 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Hydrogen bond architecture in crystal structures of N-alkylated hydrophobic amino acids†
C. H.
Görbitz
*,
A. B.
Leirvåg
and
Ø.
Jacobsen
Department of Chemistry, University of Oslo, P.O. Box 1033 Blindern, N0315 Oslo, Norway. E-mail: c.h.gorbitz@kjemi.uio.no
First published on 1st September 2014
Abstract
Herein we present the first systematic investigation of hydrogen bonding patterns and crystal packing arrangements of N-alkylated hydrophobic amino acids, including synthesis and single crystal structure determination of five new compounds.
1. Introduction
The crystal structures of hydrophobic amino acids, as chiral substances or racemates, permit systematic investigations of recurring hydrogen bonding patterns due to the absence of side chain functional groups that otherwise could have interfered with the interactions between the charged amino and carboxylate groups. All crystal structures of hydrophobic amino acids reported to date have the same basic layout. The side chains form distinct hydrophobic layers and the polar heads hydrophilic layers, Fig. 1a. The latter can in turn be divided into two distinct sheets, each incorporating two head-to-tail hydrogen-bonded chains. A total of five different types of sheets have been identified.1 The L1 sheet illustrated in Fig. 1b occurs in crystal structures of enantiomerically pure amino acids, but is also observed in structures of racemates where it is paired with its mirror image D1 sheet in the L1–D1 layer shown in Fig. 1a. The L2 sheet in Fig. 1c, with Z′ = 2, is favored for L-amino acids, while the LD sheet in Fig. 1d is reserved for racemates and quasiracemates,2 as it contains amino acid molecules with both L- and D-configuration (thus the name).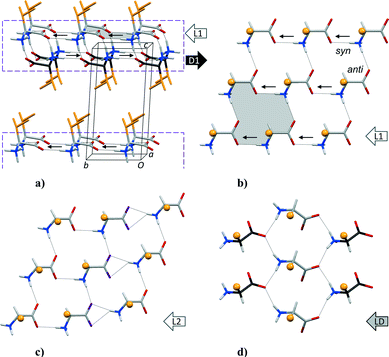 | ||
Fig. 1 (a) General packing arrangement of hydrophobic amino acids, exemplified by DL-Val.3 The C-atoms of the L-enantiomers are coloured in light grey while dark grey is used for the D-enantiomers. L1 and D1 denote the two sheets of opposite chirality constituting a hydrophilic L1–D1 layer in the structure (dashed box). Small arrows indicate the direction of the N–H bond vector in C(4) hydrogen-bonded chains13 towards carboxylate syn lone pairs. Side chains in orange make up a hydrophobic layer. (b) Detail of a L1 sheet. A 16-membered second level R44(16) ring system13 is highlighted in (a) and (b). (c) L2 layer with Z′ = 2 observed for most enantiomerically pure amino acids. (d) LD sheet with amino acids of both L- and D-configuration, as observed for racemates and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complexes between closely related amino acids (quasiracemates). Orange spheres represent side chains in (b)–(d). 1 complexes between closely related amino acids (quasiracemates). Orange spheres represent side chains in (b)–(d). | ||
The third amino H atom, which is not involved in hydrogen bonding within the sheet, serves to connect two adjacent sheets into a hydrophilic layer, Fig. 1a.3 Accordingly, alkyl substitution of this hydrogen could potentially result in a series of structures with a single-sheet architecture. Surprisingly, there have been no systematic investigations into the structures of such N-alkylated hydrophobic amino acids. An overview of N-substituted amino acids in the Cambridge Structural Database (CSD, version 5.35 of November 2013)4 is given in Table 1. Special attention has been given to structures where neither the N-substituent nor the regular side chain participates in strong hydrogen bonds. There are only five such entries in the CSD: N-methylglycine (sarcosine, CSD refcode YIHHON),5N-methyl-L-tryptophan (WAJBIS),6 which is a naturally occurring substance called abrine, N-(2-pyrimidyl-methyl)-L-alanine (QURSIG),7Nα,Nε,Nε-tri(cyanoethyl)-L-lysine (VEQZIB),8 which is a low molecular weight dendrimer with potential antimicrobial properties, and tritylglycine (LEFPUH),9 see Fig. 2.
