Open Access Article
Open Access ArticleRapid engineering of versatile molecular logic gates using heterologous genetic transcriptional modules†
Baojun
Wang
*a and
Martin
Buck
b
aCentre for Synthetic and Systems Biology, School of Biological Sciences, University of Edinburgh, Edinburgh, EH9 3JR, UK. E-mail: Baojun.Wang@ed.ac.uk; Fax: +44 (0)131 6508650; Tel: +44 (0)131 6505527
bDepartment of Life Sciences, Faculty of Natural Sciences, Imperial College London, London, SW7 2AZ, UK. E-mail: m.buck@imperial.ac.uk; Fax: +44 (0)20 75945419; Tel: +44 (0)20 75945442
First published on 21st July 2014
Abstract
We designed and constructed versatile modular genetic logic gates in bacterial cells. These function as digital logic 1-input Buffer gate, 2-input and 3-input AND gates with one inverted input and integrate multiple chemical input signals in customised logic manners. Such rapidly engineered devices serve to achieve increased sensing signal selectivity.
Biomolecular computing is an emerging computing paradigm that has the potential of combining both the sensing and processing of biochemical signals to generate autonomous programmed output readout and action.1,2 Great progress has been seen in this area which typically applies enzymes,3,4 DNA5,6 or RNA7 in an in vitro biochemical system to execute complex logic computing tasks. For example, a DNA and restriction enzyme-based in vitro diagnostic automaton detected multiple biochemical disease indicators where the final positive state only forms under a predesigned logic combination of the inputs.8 More recently, live cells have hosted various in vivo logic computing tasks such as AND9–11 and XOR12,13 gates functionalities owing to the vast array of functions available to a living cell.14 However, the present capabilities for implementing complex logic computing functions in live cells are rather limited, since few well-characterised and versatile genetic building blocks are available.14 In addition, most of the devices constructed so far are designed to use specific inputs to drive specific outputs, which make them difficult to incorporate and reuse in other cellular circuit programs.
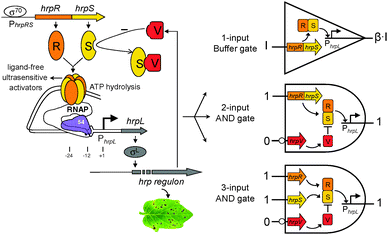
To address these bottlenecks, we present the rapid engineering of versatile modular molecular logic gates in model Escherichia coli bacteria via repurposing some heterologous genetic transcriptional modules. These modules exist in vast numbers amongst many different bacterial species, offering a largely untapped resource. As an example, we designed various modular logic devices using transcriptional modules from the hrp (hypersensitive response and pathogenicity) gene regulation system for Type III secretion in Pseudomonas syringae (Fig. 1).15,16 In the hrp gene regulatory architecture (Fig. 1), the activator proteins HrpR and HrpS form a high-order co-complex which binds the upstream activator sequence of the σ54-dependent hrpL promoter to remodel the closed σ54-RNAP-hrpL transcription complex to an open one through ATP hydrolysis, while negative regulation by the HrpV occurs through its direct interaction with HrpS.17
Since the hrp system comprises multiple components that interact and regulate each other in a defined logic manner, we considered that multiple-input combinational logic devices should be constructed based on their existing regulatory relationships. Accordingly three modular logic devices, including the digital logic 1-input Buffer gate, 2-input and 3-input AND with one inverted input gates, were designed using subsets of the hrp regulatory network (Fig. 1). The device inputs and outputs are promoters. The output promoter is only turned on when a specific logic combination of input promoters is active. One distinct advantage of the device design is the modularity (reusability), i.e. the inputs are exchangeable and can be connected to different environment-responsive promoters whilst the output may be wired to drive various other useful genes. Moreover, the genetic components used are heterologous to E. coli and thus less likely to interfere with the host endogenous genetic programs which will enable these logic devices to behave more robustly in their working contexts.9
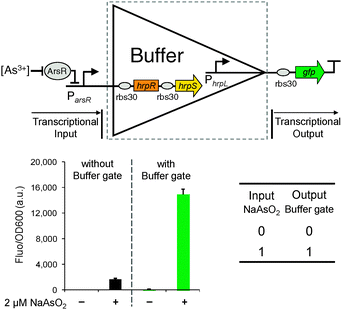
We first designed the single-input digital Buffer gate which functions as an amplifier for the input signal to achieve increased output expression capability for downstream connected logic modules.18 The Buffer gate does not change the logic relationship between the input and output, but instead acts as an isolator and enhancer that can transform the input signal to a higher amplitude signal to match the input requirement of the aligned output modules.
