Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Detection of multiple mutations in a single codon of genomic DNA†
Danishmalik Rafiq
Sayyed
a,
Satish Balasaheb
Nimse
a,
Keum-Soo
Song
b and
Taisun
Kim
*a
aInstitute for Applied Chemistry and Department of Chemistry, Hallym University, Chuncheon, 200-702, Korea. E-mail: tskim@hallym.ac.kr
bBiometrix Technology, Inc., 202 BioVenture Plaza, Chuncheon, 200-161, Korea. Fax: +82-33-256-3421
First published on 26th August 2014
Abstract
The controller DNA technology allows the detection of multiple mutations in a single codon of genomic DNA. In this technology, the controller DNA is used to control the hybridization of target DNAs with the immobilized DNAs. The controller DNA technology is rapid, specific, and cost-effective for the following reasons, (i) final results in 40 min after PCR, (ii) detection and discrimination of the six mutations in a single codon, and (iii) high sensitivity.
Antibacterial drug resistance is often correlated to the single-nucleotide polymorphisms (SNPs).1 The multidrug resistance tuberculosis (MDR-TB) strains have several mutations including the multiple mutations at codon 526 (c526). The codon CAC (c526) of the wild TB strain is altered to well-known six mutations, such as TAC, AAC, GAC, TGC, CGC, and CTC, each resulting in the MDR-TB strain. It is well known that the mutation in the codon of the genomic DNA may change the sequence of the target protein that it codes for, resulting in the poor binding of the drug to the target protein.2 Hence, the resistance to the antibacterial drugs due to these mutations is inevitable.3 Thus, monitoring the SNPs is as an alternative to monitoring the drug resistance.4 Therefore, a technique which can facilitate the detection and discrimination of multiple mutations in a specific codon with a simple experimental protocol is essential for the timely detection of the drug resistant strains and selection of drugs for the treatment.5
The conventional culture-based method is reliable only for the discrimination of wild and mutant strains. DNA sequencing is considered as a gold standard but it is labour-intensive, time consuming, expensive, and has low-throughput.6,7 Real-time PCR is another approach; however, due to the nonlinear amplification it fails to detect multiple mutations at a single codon.8 DNA microarrays, on the other hand, provide a high-throughput platform for the detection of SNPs.9 However, the requirement of elevated temperature (40–60 °C) with a longer hybridization time (3–12 h) imposes a critical limitation for the detection of multiple SNPs at a single codon.10 Furthermore, the major issues of the DNA microarrays are a low signal to background ratio (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and very low target-specific hybridizations.11
1) and very low target-specific hybridizations.11
The platform technology which can detect multiple mutations at a single codon by using a simple experimental protocol is not available for a wide range of clinical applications.
The controller DNA technology (CDT) presented in this article can identify multiple mutations at c526 of the genomic DNA of the TB strain by using a simple experimental protocol on 9G DNAChips. In the controller DNA technology, the controller DNA is a small oligo DNA that has a sequence complementary to the sequence of the genomic DNA in the probe-binding region. As shown in Scheme 1A, hybridization of the Cy5 labelled PCR product of the wild TB strain in the absence of the controller DNA results in specific as well as non-specific hybridization. The non-specific hybridization is indicated by the appearance of the spots corresponding to the mutant strains. However, as depicted in Scheme 1C, hybridization of the Cy5 labelled PCR product of the wild TB strain in the presence of the controller DNA results in the highly specific hybridization without any non-specific hybridizations. The highly specific hybridization is indicated by the appearance of the spots corresponding to the wild strain without any spots for mutant strains. Thus, controller DNA controls the hybridization of the immobilized probes with the PCR product, thereby eliminating the non-specific hybridization. The development of CDT is explained in detail. Further the sensitivity and specificity of the CDT were evaluated for its applicability in clinical samples.
Probes (Probe 1–Probe 10, ESI,† Table S1) for the detection of the multiple mutations at c526 in the TB strain were selected according to the well-established generalized probe selection method.12 The selected probes were immobilized on 9G DNAChips (ESI,† Scheme S2) according to a previously reported method.13 The 128 mer Cy5 labelled single stranded PCR product (Cy5 labelled ssDNA) was obtained by the asymmetric PCR amplification using a Cy5 labelled reverse primer and a forward primer with a ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (ESI,† Fig. S2). It is well known that the PCR product containing ssDNA shows higher hybridization efficiency than dsDNA with the immobilized probes on the DNA chips.14 It is important to note that the Cy5 labelled ssDNA in the PCR product of the wild TB strain after asymmetric PCR amplification undergoes self-dimerization15 due to the complementary base pairs in its sequence (ESI,† Scheme S1). Therefore, to separate the strands of the Cy5 labelled ssDNA in the dimerized form, a splitter DNA (5′-GCTGGGGCCTGGCGGTCTGT-3′) was used.