| Group | Numbera | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| a Numbers in bold are the sum of the subgroup numbers in plain text below. b No interfering hydrogen bonds (adding to interactions between the amino and carboxylate groups). c Amino acid other than Gly. d YIHHON. e LEFPUH. f WAJBIS. g QURSIG, VEQZIB. h AZETAC, AZPCOH, DHPROX, EQIHOC, GAYCEO, GEVMOK, GULCUM, LEGBEG, MATVAE, MATVEI, NELSEC, PROLIN (L-Pro), QAMVAB, QANRUT (DL-Pro), WAVKIN. | ||||||||||
| All | 169 | |||||||||
| Organometallic | 5 | |||||||||
| Organic | 164 | 164 | ||||||||
| Acyclic N-atom | 82 | 82 | ||||||||
| N-R-Gly | 61 | 61 | ||||||||
| N-CH3 | 31 | 31 | ||||||||
| Salt or in a complex | 30 | |||||||||
| Isolated zwitterionic | 1 | |||||||||
| NiHB | 1d | |||||||||
| N-Other | 30 | 30 | ||||||||
| Salt or in a complex | 20 | |||||||||
| Dimeric | 2 | |||||||||
| Isolated zwitterionic | 8 | |||||||||
| NiHB | 1e | |||||||||
| N-R-Xaac | 21 | 21 | ||||||||
| N-CH3 | 3 | |||||||||
| NiHB | 1f | |||||||||
| N-Other | 18 | 18 | ||||||||
| Salt or in a complex | 8 | |||||||||
| Isolated zwitterionic | 10 | |||||||||
| NiHB | 2g | |||||||||
| Endocyclic N-atom | 82 | 82 | ||||||||
| Salt or in a complex | 41 | |||||||||
| Isolated zwitterionic | 41 | |||||||||
| NiHB | 15h | |||||||||
 | ||
| Fig. 2 N-Alkylated amino acids discussed in this paper. Previously investigated molecules5–12 have been identified by their CSD4 refcodes. | ||
To evaluate the hydrogen bonding properties of more general N-alkylated hydrophobic amino acids, we have synthesized five new substances (shown in Fig. 2). These were selected to explore the effect of variable side chain bulks, including an interchange of hydrophobic groups between the amino group and the Cα carbon atom [N-isopropyl-L-phenylalanine (NiPrF) and N-benzyl-L-valine (NBnV)], and the effect of going from N-isopropyl-L-valine (NiPrV) to its racemate N-isopropyl-DL-valine (NiPrVR) as well as to the higher analogue N-isopropyl-L-leucine (NiPrL). There are 13 plain (NiHB) structures with an endocyclic N-atom in Table 1. Three of them proved to be of interest for the present investigation: L-proline itself (PROLIN)10 and the two derivatives GEVMOK11 and GULCUM12 (included in Fig. 2).
