 (m, 8H); 7.54 (m, 12H); 7.50–7.46 (m, 6H); 7.12 (m, 6H); 2.78 (m, 12H); 2.56 (s, 3H); 2.51 (s, 12H); 2.45 (s, 3H); 1.73–1.56 (m, 12H); 1.45–1.22 (m, 36H). 13C NMR (CD3CN): δ 157.8, 157.7, 155.6, 153.56, 153.51, 153.46, 152.3, 152.2, 151.2, 148.8, 148.5, 137.5, 137.4, 131.8, 131.6, 128.94, 128.89, 128.82, 128.2, 126.8, 126.7, 125.6, 124.9, 35.6, 30.9, 30.6, 30.3, 30.2, 30.16, 30.08, 30.01, 29.9, 29.8, 29.7, 21.1 and 20.8. TOF MS (ESI+): most abundant ion found for [M − 5PF6]5+, m/z 671.40. Calc. for Ru4[C168H162N24](PF6)35+, m/z 671.29; most abundant ion found for [M − 4PF6]4+, m/z 875.77. Calc. for Ru4[C168H162N24](PF6)44+, m/z 875.36; most abundant ion found for [M − 3PF6]3+, m/z 1215.32. Calc. for Ru4[C168H162N24](PF6)53+, m/z 1215.47.
The chloride salts were obtained by stirring the PF6− salts in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. The resin was removed by filtration, and the orange-red solution was evaporated and dried in oven at 70 °C for 16 h to obtain dark red [{Ru(phen)2}(μ-bbn){Ru(phen)}(μ-bbn){Ru(phen)}(μ-bbn){Ru(phen)2}]Cl8 (Rubb16-tetra), typical yields were 20–25%, based on the synthetic starting material [{Ru(phen)2}(μ-bbn){Ru(phen)Cl2}](PF6)2.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 20 mL) for 5–6 h under the nitrogen atmosphere. The reaction mixture was cooled to room temperature and the solvent evaporated to obtain an orange solid which was converted to the chloride salt by stirring in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. After filtration of the resin and removal of methanol, the solid was dissolved in water (10 mL) and loaded onto an SP-Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), eluted with water and then with 0.6 M and then 0.8 M NaCl solutions to remove the impurities. The desired non-linear tetranuclear complex was eluted with a 1 M NaCl solution containing 30% acetone. After removing the acetone, solid KPF6 was added to the eluate followed by extraction into DCM (2 × 20 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain the PF6− salt of the complex. The complex was further purified using Sephadex LH-20 with acetone as the eluent, the major first orange band was collected and the acetone evaporated to yield a bright red-orange solid of [{Ru(μ-bb16)3}{Ru(phen)2}3](PF6)8. [{Ru(μ-bb16)3}{Ru(phen)2}3](PF6)8. Anal. Calcd for C186H198N24F48P8Ru4: C, 51.5%; H, 4.61%; N, 7.8%. Found: C, 51.2%; H, 4.66%; N, 7.7%. 1H NMR (400 MHz, CD3CN): δ 8.67–8.62 (m, 8H); 8.57–8.51 (m, 8H); 8.44–8.36 (m, 10H); 8.26–8.22 (m, 10H); 8.21–8.18 (m, 8H); 7.92–7.84 (m, 8H); 7.82–7.76 (m, 8H); 7.58–7.52 (m, 12H); 7.49–7.45 (m, 6H); 7.14–7.07 (m, 6H); 2.74 (m, 12H); 2.64 (s, 3H); 2.54 (s, 12H); 2.44 (s, 3H); 1.73–1.57 (m, 12H); 1.44–1.16 (m, 72H). 