Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Compact multi-enzyme pathways in P. pastoris†
Martina
Geier‡
a,
Pia
Fauland‡
a,
Thomas
Vogl
b and
Anton
Glieder
*b
aAustrian Centre of Industrial Biotechnology (ACIB GmbH), Petersgasse 14, 8010 Graz, Austria
bInstitute of Molecular Biotechnology, NAWI Graz & DK Molecular Enzymology, TU Graz, Petersgasse 14, 8010 Graz, Austria. E-mail: anton.glieder@tugraz.at
First published on 3rd December 2014
Abstract
We report for the first time the functional simultaneous expression of nine genes from a single 2A peptide based polycistronic expression construct. The feasibility and arising opportunities for the biosynthetic pathway balancing for chemical production were demonstrated by the co-expression of the violacein and carotenoid biosynthesis pathways.
Implementing natural and synthetic pathways into microorganisms provides new opportunities for the production of chemical building blocks, food and feed ingredients and active pharmaceutical ingredients (APIs). Thus, an emerging challenge nowadays is not to produce single proteins only, but multiple ones in sufficient amounts and with balanced activities along the pathway in order to avoid accumulation of side products or intermediates.
Commonly, multigene expression is based on co-expression constructs harbouring the pathway genes under the separate control of the (same) promoter and terminator. This usually works sufficiently well for proof-of-concept studies. However, the transformation rates of microbial cells generally decrease with the increasing size of the expression construct, while on the other hand technological difficulties and the costs for labour and materials increase. More importantly, the repeated use of homologous sequences can result in recombination events, and thus in genetic instability.1 This is of special importance if physiologically problematic proteins or metabolites are produced at high levels and over extended periods. However, the genetic stability of production strains is essential for economically viable industrial processes and to maintain the product quality over extended production times. One strategy to reduce the loss of genes by homologous recombination is the use of different promoter and terminator sequences for each individual gene of the pathway. In contrast to the repeated use of the same promoter, this strategy also enables us to balance the individual pathway catalysts at the level of transcription in order to avoid bottlenecks in the pathway and in favour of a high direct flux to the desired product. Alternatively, the number of regulatory elements can be reduced by the expression of multiple genes from a single, polycistronic transcript. While this is simple to achieve in prokaryotes, eukaryotes generally do not express polycistronic operons. One way to achieve polycistronic expression is the use of internal ribosome entry sites (IRES).2 These sequences are able to initiate translation at internal sites within a polycistronic transcript. However, the employment of IRES in biotechnological applications is limited as the sequences are rather large (∼500 nucleotides) and, more importantly, result in as much as 10 fold lower expression of the downstream-encoded protein.3–5 Due to this fact this technology can hardly be applied for multigene expression. An attractive alternative constitutes the use of self-processing 2A sequences.6 These 2A sequences, also known as cis-acting hydrolase elements, are short peptides (up to 20 amino acids) originating from viral polyproteins. 2A peptides act co-translationally causing a ribosomal skip that terminates translation at the final proline codon of its C-terminally located conserved sequence “NPGP”.4,7 Thus, discrete proteins can be produced from a single transcript. It has to be noted that proteins located upstream to the 2A peptide keep this short stretch of amino acids as a C-terminal extension, while proteins located downstream of it contain an N-terminal proline. 2A peptides have been used for the co-expression of two genes in a variety of eukaryotic systems including yeasts, plants and mammalian cell lines8–10 and the stoichiometric coproduction of individual peptides was seen as an advantage compared to the IRES technology.5 However, careful analysis employing in vitro translation studies showed imbalances in protein levels for 2A mediated co-translation, too. Proteins that are encoded upstream of the 2A sequence accumulated to higher levels than those encoded downstream, and partial fusion protein formation was observed.4