2. Results and discussion
Crystallographic data are listed in Table 2, while the molecular structures of NiPrV, NiPrVR, NiPrL, NiPrF and NBnV are shown in Fig. 3. Torsion angles are listed in Table 3.| NiPrV | NiPrVR | NiPrL | NiPrF | NBnV | |
|---|---|---|---|---|---|
| a Computer programs used: APEX2 (Bruker, 2007), SAINT-Plus (Bruker, 2007), SHELXTL (Sheldrick, 2008), SADABS (Sheldrick, 1996). | |||||
| Crystal data | |||||
| Chemical formula | C8H17NO2 | C8H17NO2 | C9H19NO2 | C12H17NO2 | C12H17NO2 |
| M r | 159.23 | 159.23 | 173.25 | 207.27 | 207.27 |
| Crystal system, space group | Monoclinic, P21 | Orthorhombic, Pbcn | Orthorhombic, P212121 | Monoclinic, P21 | Orthorhombic, P212121 |
| a (Å) | 5.378(2) | 17.382(4) | 9.2948(18) | 10.750(2) | 5.766(3) |
| b (Å) | 9.788(4) | 10.705(2) | 10.790(2) | 9.407(2) | 12.082(5) |
| c (Å) | 8.399(3) | 9.608(2) | 19.871(4) | 11.079(2) | 16.559(7) |
| β (°) | 101.676(4) | 90.0 | 90.0 | 95.013(3) | 90.0 |
| V (Å3) | 433.0(3) | 1787.8(6) | 1993.0(7) | 1116.1(4) | 1153.7(9) |
| Z | 2 | 8 | 8 | 4 | 4 |
| Radiation | Mo Kα | Mo Kα | Mo Kα | Mo Kα | Cu Kα |
| Wavelength (Å) | 0.71073 | 0.71073 | 0.71073 | 0.71073 | 1.54178 |
| μ (mm−1) | 0.086 | 0.083 | 0.080 | 0.084 | 0.649 |
| Temperature (K) | 150(2) | 296(2) | 106(2) | 100(2) | 100(2) |
| Crystal size (mm) | 0.75 × 0.65 × 0.34 | 0.54 × 0.17 × 0.015 | 0.60 × 0.32 × 0.17 | 0.51 × 0.29 × 0.05 | 0.32 × 0.03 × 0.008 |
| Data collection | |||||
| T min, Tmax | 0.971, 0.640 | 0.998, 0.778 | 0.986, 0.852 | 0.996, 0.761 | 0.999, 0.801 |
| No. of measured, independent and observed [I > 2σ(I)] reflections | 2597, 1051, 939 | 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 807, 1569, 853 807, 1569, 853 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 428, 2069, 1819 428, 2069, 1819 |
7686, 2091, 1680 | 2252, 689, 495 |
| R int | 0.044 | 0.046 | 0.031 | 0.031 | 0.099 |
| θ max (°) | 28.45 | 25.01 | 25.28 | 25.02 | 39.82 |
| Refinement | |||||
| R[F2 > 2σ(F2)], wR(F2), S | 0.046, 0.130, 1.09 | 0.041, 0.129, 1.012 | 0.039, 0.091, 1.077 | 0.038, 0.095 | 0.066, 0.123 |
| No. of reflections | 1051 | 1569 | 2069 | 2091 | 689 |
| No. of parameters | 104 | 100 | 217 | 275 | 136 |
| Δρmax, Δρmin (e Å−3) | 0.335, −0.232 | 0.193, −0.171 | 0.207, −0.203 | 0.124, −0.190 | 0.177, −0.160 |
| Structure | NiPrV | NiPrVR | NiPrL | NiPrF | NBnV |
|---|---|---|---|---|---|
| a C6 (NiPrV, NiPrVR, NBnV), C7 (NiPrL) or C10 (NiPrF). b C7 (NiPrV, NiPrVR, NBnV), C8 (NiPrL) or C11 (NiPrF). c C8 (NiPrV, NiPrVR, NBnV), C9 (NiPrL) or C12 (NiPrF). | |||||
| N1–C2–C1–O1 | −31.3(3) | −31.1(3) | −35.1(3) | −37.2(3) | −16.4(16) |
| −40.6(3) | −43.3(3) | ||||
| N1–C2–C3–C4 | −165.9(2) | −172.5(2) | 178.09(19) | −173.2(2) | −160.3(11) |
| −175.76(19) | −169.2(2) | ||||
| N1–C2–C3–C5 | 69.9(3) | 62.2(3) | 74.1(15) | ||
| C2–C3–C4–C5 | −174.0(2) | 63.4(4) | |||
| 151.7(2) | 93.8(3) | ||||
| C2–C3–C4–C6/9 | 64.1(3) | −117.5(3) | |||
| −85.1(3) | −87.1(4) | ||||
| C1–C2–N1–Ca | −65.1(3) | −85.7(2) | −59.6(2) | −63.0(3) | −60.9(13) |
| −61.2(2) | −56.4(3) | ||||
| C2–N1–Ca–Cb | −67.7(3) | 55.4(3) | −58.4(3) | −85.8(3) | −40.2(15) |
| 179.15(19) | −62.9(3) | ||||
| C2–N1–Ca–Cc | 170.8(2) | −178.9(2) | −71.9(2) | 148.6(2) | |
| 165.64(19) | 175.0(2) | ||||
| N1–C6–C7–C8 | −63.5(16) | ||||