13C NMR (CD3CN): δ 157.8, 157.7, 156.9, 156.5, 156.3, 155.7, 154.8, 153.7, 153.57, 153.53, 153.49, 152.3, 152.2, 151.2, 148.8, 148.6, 137.5, 137.4, 131.8, 129.0, 128.9, 128.8, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.9, 30.6, 30.48, 30.40, 30.3, 30.2, 30.1, 30.04, 29.96, 29.91, 28.5, 21.1 and 20.7. TOF MS (ESI+): most abundant ion found for [M − 8PF6]8+, m/z 396.77. Calc. for Ru4[C186H198N24]8+, m/z 396.76; most abundant ion found for [M − 7PF6]7+, m/z 474.10. Calc. for Ru4[C186H198N24](PF6)17+, m/z 474.14; most abundant ion found for [M − 5PF6]5+, m/z 721.90. Calc. for Ru4[C186H198N24](PF6)35+, m/z 721.79. [{Ru(μ-bb12)3}{Ru(phen)2}3](PF6)8·4H2O. Anal. Calcd for C174H182N24F48O4P8Ru4: C, 49.3%; H, 4.33%; N, 7.9%. Found: C, 48.9%; H, 4.36%; N, 7.6%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.63 (m, 8H); 8.56–8.52 (m, 8H); 8.42–8.34 (m, 10H); 8.28–8.22 (m, 10H); 8.21–8.17 (m, 8H); 7.90–7.86 (m, 8H); 7.84–7.78 (m, 8H); 7.57–7.52 (m, 12H); 7.50–7.46 (m, 6H); 7.13–7.08 (m, 6H); 2.77 (m, 12H); 2.62 (s, 3H); 2.52 (s, 12H); 2.45 (s, 3H); 1.71–1.56 (m, 12H); 1.41–1.14 (m, 48H). 13C NMR (CD3CN): δ 157.8, 157.7, 156.8, 156.2, 156.0, 155.6, 154.5, 153.9, 153.6, 153.4, 152.3, 152.2, 151.2, 148.8, 148.6, 137.5, 137.4, 131.8, 129.0, 128.95, 128.89, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.9, 30.7, 30.4, 30.3, 30.2, 30.1, 30.06, 30.02, 29.9, 29.3, 28.3, 21.2 and 21.1. TOF MS (ESI+): most abundant ion found for [M − 8PF6]8+, m/z 375.75. Calc. for Ru4[C174H174N24]8+, m/z 375.71; most abundant ion found for [M − 7PF6]7+, m/z 450.00. Calc. for Ru4[C174H174N24](PF6)17+, m/z 450.10; most abundant ion found for [M − 6PF6]6+, m/z 549.16. Calc. for Ru4[C174H174N24](PF6)26+, m/z 549.28; most abundant ion found for [M − 5PF6]5+, m/z 688.18. Calc. for Ru4[C174H174N24](PF6)35+, m/z 688.13. [{Ru(μ-bb10)3}{Ru(phen)2}3](PF6)8·4H2O. Anal. Calcd for C168H170N24F48O4P8Ru4: C, 48.6%; H, 4.13%; N, 8.1%. Found: C, 48.5%; H, 4.37%; N, 7.7%. 1H NMR (400 MHz, CD3CN): δ 8.69–8.62 (m, 8H); 8.57–8.52 (m, 8H); 8.44–8.31 (m, 10H); 8.29–8.22 (m, 10H); 8.21–8.16 (m, 8H); 7.91–7.85 (m, 8H); 7.82–7.76 (m, 8H); 7.58–7.52 (m, 12H); 7.51–7.46 (m, 6H); 7.13–7.07 (m, 6H); 2.76 (m, 12H); 2.60 (s, 3H); 2.54 (s, 12H); 2.44 (s, 3H); 1.72–1.60 (m, 12H); 1.42–1.20 (m, 36H). 13C NMR (CD3CN): δ 157.8, 157.7, 157.0, 156.9, 156.4, 155.9, 155.6, 155.3, 153.57, 153.52, 153.4, 153.3, 152.3, 152.1, 151.2, 150.8, 148.8, 148.5, 137.5, 137.4, 131.8, 128.96, 128.90, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.96, 30.91, 30.6, 30.2, 30.08, 30.01, 29.8, 29.3, 28.3, 21.2 and 20.9. TOF MS (ESI+): most abundant ion found for [M − 5PF6]5+, m/z 671.36. Calc. for Ru4[C168H162N24](PF6)35+, m/z 671.29; most abundant ion found for [M − 4PF6]4+, m/z 875.45. Calc. for Ru4[C168H162N24](PF6)44+, m/z 875.36; most abundant ion found for [M − 3PF6]3+, m/z 1215.60. Calc. for Ru4[C168H162N24](PF6)53+, m/z 1215.47; most abundant ion found for [M − 2PF6]2+, m/z 1895.85. Calc. for Ru4[C168H162N24](PF6)62+, m/z 1895.69.