The Buffer gate was built to express from an operon the cooperative activator proteins HrpR and HrpS with hrpL as the output promoter (Fig. 2). The high order HrpRS assemblies synergistically activate the downstream tightly controlled σ54-dependent19hrpL promoter, thus assisting amplification20,21 of the transcriptional input signal. To characterise the device, the input was connected to an arsenic responsive promoter22 and the output to a gfp (green fluorescent protein) reporter. The input promoter alone was first systematically characterised with gfp as its output, in the presence of arsenite and produced a mild output response (Fig. S1 and S2, ESI†). In contrast, when the Buffer gate was applied, the output significantly increased to a much higher level (∼9 fold) under the same 2 μM NaAsO2 input induction (Fig. 2). The characterised responses under different arsenite levels help establish that the device behaves well as a digital logic Buffer gate (Fig. S1, ESI†). The Buffer gate can be used to increase the sensitivity of transcription-based biosensors (e.g. the arsenic sensor in this study) as well as to modulate the amplitude of an input signal to facilitate the coupling and matching of genetic modules with vastly different input–output strengths.23
We next designed the double-input AND gate with one inverted input, a digital logic device that only produced an observable output when the 1st input is present and the 2nd input is absent (Fig. 3). The function is equivalent to the combination of connecting a NOT gate to one input of a canonical AND gate. The device can be used to improve the selectivity of biochemical sensors such that they will then only generate a valid readout under the specific logic combination of the two chosen sensing chemicals.
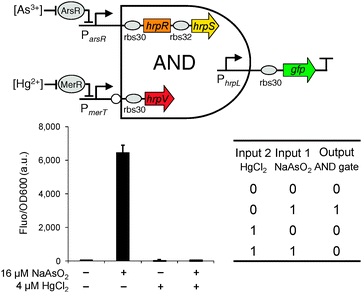 | ||
| Fig. 3 The design and characterisation of the double-input modular AND gate with one inverted input. Error bars, s.d. (n = 3). | ||
The double-input AND gate above was built by expressing from an operon HrpR and HrpS, under the control of the non-inverted input and expressing separately the inhibitor HrpV under the inverted input (Fig. 1). To characterise the device, we connected the non-inverted input to the arsenic responsive promoter (Fig. S2, ESI†) and the inverted input to a mercury responsive promoter22 (Fig. S3, ESI†) whilst the final output was the gfp reporter. To achieve a close to zero off-state behaviour, we used a weak RBS9 (ribosome binding site, rbs32, Table S1, ESI†) ahead of the activator hrpS gene but a strong RBS (rbs30) ahead of the inhibitor hrpV gene. As a result, HrpV inhibitor will be expressed in excess over the activator HrpRS complex when both inputs are activated, leading to complete inhibition of the output hrpL promoter. The characterisation result shows that the device only produced an observable output when NaAsO2 (16 μM) is present and HgCl2 (4 μM) is absent among the four possible logic combinations of the two chemical inputs (Fig. 3). These outcomes establish the effectiveness of the digital logic AND gate having one inverted input function.
Extending from the double-input AND gate, we designed the triple-input AND gate with one inverted input, a combinational digital logic device that should only produce an output when the first two inputs are present and the third input is absent (Fig. 4). The predicted function is equivalent to that of connecting the output of a canonical AND gate to the non-inverted input of the above built double-input AND gate. Similarly, the device can be used to increase the selectivity of biochemical sensors to only produce a significant readout under the specific logic combination of the three target sensing chemicals.
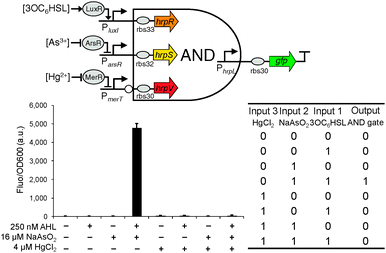 | ||
| Fig. 4 The design and characterisation of the triple-input modular AND gate with one inverted input. Error bars, s.d. (n = 3). | ||
The triple-input AND gate was then built by expressing the two cooperative activator proteins HrpR and HrpS under the control of the two non-inverted inputs while expressing the inhibitor HrpV under the control of the third inverted input (Fig. 1). To characterise the device, we connected the two non-inverted inputs to the arsenic (Fig. S2, ESI†) and AHL22 (Fig. S4, ESI†) responsive promoters respectively whereas the inverted input connected to the mercury responsive promoter (Fig. S3, ESI†). Similarly, to achieve a close to zero off-state behaviour, we used two weak RBSs (rbs33 and rbs32, Table S1, ESI†)9 ahead of the activator hrpR and hrpS gene but a strong RBS (rbs30) ahead of the inhibitory hrpV gene. Characterisation shows that the device only produced an observable output when both 3OC6HSL (250 nM) and NaAsO2 (16 μM) are present and HgCl2 (4 μM) is absent among the eight possible logic combinations of the three chemical inputs (Fig. 4), proving the correct logic function of this triple-input AND gate with one inverted input.