1 (ESI,† Fig. S2). It is well known that the PCR product containing ssDNA shows higher hybridization efficiency than dsDNA with the immobilized probes on the DNA chips.14 It is important to note that the Cy5 labelled ssDNA in the PCR product of the wild TB strain after asymmetric PCR amplification undergoes self-dimerization15 due to the complementary base pairs in its sequence (ESI,† Scheme S1). Therefore, to separate the strands of the Cy5 labelled ssDNA in the dimerized form, a splitter DNA (5′-GCTGGGGCCTGGCGGTCTGT-3′) was used.
The immobilized probes on 9G DNAChips were allowed to hybridize with 100 fmol of the Cy5 labelled ssDNA of the wild TB strain in the presence of 100 pmol of a splitter DNA at 25 °C for 30 min. The result of hybridization is depicted in Fig. 1B (No controller, NOC). Probe 1 corresponding to the wild strain is the only one which should have shown hybridization with the Cy5 labelled ssDNA. Unfortunately, all probes corresponding to the mutant strains showed non-specific hybridization with the Cy5 labelled ssDNA of the wild TB strain. The ratio of the target specific to the non-specific hybridization was found to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Similar non-specific hybridizations were also observed upon hybridization of the Cy5 labelled ssDNA of the mutant strain (c526 CAC → CGC, ESI,† Fig. S3). From these results it was clear that there was no control on the hybridization of the immobilized probes with the corresponding Cy5 labelled ssDNA.
1. Similar non-specific hybridizations were also observed upon hybridization of the Cy5 labelled ssDNA of the mutant strain (c526 CAC → CGC, ESI,† Fig. S3). From these results it was clear that there was no control on the hybridization of the immobilized probes with the corresponding Cy5 labelled ssDNA.
Therefore, to control the hybridization of the immobilized probes with the Cy5 labelled ssDNA, a controller DNA (CD) was designed. It was assumed that the controller DNA can strongly compete with the immobilized probes for hybridization with the Cy5 labelled ssDNA if it has a complementary sequence with Cy5 labelled ssDNA in the probe binding region.
Therefore, to choose the optimum binding position and the optimum length of the controller DNAs, a nucleotide in the Cy5 labelled ssDNA, complementary to the first nucleotide in the immobilized probe, was assigned number 1. The other complementary nucleotides were assigned numbers in the ascending order. To optimize the binding position of the controller DNA, 11 DNAs (CD1–CD11) with the length of 10 nucleotides (ESI† Table S2) were chosen, as shown in Fig. 1A. CD1–CD10 and CD11 leave 1–10 and 12 nucleotides open, respectively, on the Cy5 labelled ssDNA for hybridization with the immobilized probes, as demonstrated in Fig. 1A. The results of hybridizations of the immobilized probes with the Cy5 labelled ssDNA in the presence of 100 pmol of these controller DNAs are depicted in Fig. 1B and C.
As demonstrated in Fig. 1B and C, CD1 leaving only one nucleotide open with the binding position 2, completely prohibits the hybridization of the immobilized probes with the Cy5 labelled ssDNA. However, CD2, CD3, CD4, and CD5 with the binding positions 3, 4, and 5, respectively, allow controlled hybridization of immobilized probes with the Cy5 labelled ssDNA. The controlled hybridization is indicated by the absence of non-specific hybridizations. The controlled hybridization is also indicated by the low fluorescence intensity in the case of CD2, CD3, and CD4. It is very important to note that, if controller DNA covers most of the nucleotides in the probe binding region (CD1, CD2, CD3) it is very difficult for the immobilized probe to displace the controller DNA bound to the Cy5 labelled ssDNA. Therefore, the probes show only specific hybridization with very low fluorescence intensity.