For NiPrL and NiPrF there are two molecules in the asymmetric unit. These differ primarily with regards to the orientation of the regular side chain. In the crystal structure of NiPrL the side chain of molecule A adopts the most common conformation for the side chain of L-Leu, with N1A–C2A–C3A–C4A (χ1) = trans and C2A–C3A–C4A–C5A/C6A (χ2,1/χ2,2) = trans/gauche+, as seen for 14 out of 15 zwitterionic L-Leu molecules (after inversion of some D-Leu molecules) in the CSD. Molecule B on the other hand has a unique, χ1 = trans, χ2,1/χ2,2 = trans/gauche−, conformation not previously observed for unprotected, zwitterionic L-Leu. Large deviations from the ideal staggered geometry (dihedral angles of ±60 and 180°) point to a strained molecular geometry caused by packing interactions, as will be discussed in more detail below. The two molecules in NiPrF adopt the same overall conformation, but with a >30° difference between molecules A and B for the dihedral angle C2–C3–C4–C5. The Val residues in NiPrV, NiPrVR and NBnV share a conformation with N1–C2–C3–C4/C5 (χ1,1/χ1,2) = trans/gauche+, which in crystal structures of L-Val occur with about the same frequency as trans/gauche− (eight and seven observations, respectively). The third rotamer, gauche+/gauche−, is less common with three previous observations. The N-isopropyl group has the same orientation in all molecules in Fig. 3 except NiPrV.
The unit cells and crystal packing arrangements of NiPrV, NiPrVR, NiPrL, and NiPrF are depicted in Fig. 4.
 | ||
| Fig. 4 Unit cell and crystal packing arrangement of (a) NiPrV, (b) NiPrVR, (c) NiPrL and (d) NiPrF. H atoms bonded to C atoms have been omitted for clarity. | ||
Together with WAJBIS, QURSIG and VEQZIB these compounds form the anticipated molecular monolayers. In principle, one of the familiar hydrogen bonded sheets seen in crystal structures of the regular amino acids, such as L1 and L2 illustrated in Fig. 1, could have been retained for their N-alkylated counterparts, which would have led to molecular monolayers with the regular side chains and N-alkyl groups positioned on opposing faces, Fig. 5.
 | ||
| Fig. 5 Hypothetical structure of a molecular monolayer of an N-alkylated amino acid as it would have appeared if a L1 hydrogen bonding pattern had been retained. Color coding as in Fig. 2, alkyl group appear as small spheres. | ||
Instead, we find that a slightly modified type of sheet is used in these structures, Fig. 6.
 | ||
| Fig. 6 Hydrogen-bonded sheets in (a) NiPrV, (b) NiPrVR, (c) NiPrF, (d) NiPrL, (e) WAJBIS6 and (f) GEVMOK.11 Hydrogen bonds shorter than 2.6 Å are indicated by dotted lines. The weaker component in the three-center interaction indicated for WAJBIS (arrow) is 2.46 Å (after normalization of the N–H distance to 0.92 Å). In other structures the corresponding distances are >2.67 Å. Two C(4) chains have been highlighted in (a), arrows indicate the directions of the N–H bond vectors. The blue shade gives the size of the repeating pattern of NiPrV (in this case equivalent to the footprint of the unit cell). The same shade (on the same scale) appears in (e). A R44(16) ring system is shaded in (b). | ||
Compared to the sheets in Fig. 1 every second N–H⋯O (syn) head-to-tail chain is here flipped in the opposite direction. This has the effect of positioning both side chains and N-alkyl groups on alternating sides of the hydrogen-bonded sheet, unlike the model shown in Fig. 5. At the same time are not only two chains with first level graph set13C(4) chains retained, but also second level R44(16) sixteen-membered rings that are related, but not identical to those of the L1 sheet. As in the L1 and L2 sheets, N–H⋯O (anti) head-to-tail chains are parallel in Fig. 6. This means that all such chains are in fact running in the same direction in the monoclinic structures of NiPrV and NiPrL in Fig. 4, while in the orthorhombic structure of NiPrF directions are opposite in adjacent layers.