In a few cases the viral 2A co-translation strategy was successfully employed to express short biosynthetic pathways.11,12 Up to four genes have been expressed in a polycistronic format to demonstrate possible applications for gene therapies.13,14 However, additional necessary genes so far have been expressed either by employing an additional promoter or from a separate expression cassette. As many interesting natural biosynthetic routes consist of more than four genes, our intention was to investigate whether 2A peptides are also suitable for the coordinate expression of longer pathways. The expression host for our study was the methylotrophic yeast Pichia pastoris (Komagataella phaffii), which might be especially suitable to accept multiple short 2A peptide sequences in close proximity due to its low homologous recombination activity compared to baker's yeast. In addition, it is simple to grow this yeast to high cell densities and it is capable to express rather complex proteins functionally.15
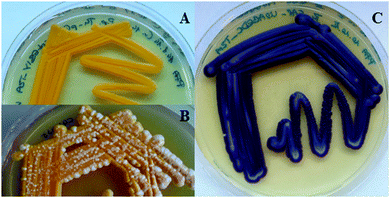
In the first step, a set of different 2A sequences was tested for functionality in P. pastoris. In addition to the FMDV2A sequence of the foot-and-mouth disease virus, which was already shown to be useful in P. pastoris,16 the P2A sequence of porcine teschovirus-1 and the T2A sequence of Thosea asigna virus were employed for polycistronic expression trials.13 Therefore, expression constructs harbouring the fluorescent proteins eGFP and sTomato separated by the individual 2A sequences as well as an N-terminal His-tag for Western blot analysis of the resulting gene products were generated (Fig. S1, ESI†). The observed total expression levels were in the same order of magnitude independent of the employed 2A peptide. In addition, the coordinate expression of the fluorescent proteins was not influenced by the position of the corresponding genes within the polycistron as deduced from fluorescence measurements (Fig. S2, ESI†). However, the gene position might be a critical parameter if more complex or more than 2 proteins are produced using the 2A peptides. Western blot analysis revealed that the T2A and P2A based constructs resulted in a substantial amount of discrete protein products, but with a small portion of the fused proteins (Fig. S3, ESI†). The amount of fusion protein left slightly varied with the order of the genes coding for the two fluorescent proteins. On the other hand, the efficiency of the FMDV2A sequence to produce individual proteins varied strongly yielding also recombinant strains that only produced the protein fusion. These findings were in accordance with reports from the literature.16 Consequently, the T2A peptide from Thosea asigna was chosen to set up the carotenoid biosynthesis pathway from P. ananatis consisting of four genes (the T2A coding sequence was employed in different nucleotide sequence versions to avoid sequence identities). Similar to the 2A peptide-based expression of a three gene carotenoid pathway in S. cerevisiae,11 the polycistronic expression of the four genes in Pichia resulted in strains that displayed a stable orange phenotype (Fig. 1A), an important feature for industrial strain construction. In contrast, the majority of strains based on co-expression constructs harbouring each carotenoid pathway gene under the separate control of the same regulatory elements showed a heterogeneous phenotype, i.e. orange cells were overgrown by white ones (Fig. 1B). Strain analysis by PCR revealed that these white Pichia transformants have lost either one or several pathway genes indicating severe issues with the genetic stability.
The 2A peptide based expression concept was also successfully transferred to the violacein pathway from C. violaceum which required the co-expression of five genes in P. pastoris for the first time (Fig. 1C).