CD6 with the binding position 7 leaving six nucleotides open demonstrates highest fluorescence intensity of 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 without any non-specific interaction. The competitive control of CD6 on the hybridization of immobilized probes with the Cy5 labelled ssDNA results in a target specific to non-specific hybridization ratio of more than 100
000 without any non-specific interaction. The competitive control of CD6 on the hybridization of immobilized probes with the Cy5 labelled ssDNA results in a target specific to non-specific hybridization ratio of more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
However, CD7 with the binding position 8 leaving seven nucleotides open loses control over hybridization, which results in the non-specific hybridization. Moreover, CD8, CD9, and CD10 with the binding positions 9, 10, and 11, respectively, constantly lose control over hybridization resulting in an increase in non-specific hybridization intensity from 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 50
000 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. CD11 with the binding position 13 leaving twelve nucleotides open, completely loses control over hybridization resulting in a target specific to non-specific hybridization ratio of 1
000. CD11 with the binding position 13 leaving twelve nucleotides open, completely loses control over hybridization resulting in a target specific to non-specific hybridization ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. From this data it is clear that if the controller DNA leaves most of the nucleotides open (CD7–CD11, 7–12 nucleotides, respectively) in the probe binding region, the immobilized probes can easily displace the controller DNA for hybridization with the Cy5 labelled ssDNA. Therefore, the probes corresponding to the wild as well as mutated strains can hybridize with the Cy5 labelled ssDNAs resulting in specific as well as non-specific hybridizations. However, when the controller DNA leaves only six nucleotides open (CD6) for hybridization of immobilized probes with the Cy5 labelled ssDNAs, both the controller as well as probes start to have a competition for the hybridization with the Cy5 labelled ssDNAs resulting only in the target specific hybridizations. Therefore, CD6 was considered to be optimum and used for further experiments.
1. From this data it is clear that if the controller DNA leaves most of the nucleotides open (CD7–CD11, 7–12 nucleotides, respectively) in the probe binding region, the immobilized probes can easily displace the controller DNA for hybridization with the Cy5 labelled ssDNA. Therefore, the probes corresponding to the wild as well as mutated strains can hybridize with the Cy5 labelled ssDNAs resulting in specific as well as non-specific hybridizations. However, when the controller DNA leaves only six nucleotides open (CD6) for hybridization of immobilized probes with the Cy5 labelled ssDNAs, both the controller as well as probes start to have a competition for the hybridization with the Cy5 labelled ssDNAs resulting only in the target specific hybridizations. Therefore, CD6 was considered to be optimum and used for further experiments.
To optimize the length of the controller DNA, 7 DNAs (CD12–CD18) starting from the binding position 7 with the lengths 5 mer, 6 mer, 8 mer, 9 mer, 12 mer, 14 mer, and 16 mer, respectively, (ESI† Table S3) were chosen, as demonstrated in Fig. 1D. The results of the hybridizations with the Cy5 labelled ssDNA in the presence of 100 pmol of these controller DNAs (CD12–CD18) are depicted in Fig. 1E and F.
As demonstrated in Fig. 1E and F, CD12 and CD13 with the lengths of 5 mer and 6 mer, respectively, did not show any control over hybridization. CD12 and CD13 are too short to compete with the immobilized probes for hybridization with Cy5 labelled ssDNA resulting in very high non-specific hybridization. The target specific to non-specific hybridization ratio was only 1.2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. However, with the increase in the length of controller DNA to 8 mer and 9 mer as in the case of CD14 and CD15, respectively, the target specific to non-specific hybridization ratio increased to 2
1. However, with the increase in the length of controller DNA to 8 mer and 9 mer as in the case of CD14 and CD15, respectively, the target specific to non-specific hybridization ratio increased to 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 6
1 and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The data for CD14 and CD15 demonstrate that they show partial control over hybridization of immobilized probes with the Cy5 labelled ssDNA as compared to CD12 and CD13.
1. The data for CD14 and CD15 demonstrate that they show partial control over hybridization of immobilized probes with the Cy5 labelled ssDNA as compared to CD12 and CD13.
Interestingly, controllers CD16 and CD17 with the lengths of 10 mer and 12 mer, respectively, gain complete control over the hybridization of the immobilized probes with the Cy5 labelled ssDNA. The use of these two controller DNAs allowed us to achieve a target specific to non-specific hybridization ratio of more than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.
1.
Thus, it is clear that the longer controller DNAs effectively compete with immobilized probes for hybridization with the Cy5 labelled ssDNA resulting in a decrease in non-specific hybridizations.
However, the controller DNAs CD18 and CD19 with the lengths of 14 mer and 16 mer, respectively, competitively prohibit the binding of the immobilized probes with the Cy5 labelled ssDNA. The competitive prohibition of hybridization was indicated by a decrease in fluorescence intensity for target specific hybridization from 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 10
000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. Therefore, the controller DNA CD16 with the length of 10 mer was considered optimum and used for further experiments.
000. Therefore, the controller DNA CD16 with the length of 10 mer was considered optimum and used for further experiments.