1, 20 mL) for 5–6 h under the nitrogen atmosphere. The reaction mixture was cooled to room temperature and the solvent evaporated to obtain an orange solid which was converted to the chloride salt by stirring in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. After filtration of the resin and removal of methanol, the solid was dissolved in water (10 mL) and loaded onto an SP-Sephadex C-25 cation exchange column (2 cm diam. × 25 cm), eluted with water and then with 0.6 M and then 0.8 M NaCl solutions to remove the impurities. The desired non-linear tetranuclear complex was eluted with a 1 M NaCl solution containing 30% acetone. After removing the acetone, solid KPF6 was added to the eluate followed by extraction into DCM (2 × 20 mL). The organic layer was washed with water (20 mL), dried over anhydrous Na2SO4, and evaporated to dryness to obtain the PF6− salt of the complex. The complex was further purified using Sephadex LH-20 with acetone as the eluent, the major first orange band was collected and the acetone evaporated to yield a bright red-orange solid of [{Ru(μ-bb16)3}{Ru(phen)2}3](PF6)8. [{Ru(μ-bb16)3}{Ru(phen)2}3](PF6)8. Anal. Calcd for C186H198N24F48P8Ru4: C, 51.5%; H, 4.61%; N, 7.8%. Found: C, 51.2%; H, 4.66%; N, 7.7%. 1H NMR (400 MHz, CD3CN): δ 8.67–8.62 (m, 8H); 8.57–8.51 (m, 8H); 8.44–8.36 (m, 10H); 8.26–8.22 (m, 10H); 8.21–8.18 (m, 8H); 7.92–7.84 (m, 8H); 7.82–7.76 (m, 8H); 7.58–7.52 (m, 12H); 7.49–7.45 (m, 6H); 7.14–7.07 (m, 6H); 2.74 (m, 12H); 2.64 (s, 3H); 2.54 (s, 12H); 2.44 (s, 3H); 1.73–1.57 (m, 12H); 1.44–1.16 (m, 72H). 13C NMR (CD3CN): δ 157.8, 157.7, 156.9, 156.5, 156.3, 155.7, 154.8, 153.7, 153.57, 153.53, 153.49, 152.3, 152.2, 151.2, 148.8, 148.6, 137.5, 137.4, 131.8, 129.0, 128.9, 128.8, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.9, 30.6, 30.48, 30.40, 30.3, 30.2, 30.1, 30.04, 29.96, 29.91, 28.5, 21.1 and 20.7. TOF MS (ESI+): most abundant ion found for [M − 8PF6]8+, m/z 396.77. Calc. for Ru4[C186H198N24]8+, m/z 396.76; most abundant ion found for [M − 7PF6]7+, m/z 474.10. Calc. for Ru4[C186H198N24](PF6)17+, m/z 474.14; most abundant ion found for [M − 5PF6]5+, m/z 721.90. Calc. for Ru4[C186H198N24](PF6)35+, m/z 721.79. [{Ru(μ-bb12)3}{Ru(phen)2}3](PF6)8·4H2O. Anal. Calcd for C174H182N24F48O4P8Ru4: C, 49.3%; H, 4.33%; N, 7.9%. Found: C, 48.9%; H, 4.36%; N, 7.6%. 1H NMR (400 MHz, CD3CN): δ 8.68–8.63 (m, 8H); 8.56–8.52 (m, 8H); 8.42–8.34 (m, 10H); 8.28–8.22 (m, 10H); 8.21–8.17 (m, 8H); 7.90–7.86 (m, 8H); 7.84–7.78 (m, 8H); 7.57–7.52 (m, 12H); 7.50–7.46 (m, 6H); 7.13–7.08 (m, 6H); 2.77 (m, 12H); 2.62 (s, 3H); 2.52 (s, 12H); 2.45 (s, 3H); 1.71–1.56 (m, 12H); 1.41–1.14 (m, 48H). 