NiPrL and NiPrF both have Z′ = 2, but display the same hydrogen bonding pattern as NiPrV and other structures in the group. The difference between a L1 layer with Z′ = 1 and a L2 layer with Z′ = 2 for regular amino acids in Fig. 1b and c is thus not reproduced here. This means that the increase from Z′ = 1 to Z′ = 2 for NiPrL and NiPrF is due to better stacking of hydrophobic groups rather than an improved hydrogen bonding arrangement. Upon closer inspection, the pattern of A and B molecules in the crystal structures of NiPrL and NiPrF in Fig. 6c and d are dissimilar, so these molecules adapt to the challenges of stacking their side chains in slightly different manners, in addition to the shift from a monoclinic to an orthorhombic space group.
Although we had expected NBnV to belong to the same structural family as the other four substances investigated here, already the crystallization behavior (see below) suggested that this was not the case. The crystal structure shown in Fig. 7a may at first seem to share some features with e.g. its “retroanalogue” NiPrF, but in fact there is no hydrogen bonded sheet in NBnV, only a hydrogen bonded tape composed of a series of fused 11-membered rings, Fig. 7b.
 | ||
| Fig. 7 (a) Unit cell and crystal packing of NBnV. (b) Hydrogen bonding pattern of NBnV. Color coding and style as in Fig. 4 and 5. | ||
The density of NBnV, 1.193 g cm−3, is slightly lower than 1.234 g cm−3 for NiPrF, so the shift of hydrogen bonding pattern is apparently not driven by a more efficient crystal packing arrangement. In this connection, compounds with endocyclic N-atoms in five-membered rings provide interesting, additional information. Together with GEVMOK11 and GULCUM,12L-proline (PROLIN),10 take on the same hydrogen bonding pattern as the acyclic group, Fig. 6f. If, however, the –CγH2– group is substituted with –S– (thioproline, NELSEC),14 the pattern is shifted to that of NBnV (apparently the only other occurrence of this kind of tape together with (2R,1′S)-2-(1′-benzyl-2′-hydroxyethylamino)-4-phenylbutanoic acid (AZABEI)).15 Methylene and sulfur are of comparable sizes, showing that minute changes to molecular structure can have a profound impact on the overall crystal packing arrangement.
For PROLIN, GEVMOK and GULCUM we furthermore note that the ring system forces the side chain and the N-alkyl substituent of an individual molecule to be on the same side of a hydrogen-bonded sheet, Fig. 8e, and not on opposite sides as in Fig. 8b–d. We believe this inherent rigidity is responsible for the fact that 10 out of 13 compounds with an endocyclic N-atom in Table 1 form other types of hydrogen bonding patterns.
 | ||
Fig. 8 Crystal packing of (a) L-Ile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D-Ala (FITHIZ),16 (b) N-methyl-L-tryptophan (WAJBIS),6 (c) N-(2-pyrimidylmethyl)-L-alanine (QURSIG),7Nα,Nε,Nε-tri(cyanoethyl)-L-lysine (VEQZIB),8 and (e) L-proline (PROLIN).10 Color coding as in Fig. 3. The C-atoms in the ring system of L-proline appear in light green. D-Ala (FITHIZ),16 (b) N-methyl-L-tryptophan (WAJBIS),6 (c) N-(2-pyrimidylmethyl)-L-alanine (QURSIG),7Nα,Nε,Nε-tri(cyanoethyl)-L-lysine (VEQZIB),8 and (e) L-proline (PROLIN).10 Color coding as in Fig. 3. The C-atoms in the ring system of L-proline appear in light green. | ||
Two previous structures identified in Table 1 also do not share the hydrogen bonding pattern shown in Fig. 6. Sarcosine (YIHHON),5 which has a single methyl group only, is able to generate a high density crystal structure (1.309 g cm−3) with a three-dimensional hydrogen bonding pattern. LEFPUH,9 on the other hand, has three bulky phenyl groups that are incompatible with formation of even a 2-D hydrogen bonded sheets. Hydrogen bonding is in this case reduced to a single 1-D chain that leaves one of the two NH hydrogen bond donors unused (instead, the aromatic groups serve as acceptors in weaker interactions).