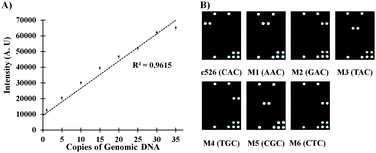
To determine the sensitivity of the CDT, the hybridization solution containing the PCR products of the 1–35 copies of the wild TB strain were used for hybridization. Fig. 2A (ESI,† Fig. S4 and S5) clearly indicates that the present technology can distinguish the SNP target to as low as 1 copy of the genomic DNA. Similar results were also obtained for the mutation sample. The sensitivity of the CDT is comparable with that of the reported methods.16
Fig. 2B demonstrates that the use of controller oligo DNAs allows highly sensitive and highly specific detection of all the six mutations in the c526 of the genomic DNA of TB. To evaluate the clinical applicability of the CDT in the detection of multiple mutations at c526 of the genomic DNA of TB, known clinical samples comprising wild and six mutant strains were examined (ESI,† Fig. S6–S15). These results showed 100% agreement with the results of sequencing analysis for each clinical sample.
The controller DNA technology allows rapid, specific, and cost-effective detection of multiple SNPs in the clinical samples for the following reasons, (i) detection and discrimination of the multiple mutations at a single codon, (ii) high sensitivity in terms of the specific to non-specific hybridization ratio higher than 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and (iii) final results in 40 min after PCR (ESI,† Scheme S3). 100% agreement of the clinical results with sequencing analysis demonstrates high sensitivity and specificity of the CDT. In turn, this technology can boost the ongoing efforts on monitoring the drug resistance for the rapid cure and prevention of the spread of the disease.
1, and (iii) final results in 40 min after PCR (ESI,† Scheme S3). 100% agreement of the clinical results with sequencing analysis demonstrates high sensitivity and specificity of the CDT. In turn, this technology can boost the ongoing efforts on monitoring the drug resistance for the rapid cure and prevention of the spread of the disease.
This study was supported by a grant of the Korean Health Technology R&D Project, Ministry of Health & Welfare, Republic of Korea (Project No. HI13C0858).
Notes and references
- S. H. Abbadi, et al. , Int. J. Infect. Dis., 2009, 13, 673 CrossRef CAS PubMed.
- P. C. Ng and S. Henikoff, Nucleic Acids Res., 2003, 31, 3812 CrossRef CAS PubMed.
- N. Woodford and M. J. Ellington, Clin. Microbiol. Infect., 2007, 13, 5 CrossRef CAS PubMed.
- H. Wang, et al. , Anal. Chem., 2011, 83, 1883 CrossRef CAS PubMed; Y. Zhang, et al. , Nanoscale, 2013, 5, 5027 RSC.
- J. A. Caminero, et al. , Lancet Infect. Dis., 2010, 10, 621 CrossRef CAS; M. Yagui, et al. , Int. J. Tuberc. Lung. Dis., 2006, 10, 838 Search PubMed.
- K. Sato, et al. , Anal. Biochem., 2006, 350, 162 CrossRef CAS PubMed.
- B. Veigas, et al. , Nanotechnology, 2010, 21, 1 CrossRef PubMed.
- L. van der Weyden and D. J. Adams, Biochim. Biophys. Acta, 2007, 1776, 58 CAS.
- S. B. Nimse, et al. , Chem. Soc. Rev., 2013, 42, 366 RSC.
- L. Wang and P. C. H. Li, J. Agric. Food Chem., 2007, 55, 10509 CrossRef CAS PubMed; A. Sedighi and P. C. H. Li, Int. J. Mater. Sci. Eng., 2013, 1, 45 Search PubMed.
- Y. D. Choi, et al. , Gynecol. Oncol., 2007, 98, 369 CrossRef PubMed.
- S. B. Nimse, et al. , Chem. Commun., 2011, 47, 12444 RSC.
- K. Song, et al. , Chem. Commun., 2011, 47, 7104 RSC; S. B. Nimse, et al. , Tetrahedron Lett., 2010, 51, 6156 CrossRef CAS PubMed; S. B. Nimse, et al. , Tetrahedron Lett., 2009, 50, 7346 CrossRef PubMed.
- X. Tang, et al. , BioTechniques, 2006, 40, 759 CrossRef CAS.
- M. Deng, et al. , J. Clin. Microbiol., 2012, 50, 3443 CrossRef CAS PubMed.
- W. Shen, et al. , Chem. Commun., 2013, 49, 8114 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and experimental methods. See DOI: 10.1039/c4cc06220e |
| This journal is © The Royal Society of Chemistry 2014 |