13C NMR (CD3CN): δ 157.8, 157.7, 156.8, 156.2, 156.0, 155.6, 154.5, 153.9, 153.6, 153.4, 152.3, 152.2, 151.2, 148.8, 148.6, 137.5, 137.4, 131.8, 129.0, 128.95, 128.89, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.9, 30.7, 30.4, 30.3, 30.2, 30.1, 30.06, 30.02, 29.9, 29.3, 28.3, 21.2 and 21.1. TOF MS (ESI+): most abundant ion found for [M − 8PF6]8+, m/z 375.75. Calc. for Ru4[C174H174N24]8+, m/z 375.71; most abundant ion found for [M − 7PF6]7+, m/z 450.00. Calc. for Ru4[C174H174N24](PF6)17+, m/z 450.10; most abundant ion found for [M − 6PF6]6+, m/z 549.16. Calc. for Ru4[C174H174N24](PF6)26+, m/z 549.28; most abundant ion found for [M − 5PF6]5+, m/z 688.18. Calc. for Ru4[C174H174N24](PF6)35+, m/z 688.13. [{Ru(μ-bb10)3}{Ru(phen)2}3](PF6)8·4H2O. Anal. Calcd for C168H170N24F48O4P8Ru4: C, 48.6%; H, 4.13%; N, 8.1%. Found: C, 48.5%; H, 4.37%; N, 7.7%. 1H NMR (400 MHz, CD3CN): δ 8.69–8.62 (m, 8H); 8.57–8.52 (m, 8H); 8.44–8.31 (m, 10H); 8.29–8.22 (m, 10H); 8.21–8.16 (m, 8H); 7.91–7.85 (m, 8H); 7.82–7.76 (m, 8H); 7.58–7.52 (m, 12H); 7.51–7.46 (m, 6H); 7.13–7.07 (m, 6H); 2.76 (m, 12H); 2.60 (s, 3H); 2.54 (s, 12H); 2.44 (s, 3H); 1.72–1.60 (m, 12H); 1.42–1.20 (m, 36H). 13C NMR (CD3CN): δ 157.8, 157.7, 157.0, 156.9, 156.4, 155.9, 155.6, 155.3, 153.57, 153.52, 153.4, 153.3, 152.3, 152.1, 151.2, 150.8, 148.8, 148.5, 137.5, 137.4, 131.8, 128.96, 128.90, 128.2, 126.8, 126.7, 125.7, 124.9, 35.6, 30.96, 30.91, 30.6, 30.2, 30.08, 30.01, 29.8, 29.3, 28.3, 21.2 and 20.9. TOF MS (ESI+): most abundant ion found for [M − 5PF6]5+, m/z 671.36. Calc. for Ru4[C168H162N24](PF6)35+, m/z 671.29; most abundant ion found for [M − 4PF6]4+, m/z 875.45. Calc. for Ru4[C168H162N24](PF6)44+, m/z 875.36; most abundant ion found for [M − 3PF6]3+, m/z 1215.60. Calc. for Ru4[C168H162N24](PF6)53+, m/z 1215.47; most abundant ion found for [M − 2PF6]2+, m/z 1895.85. Calc. for Ru4[C168H162N24](PF6)62+, m/z 1895.69.
The chloride salts were obtained by stirring the PF6− salts in methanol with Amberlite IRA-402 (chloride form) anion-exchange resin. The resin was removed by filtration, and the orange-red solution was evaporated and dried in oven at 70 °C for 16 h to obtain dark red [{Ru(μ-bbn)3}{Ru(phen)2}3}]Cl8 (Rubbn-tetra-nl), typical yields were 25–35%, based on the synthetic starting material [Ru(DMSO)4Cl2].
Acknowledgements
We gratefully thank Ms Anitha Jeyasingham from the Mass Spectrometry Facility at the Research School of Chemistry, Australian National University, for the mass spectral data.References
- H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards Jr., D. Gilbert, L. B. Rice, M. Scheld, B. Spellberg and J. Bartlett, IDSA Report on Development Pipeline, 2009, 48, 1.
- E. Wong and C. M. Giandomenico, Chem. Rev., 1999, 99, 2451 CrossRef CAS PubMed
.
-
N. Farrell, in Platinum-Based Drugs in Cancer Therapy, ed. L. R. Kelland and N. Farrell, Humana Press, Totowa, NJ, 2000, pp. 321–338 Search PubMed
.
- N. J. Wheate and J. G. Collins, Coord. Chem. Rev., 2003, 241, 133 CrossRef CAS
.
- A. D. Richards, A. Rodger, M. J. Hannon and A. Bolhuis, Intl. J. Antimicrobial Agents, 2009, 33, 469 CrossRef CAS PubMed
.
- A. Anthonysamy, S. Balasubramanian, V. Shanmugaiah and N. Mathivanan, Dalton Trans., 2008, 2136 RSC
.