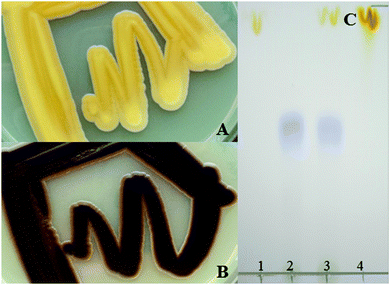
Encouraged by the surprisingly efficient transcription and translation of these long constructs the carotenoid and violacein pathways were combined to yield a polycistronic expression construct of nine genes. Intrigued by our observation that the order of the sequences influenced the efficiency of the 2A strategy to produce individual proteins and by reported nonstoichiometric peptide production we were interested to see if this can be used to engineer optimized and more balanced pathways. As a key experiment two different constructs were generated for the 9 target proteins, harbouring either the carotenoid pathway genes upstream of the violacein pathway or the other way round. The size of the resulting polycistronic transcript was ∼12 kb. The functional expression of both biosynthetic pathways was indicated by a brownish appearance of the yeast cells due to the accumulation of the orange and purple products of the two pathways, β-carotene and violacein (Fig. 2). However, the order of the pathway genes in the polycistronic construct had a significant effect on the pathway efficiency. If the violacein pathway genes were placed first, the resulting strains showed a clear brown phenotype already after 60 h of incubation, while the phenotype of strains harbouring the construct with the carotenoid pathway genes first was not that pronounced at this time, needing longer time to reach a similar brown colouring. A proportion of ribosomes might completely terminate translation at the 2A sequence thus omitting the translation of the remaining polycistronic transcript.4 This might occur more often when an increasing number of 2A sequences have to be read over. Partial fusion protein formation, as we had observed with the reporter proteins, might enhance this effect and the difference between alternative arrangements of individual components of the expression constructs. Whether premature termination of transcription is also a major factor to cause or enhance this positional effect is still under investigation. Nevertheless, for the first time this study shows that efficient polycistronic multigene expression is possible by positioning individual genes at different positions to fine tune the pathway expression. This is an attractive simple alternative to transcriptional regulation by different individual promoters. The short DNA sequences coding for 2A peptide sequences also provide future opportunity to serve as universal linkers for random combinatorial assembly of the individual coding sequences to optimize the order for improved and balanced expression of individual pathway components.
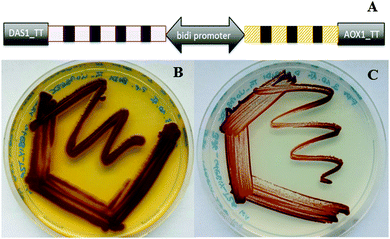
In an unprecedented evaluation of alternative compact pathway construction with even more genes and as an additional innovative strategy to tune the efficiency of individual pathway components we also combined the concept of polycistronic pathway expression with bidirectional promoters, short DNA sequences driving gene expression in both directions.17 Constructs harbouring the violacein and carotenoid biosynthesis pathways in a bidirectional polycistronic format were generated. Also this new expression strategy resulted in strains successfully producing the target products of both pathways (Fig. 3).
In conclusion, we have successfully expressed nine genes from a single polycistronic construct employing 2A peptides in the yeast, P. pastoris. To the best of our knowledge this is the highest number of genes expressed in a coordinated fashion so far. By further combining these 2A peptides with bidirectional promoters this number might easily be exceeded. Thus, the presented expression strategy represents a valuable tool for the quick and simple establishment of multi-enzyme pathways. The corresponding expression cassettes can be designed in a compact manner facilitating not only their construction, but all further molecular biologic steps. A further advantage is that 2A peptides allow the generation of genetically stable strains as the repetitive use of long homologous sequences is avoided. Various factors lead to partial generation of fusion proteins and non-stoichiometric production of the co-translationally produced proteins, and these effects imply useful strategies to optimize and balance pathway efficiencies by different sequential arrangements. Therefore, the 2A based expression concept not only simplifies and speeds up the discovery and feasibility of multi-enzyme pathways, but also their engineering. It remains to be shown whether multiplication of individual protein coding regions can be used for additional improvements. By applying existing recombination techniques, the 2A sequences can also be exploited as linkers to generate shuffled libraries containing the pathway genes in variable order and copies or functional homologs of individual pathway proteins. As the pathway genes are not separated by any additional regulatory sequence elements, the polycistronic expression cassette can directly be subjected to methods for random library generation such as error-prone PCR. However, as one deleterious mutation in one gene can cause the shut-down of the whole pathway, suitable high-throughput screenings are essential.