The hydrogen pattern in Fig. 6 is very flexible and can accommodate a wide range of side chains and N-substituents. The periodicity along the N–H⋯O (syn) interaction is almost constant, with a very limited 0.06 Å range for the values listed in Table 4, but the periodicity along the N–H⋯O (anti) interaction varies considerably over a 1.19 Å range. Obviously, hydrogen bond lengths in the two perpendicular directions are affected, as seen from the list of N⋯O distances in Table 4 (complete hydrogen bond geometries are available as ESI†). The explanation for this variability lies in the size and shape of the hydrophobic moieties.
| Structure | Anti chain/Å | Syn chain/Å | Area/Å2 | N⋯O (anti) | N⋯O (anti) |
|---|---|---|---|---|---|
| a Sorted on decreasing length for the axis length in the anti direction, except for proline and its two derivatives with endocyclic N-atoms. b Blue rectangles in Fig. 6. c Unit cell length divided by 2. d No s.u.'s provided. | |||||
| NiPrV | 9.787(4) | 5.378(2) | 52.63b | 2.951(3) | 2.736(3) |
| NiPrVR | 9.608(2) | 5.353(1)c | 51.53 | 2.909(3) | 2.799(2) |
| NiPrF | 9.407(2) | 5.375(1)c | 50.56 | 2.854(3) | 2.758(3) |
| 2.805(3) | 2.735(3) | ||||
| NiPrL | 9.2948(18) | 5.395(1)c | 50.15 | 2.794(3) | 2.730(2) |
| 2.800(2) | 2.772(2) | ||||
| QURSIG | 8.7009(6) | 5.3353(4) | 46.42 | 2.782(1) | 2.719(1) |
| VEQZIB | 8.6252(10) | 5.3589(5)c | 46.22 | 2.690(1) | 2.759(1) |
| 2.729(1) | 2.742(1) | ||||
| WAJBIS | 8.595(1) | 5.372(1) | 46.17b | 2.676(5) | 2.743(5) |
| — | |||||
| PROLIN | 9.020 | 5.200 | 46.90 | 2.710 | 2.686 |
| GEVMOK | 9.752(3) | 5.2193(17) | 50.90 | 2.764(2) | 2.736(2) |
| GULCUM | 8.7431(8) | 5.3140(6) | 46.46 | 2.822d | 2.738d |
WAJBIS
6 and QURSIG7 are interesting in that both compounds have methyl groups. For regular, hydrophobic amino acids methyl groups are usually too small to participate in layer formation. Our own efforts to prepare quasiracemic L![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D amino acid complexes with Ala as one of the components were uniformly unsuccessful except for L-Ile
D amino acid complexes with Ala as one of the components were uniformly unsuccessful except for L-Ile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D-Ala (FITHIZ),16 where a nice fit is observed between surfaces where Ile ridges fit into Ala grooves, Fig. 8a. For WAJBIS, which represents an extreme value for the periodicity along the N–H⋯O (anti) interaction at 8.595(1) Å, Fig. 6e, the methyl groups serve only to fill voids close to the polar heads, the surface of a molecular monolayer essentially being formed by the bulky Trp indole groups, Fig. 8b. For QURSIG, Fig. 8c, the situation is rather similar, although the roles of the two hydrophobic groups have been reversed. The side chain methyl group is, however, more exposed than the N-methyl group of WAJBIS and forms intermolecular contacts across the hydrophobic layer as in the L-Ile
D-Ala (FITHIZ),16 where a nice fit is observed between surfaces where Ile ridges fit into Ala grooves, Fig. 8a. For WAJBIS, which represents an extreme value for the periodicity along the N–H⋯O (anti) interaction at 8.595(1) Å, Fig. 6e, the methyl groups serve only to fill voids close to the polar heads, the surface of a molecular monolayer essentially being formed by the bulky Trp indole groups, Fig. 8b. For QURSIG, Fig. 8c, the situation is rather similar, although the roles of the two hydrophobic groups have been reversed. The side chain methyl group is, however, more exposed than the N-methyl group of WAJBIS and forms intermolecular contacts across the hydrophobic layer as in the L-Ile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) D-Ala complex.