Here, we have constructed a set of modular genetic logic gates with versatile digital logic functions and demonstrated that use of heterologous genetic transcriptional modules in different bacterial species can guide the design and rapid engineering of novel biological devices. The method presented here should enable future design and construction of many orthogonal genetic logic devices from diverse heterologous building blocks to expand the currently limited parts kit for gene circuit engineering.14
The genetic logic devices described here have particular advantages including the tight unactivated off state attributed to the underlying σ54-dependent strict transcriptional activation requirement.19 The high-order co-dependent HrpRS complexes employed in the system contribute to the sharp switching between the off and on states of the devices, resulting in digital-like response behaviour. Moreover, these devices are modular and can, potentially, be easily connected to other modules to construct larger and more complex systems with advanced functions.
This work was supported by a BBSRC project grant [BB/K016288/1] and a Royal Society research grant award [RG120527] to B.W.
Notes and references
- Y. Benenson, Nat. Rev. Genet., 2012, 13, 455 CrossRef CAS PubMed.
- J. Wang and E. Katz, Anal. Bioanal. Chem., 2010, 398, 1591 CrossRef CAS PubMed.
- J. Zhou, M. A. Arugula, J. Halámek, M. Pita and E. Katz, J. Phys. Chem. B, 2009, 113, 16065 CrossRef CAS PubMed.
- E. Katz and V. Privman, Chem. Soc. Rev., 2010, 39, 1835 RSC.
- G. Seelig, D. Soloveichik, D. Y. Zhang and E. Winfree, Science, 2006, 314, 1585 CrossRef CAS PubMed.
- L. Qian and E. Winfree, Science, 2011, 332, 1196 CrossRef CAS PubMed.
- D. Faulhammer, A. R. Cukras, R. J. Lipton and L. F. Landweber, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1385 CrossRef CAS.
- B. Gil, M. Kahan-Hanum, N. Skirtenko, R. Adar and E. Shapiro, Nano Lett., 2011, 11, 2989 CrossRef CAS PubMed.
- B. Wang, R. I. Kitney, N. Joly and M. Buck, Nat. Commun., 2011, 2, 508 CrossRef PubMed.
- Z. Li, M. A. Rosenbaum, A. Venkataraman, T. K. Tam, E. Katz and L. T. Angenent, Chem. Commun., 2011, 47, 3060 RSC.
- M. A. Arugula, N. Shroff, E. Katz and Z. He, Chem. Commun., 2012, 48, 10174 RSC.
- J. Bonnet, P. Yin, M. E. Ortiz, P. Subsoontorn and D. Endy, Science, 2013, 340, 599 CrossRef CAS PubMed.
- A. Tamsir, J. J. Tabor and C. A. Voigt, Nature, 2011, 469, 212 CrossRef CAS PubMed.
- B. Wang and M. Buck, Trends Microbiol., 2012, 20, 376 CrossRef CAS PubMed.
- Q. Jin, R. Thilmony, J. Zwiesler-Vollick and S.-Y. He, Microbes Infect., 2003, 5, 301 CrossRef CAS.
- D. Buttner and U. Bonas, Curr. Opin. Microbiol., 2006, 9, 193 CrossRef PubMed.
- M. Jovanovic, E. H. James, P. C. Burrows, F. G. M. Rego, M. Buck and J. Schumacher, Nat. Commun., 2011, 2, 177 CrossRef PubMed.
- C.-H. Chuang and C.-L. Lin, BMC Syst. Biol., 2014, 8, 63 CrossRef PubMed.
- M. Buck, D. Bose, P. Burrows, W. Cannon, N. Joly, T. Pape, M. Rappas, J. Schumacher, S. Wigneshweraraj and X. Zhang, Biochem. Soc. Trans., 2006, 34, 1067 CrossRef CAS PubMed.
- Q. Zhang, S. Bhattacharya and M. E. Andersen, Open Biol., 2013, 3, 130031 CrossRef PubMed.
- B. Wang, M. Barahona and M. Buck, Nucleic Acids Res., 2014 DOI:10.1093/nar/gku593.
- B. Wang, M. Barahona and M. Buck, Biosens. Bioelectron., 2013, 40, 368 CrossRef CAS PubMed.
- D. Del Vecchio, A. J. Ninfa and E. D. Sontag, Mol. Syst. Biol., 2008, 4, 161 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Supplementary methods, gene regulatory sequences used, the characterised dose responses of the single-input Buffer gate and the three promoter inputs under various cognate chemical induction levels. See DOI: 10.1039/c4cc05264a |
| This journal is © The Royal Society of Chemistry 2014 |