The research leading to these results received funding from the European Community's Seventh Framework Programme (FP7/2007-2013) and EFPIA companies' in kind contribution for the Innovative Medicine Initiative under Grant Agreement No. 115360 (Chemical manufacturing methods for the 21st century pharmaceutical industries, CHEM21). In addition, the work was supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol and ZIT – Technology Agency of the City of Vienna through the COMET-Funding Program managed by the Austrian Research Promotion Agency FFG. We also gratefully acknowledge the Austrian Science Fund (FWF) project number W901 (DK ‘Molecular Enzymology’ Graz) for funding to T.V.
Notes and references
- T. Zhu, M. Guo, C. Sun, J. Qian, Y. Zhuang, J. Chu and S. Zhang, Biotechnol. Lett., 2009, 31, 679–684 CrossRef CAS PubMed.
- R. Morgan, L. Couture, O. Elroy-Stein, J. Ragheb, B. Moss and W. Anderson, Nucleic Acids Res., 1992, 20, 1293–1299 CrossRef CAS PubMed.
- V. Douin, S. Bornes, L. Creancier, P. Rochaix, G. Favre, A.-C. Prats and B. Couderc, BMC Biotechnol., 2004, 4, 16 CrossRef PubMed.
- P. de Felipe, G. A. Luke, L. E. Hughes, D. Gani, C. Halpin and M. D. Ryan, Trends Biotechnol., 2006, 24, 68–75 CrossRef CAS PubMed.
- G. A. Luke, in Innovations in Biotechnology, ed. A. E. Rijeka, Croatia, 2012, pp. 161–186 Search PubMed.
- M. D. Ryan, A. M. King and G. P. Thomas, J. Gen. Virol., 1991, 72, 2727–2732 CrossRef CAS.
- M. L. Donnelly, G. Luke, A. Mehrotra, X. Li, L. E. Hughes, D. Gani and M. D. Ryan, J. Gen. Virol., 2001, 82, 1013–1025 CAS.
- C. Halpin, S. E. Cooke, A. Barakate, A. El Amrani, M. D. Ryan and Z. Seeds, Plant J., 1999, 17, 453–459 CrossRef CAS.
- P. de Felipe, L. E. Hughes, M. D. Ryan and J. D. Brown, J. Biol. Chem., 2003, 278, 11441–11448 CrossRef CAS PubMed.
- S. Furler, J. Paterna, M. Weibel and H. Bu, Gene Ther., 2001, 8, 864–873 CrossRef CAS PubMed.
- J. Beekwilder, H. M. van Rossum, F. Koopman, F. Sonntag, M. Buchhaupt, J. Schrader, R. D. Hall, D. Bosch, J. T. Pronk, A. J. van Maris and J.-M. Daran, J. Biotechnol., 2014, 1–10 Search PubMed.
- S. Unkles, V. Valiante, D. Mattern and A. Brakhage, Chem. Biol., 2014, 21, 502–508 CrossRef CAS PubMed.
- A. L. Szymczak, C. J. Workman, Y. Wang, K. M. Vignali, S. Dilioglou, E. F. Vanin and D. A. Vignali, Nat. Biotechnol., 2004, 22, 589–594 CrossRef CAS PubMed.
- C. Ma and A. Mitra, Mol. Breed., 2002, 9, 191–200 CrossRef CAS.
- J. M. Cregg, J. L. Cereghino, J. Shi and D. R. Higgins, Mol. Biotechnol., 2000, 16, 23–52 CrossRef CAS PubMed.
- S. Wang, Q. Yao, J. Tao, Y. Qiao and Z. Zhang, Appl. Microbiol. Biotechnol., 2007, 77, 891–899 CrossRef CAS PubMed.
- EP14175932.4, unpublished.
Footnotes |
| † Electronic supplementary information (ESI) available: Western blot analysis of the preliminary experiments and experiments including a ubiquitin tag to demonstrate the feasibility to create authentic N-termini as well as experimental details and sequence information. See DOI: 10.1039/c4cc08502g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2015 |