D-Ala complex.
The isopropyl groups of NiPrV evidently build considerable strain into the network, as the periodicity along the N–H⋯O (anti) reaches the maximum value of 9.788(4) Å, Fig. 6a. A further increase in side chain bulk to sec-butyl in NiPrL or benzyl in NiPrF forces a Z′ = 2 system. VEQZIB,8 also with Z′ = 2, illustrates how the separation between hydrophilic layers can be increased dramatically when the combined size of the two hydrophobic groups in the molecule becomes very large, Fig. 8d.
A most interesting comparison can be made between the structure of NiPrV and its racemate NiPrVR. Although examples exist, such as L-Ala and DL-Ala,17 it is very uncommon for a racemate to share hydrogen bonding pattern and general packing arrangement with the corresponding enantiomerically pure compound. This is the situation for NiPrV and NiPrVR, as can be seen from Fig. 4a and b and Fig. 6a and b, respectively. Compared to the structure of NiPrV, the roles of the side chain and the N-isopropyl group with respect to space filling are reversed in the structure of NiPrVR, rendered possible also by a change in N-isopropyl conformation as described above. It follows that the display of isopropyl groups on both sides of a molecular monolayer gives two very similar surfaces, Fig. 9a and b. NiPrL is quite similar. The transition to Z′ = 2 can be seen (Fig. 9c) as the result of an alternating set of conformations for the side chains. The surface of NiPrV is also closely reminiscent of the hydrophobic surface of a molecular double-layer in DL-Val, Fig. 1a and 9d, while the corresponding surface for L-Val18 shows yet another way of arranging these groups (illustration available as ESI†).
3. Experimental
NiPrV, NiPrL and NiPrF were prepared by reductive amination of acetone with the corresponding proteinogenic amino acids, using sodium cyanoborohydride (NaBH3CN) as reducing agent. NiPrVR was prepared by mixing equimolar amounts of L-NiPrV and D-NiPrV. NBnV was prepared by reductive amination of benzaldehyde with L-Val. See the ESI† for experimental details. Single crystals of NiPrV, NiPrVR, NiPrL and NBnV were obtained by dissolving about 0.2–0.5 mg of each compound in 30 μl of water in a small test tube which subsequently was sealed with Parafilm®. After pricking a hole in the film with a needle, the tube was set to equilibrate inside a larger tube filled with about 1 ml of acetonitrile. The same method was used for NiPrF, but with hexafluoro-2-propanol as the solvent and water as the precipitating agent. Well-shaped crystals formed within a week for all substances except NBnV, which yielded long (several mm) but exceedingly thin needles that easily bent and fractured. These also had a pronounced right-handed twist. One needle was cut with a scalpel to 0.30 mm length for collection of X-ray data on a Bruker SMART APEX II CCD diffractometer equipped with an Oxford Cryostream low temperature device. Data integration/reduction and absorption correction were carried out by the programs SAINT and SADABS,19 respectively, while SHELXTL20 was used for refinements. No structural disorder was discovered. Heavy atoms were refined with anisotropic displacement parameters, whereas all hydrogen atoms were constrained to theoretical positions. Some restraints were used in the refinement of NBnV; details are in the SHELXL .res file which is part of the submitted cif. Molecular images and packing graphics were generated using the program Mercury.4,21Conclusions
In summary, the available set of ten related structures, including six compounds studied previously and four out of the five studied here, demonstrates a remarkably robust and versatile hydrogen bonding arrangement for N-alkylated amino acids that is compatible with a wide range of hydrophobic side chains and N-alkyl groups.Notes and references
- C. H. Görbitz, K. Vestli and R. Orlando, Acta Crystallogr., Sect. B: Struct. Sci., 2009, 65, 393–400 Search PubMed.