- K. A. Kumar, K. L. Reddy, S. Vidhisha and S. Satyanarayana, Appl. Organomet. Chem., 2009, 23, 409 CrossRef CAS
.
- B. Biersack, R. Diestel, C. Jagusch, F. Sasse and R. Schobert, J. Inorg. Biochem., 2009, 103, 72 CrossRef CAS PubMed
.
- A. Bolhuis, L. Hand, J. E. Marshall, A. D. Richards, A. Rodger and J. Aldrich-Wright, Eur. J. Pharm. Sci., 2011, 42, 313 CrossRef CAS PubMed
.
- P. L. Lam, G.-L. Lu, K.-M. Hon, K.-W. Lee, C.-L. Ho, X. Wang, J. C.-O. Tang, K.-H. Lam, R. S.-M. Wong, S. H.-L. Kok, Z.-X. Bian, H. Li, K. K.-H. Li, R. Gambari, C.-H. Chui and W.-Y. Wong, Dalton Trans., 2014, 43, 3949 RSC
.
- F. P. Dwyer, E. C. Gyarfas, W. P. Rogers and J. H. Koch, Nature, 1952, 170, 190 CrossRef CAS
.
- F. P. Dwyer, I. K. Reid, A. Shulman, G. M. Laycock and S. Dixson, Aust. J. Exp. Biol. Med. Sci., 1969, 47, 203 CrossRef CAS
.
- F. Li, Y. Mulyana, M. Feterl, J. Warner, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 5032 RSC
.
- F. Li, M. Feterl, Y. Mulyana, J. M. Warner, J. G. Collins and F. R. Keene, J. Antimicrob. Chemother., 2012, 67, 2686 CrossRef CAS PubMed
.
- M. Pandrala, F. Li, M. Feterl, Y. Mulyana, J. M. Warner, L. Wallace, F. R. Keene and J. G. Collins, Dalton Trans., 2013, 42, 4686 RSC
.
- F. Li, E. J. Harry, A. L. Bottomley, M. D. Edstein, G. W. Birrell, C. E. Woodward, F. R. Keene and J. G. Collins, Chem. Sci., 2014, 5, 685 RSC
.
- M. Pandrala, F. Li, L. Wallace, P. J. Steel, B. Moore II, J. Autschbach, J. G. Collins and F. R. Keene, Aust. J. Chem., 2013, 66, 1065 CrossRef CAS
.
- M. J. Hannon, Pure Appl. Chem., 2007, 79, 2243 CrossRef CAS
.
- B. M. Zeglis, V. C. Pierre and J. K. Barton, Chem. Commun., 2007, 4565 RSC
.
- F. R. Keene, J. A. Smith and J. G. Collins, Coord. Chem. Rev., 2009, 253, 2021 CrossRef CAS PubMed
.
- M. R. Gill and J. A. Thomas, Chem. Soc. Rev., 2012, 41, 3179 RSC
.
- P. Lincoln and B. Nordén, Chem. Commun., 1996, 2145 RSC
.
- J. Malina, M. J. Hannon and V. Brabec, Chem. – Eur. J., 2008, 14, 10408 CrossRef CAS PubMed
.
- M. R. Gill, J. Garcia-Lara, S. J. Foster, C. Smythe, G. Battaglia and J. A. Thomas, Nat. Chem., 2009, 1, 662 CrossRef CAS PubMed
.
- U. McDonnell, J. M. C. A. Kerchoffs, R. P. M. Castineiras, M. R. Hicks, A. C. G. Hotze, M. J. Hannon and A. Rodger, Dalton Trans., 2008, 667 RSC
.
- X.-L. Zhao, Z.-S. Li, Z.-B. Zheng, A.-G. Zhang and K.-Z. Wang, Dalton Trans., 2013, 42, 5764 RSC
.
- D. R. Boer, L. Wu, P. Lincoln and M. Coll, Angew. Chem., Int. Ed., 2014, 53, 1949 CrossRef CAS PubMed
.
- A. K. Renfrew, Metallomics, 2014, 6, 1324 RSC
.
- B. Therrien, G. Süss-Fink, P. Govindaswamy, A. K. Renfrew and P. J. Dyson, Angew. Chem., Int. Ed., 2008, 47, 3773 CrossRef CAS PubMed
.
- G. Süss-Fink, Dalton Trans., 2010, 39, 1673 RSC
.
- X. Li, X.-J. Li, Z.-Y. Wu and C.-W. Yan, New J. Chem., 2012, 36, 2472 RSC
.
- W. Luo, X.-G. Meng, G.-Z. Chen and Z.-P. Ji, Inorg. Chim. Acta, 2009, 362, 551 CrossRef CAS PubMed
.
- Y. Mulyana, D. K. Weber, P. D. Buck, C. A. Motti, J. G. Collins and F. R. Keene, Dalton Trans., 2011, 40, 1510 RSC
.
- N. C. Fletcher, M. Nieuwenhuyzen and S. Rainey, J. Chem. Soc., Dalton Trans., 2001, 2641 RSC
.
- P. M. van Vliet, J. G. Haasnoot and J. Reedijk, Inorg. Chem., 1994, 33, 1934 CrossRef CAS
.
-
E. A. Seddon and K. R. Seddon, The Chemistry of Ruthenium, Elsevier, Amsterdam, 1984 Search PubMed
.
- M. G. Sauaia, E. Tfouni, R. H. de Almeida Santos, M. T. do Prado Gambardella, M. P. F. M. Del Lama, L. F. Guimaraes and R. S. da Silva, Inorg. Chem. Commun., 2003, 6, 864 CrossRef
.
- A. Juris, V. Balzani, F. Barigelletti, S. Campagna, P. Belser and A. Von Zelewsky, Coord. Chem. Rev., 1988, 84, 85 CrossRef CAS
.
- Y. Kawanishi, N. Kitamura and S. Tazuke, Inorg. Chem., 1989, 28, 2968 CrossRef CAS
.
- A. B. P. Lever, Inorg. Chem., 1990, 29, 1271 CrossRef CAS
.
- A. Macatangay, G. Y. Zheng, D. P. Rillema, D. C. Jackman and J. W. Merkert, Inorg. Chem., 1996, 35, 6823 CrossRef CAS PubMed
.
- A. Bouskila, B. Drahi, E. Amouyal, I. Sasaki and A. Gaudemer, J. Photochem. Photobiol., A, 2004, 163, 381 CrossRef CAS PubMed
.
- M. Furue, N. Kuroda and S. Nozakura, Chem. Lett., 1986, 1209 CrossRef CAS
.
- M. Cavazzini, S. Quici, C. Scalera, F. Puntoriero, G. La Ganga and S. Campagna, Inorg. Chem., 2009, 48, 8578 CrossRef CAS PubMed
.
- M. Staffilani, E. Höss, U. Giesen, E. Schneider, F. Hartl, H.-P. Josel and L. De Cola, Inorg. Chem., 2003, 42, 7789 CrossRef CAS PubMed
.
- F. Li, M. Feterl, J. M. Warner, F. R. Keene and J. G. Collins, J. Antimicrob. Chemother., 2013, 68, 2825 CrossRef CAS PubMed
.
-
F. Li, PhD thesis, Inert dinuclear polypyridylruthenium(II) complexes as antimicrobial agents, University of New South Wales, Australia, 2013 Search PubMed
.
- J. L. Morgan, C. B. Spillane, J. A. Smith, P. D. Buck, J. G. Collins and F. R. Keene, Dalton Trans., 2007, 4333 RSC
.
- X. Hua and A. V. Zelewsky, Inorg. Chem., 1995, 34, 5791 CrossRef CAS
.
- T. Tagano, Inorg. Chim. Acta, 1992, 195, 221 CrossRef
.
- I. P. Evans, A. Spencer and G. Wilkinson, J. Chem. Soc., Dalton Trans., 1973, 204 RSC
.
-
Clinical and Laboratory Standards Institute, Performance Standards for Antimicrobial Susceptibility Testing: Nineteenth Informational Supplement M100-S19, CLSI, Wayne, PA, USA, 2009 Search PubMed
.
-
M. Motyl, K. Dorso, J. Barrett and R. Giacobbe, Current Protocols in Pharmacology, John Wiley & Sons, New York, 2005, 13A.3.1–13A.3.22 Search PubMed
.
- D. Bao, B. Millare, W. Xia, B. G. Steyer, A. A. Gerasimenko, A. Ferreira, A. Contreras and V. I. Vullev, J. Phys. Chem. A, 2009, 113, 1259 CrossRef CAS PubMed
.
| This journal is © The Royal Society of Chemistry 2014 |