- C. H. Görbitz, K. Rissanen, A. Valkonen and Å. Husabø, Acta Crystallogr., Sect. C: Cryst. Struct. Commun., 2009, 65, o267–o272 Search PubMed.
- R. Flaig, T. Koritsanszky, B. Dittrich, A. Wagner and P. Luger, J. Am. Chem. Soc., 2002, 124, 3407–3417 CrossRef CAS PubMed.
- F. Allen, Acta Crystallogr., Sect. B: Struct. Sci., 2002, 58, 380–388 CrossRef PubMed.
- B. Dittrich and M. A. Spackman, Acta Crystallogr., Sect. A: Found. Crystallogr., 2007, 63, 426–436 CAS.
- J. Seetharaman, S. S. Rajan and R. Srinivasan, J. Crystallogr. Spectrosc. Res., 1993, 23, 167–170 CrossRef CAS.
- X. Li, T. Liu, B. Hu, G. Li, H. Zhang and R. Cao, Cryst. Growth Des., 2010, 10, 3051–3059 CAS.
- J. Janiszewska and Z. Urbańczyk-Lipkowska, J. Mol. Struct., 2006, 789, 112–117 CrossRef CAS PubMed.
- M. Rieckhoff, M. Noltemeyer, F. T. Edelman, I. Haiduc and I. Silaghi-Dumitrescu, J. Organomet. Chem., 1993, 469, C19–C21 CrossRef.
- R. L. Kayushina and B. K. Vainshtein, Kristallografiya, 1965, 10, 833–844 Search PubMed.
- S. Hanessian, Z. Shao and J. S. Warrier, Org. Lett., 2006, 8, 4787–4790 CrossRef CAS PubMed.
- E. Cini, G. Giorgi, M. Rodriquez and M. Taddei, Synlett, 2009, 1562–1566 CAS.
- M. C. Etter, J. C. MacDonald and J. Bernstein, Acta Crystallogr., Sect. B: Struct. Sci., 1990, 46, 256–262 CrossRef.
- N. Grant, M. F. Ward, M. Jaspars and W. T. A. Harrison, Acta Crystallogr., Sect. E: Struct. Rep. Online, 2001, 57, o697–o699 CAS.
- D. Berkeš, A. Kolarovič, R. G. Raptis and P. Baran, J. Mol. Struct., 2004, 697, 101–107 CrossRef PubMed.
- B. Dalhus and C. H. Görbitz, Acta Crystallogr., Sect. B: Struct. Sci., 1999, 55, 424–431 CrossRef PubMed.
- R. Destro, R. Soave and M. Barzaghi, J. Phys. Chem. B, 2008, 112, 5163–5174 CrossRef CAS PubMed.
- B. Dalhus and C. H. Görbitz, Acta Chem. Scand., 1996, 50, 544–548 CrossRef CAS PubMed.
- Bruker APEX2, SAINT-Plus and SADABS (2007), Bruker AXS Inc., Madison, Wisconsin, USA Search PubMed.
- G. Sheldrick, Acta Crystallogr., Sect. A: Found. Crystallogr., 2008, 64, 112–122 CrossRef CAS PubMed.
- C. F. Macrae, I. J. Bruno, J. A. Chisholm, P. R. Edgington, P. McCabe, E. Pidcock, L. Rodriguez-Monge, R. Taylor, J. van de Streek and P. A. Wood, J. Appl. Crystallogr., 2008, 41, 466–470 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Additional illustrations of hydrogen bonding and crystal packing, synthetic details. CCDC 985300–985304. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c4ce01412j |
| This journal is © The Royal Society of Chemistry 2014 |


