Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
C5-Amino acid functionalized LNA: positively poised for antisense applications†
Dale C.
Guenther
,
Pawan
Kumar
,
Brooke A.
Anderson
and
Patrick J.
Hrdlicka
*
Department of Chemistry, University of Idaho, 875 Perimeter Drive MS2343, Moscow, ID 83844-2343, USA. E-mail: hrdlicka@uidaho.edu
First published on 10th June 2014
Abstract
Incorporation of positively charged C5-amino acid functionalized LNA uridines into oligodeoxyribonucleotides (ONs) results in extraordinary RNA affinity, binding specificity and stability towards 3′-exonucleases.
Modulation of gene expression by antisense-based approaches continues to stimulate the development of novel chemically modified nucleotides in search of oligonucleotides with improved RNA affinity, binding specificity, and pharmacokinetic profiles.1 Of the many modifications studied for antisense applications, conformationally restricted nucleotides,2 and locked nucleic acid (LNA) in particular,3 have emerged as particularly promising chemistries toward this end. Over the past 15 years, major efforts have been devoted to increase the chemical diversity of LNA nucleotides through additional modification of the sugar ring, with the aim of improving properties for antisense applications.3,4 Modification of the nucleobase of LNA, on the other hand, is an underexplored strategy for optimization of pharmacodynamic and pharmacokinetic properties.
Modification of the C5-position of pyrimidines is particularly interesting due to synthetic feasibility and functional group tolerance, as attached moieties are directed into the major groove with minimal perturbation of the duplex structure.5,6 We have previously shown that attachment of small hydrophilic moieties to the C5-position of LNA uridine (U) such as 3-aminopropyn-1-yl promotes even greater target affinity than canonical LNA, while attachment of large hydrophobic substituents such as cholesterol confers complete stability against exonucleases, albeit at the expense of decreased target affinity.7 At the onset of the present study, we hypothesized that hydrophilic moieties of intermediate size would contribute to both additionally improved affinity and nuclease stability. Furthermore, positively charged moieties often improve cellular uptake through electrostatic interactions.8,9 Amino acids are a class of molecules that fit the size, hydrophilicity and charge criteria.10 Driven by the desire to exploit the properties of both LNA and basic amino acid residues, we report the synthesis of three C5-amino acid functionalized LNA-U phosphoramidites and the biophysical characterization of ONs modified with these units.
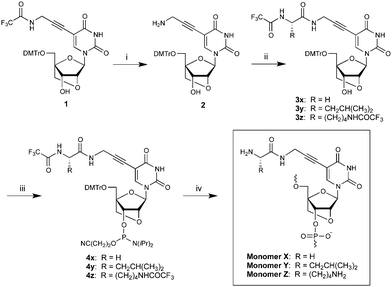
Synthesis of phosphoramidites 4x/y/z initiates from known nucleoside 1 (Scheme 1).7 Deprotection of the trifluoroacetamide (TFA) group using saturated methanolic ammonia reveals the aminopropynyl group at the C5-position of 2 in 97% yield.‡ TSTU-mediated coupling of TFA-protected glycine, leucine or lysine gives 3x/y/z in moderate yields (48–58%).§ Subsequent O3′-phosphitylation affords phosphoramidites 4x/y/z, which were used for incorporation of monomers X/Y/Z into ONs by automated DNA solid-phase synthesis (15 min, 4,5-dicyanoimidazole as activator, average coupling yield >90%). The composition and purity of all modified ONs was ascertained by MALDI-MS analysis (Table S1, ESI†) and ion-pair reversed-phase HPLC, respectively.
ONs modified with C5-amino acid functionalized LNA-U monomers X/Y/Z display higher affinity toward complementary RNA than ONs modified with canonical LNA thymidine (monomer L) (Table 1). Incorporation of glycine-conjugated monomer X consistently increases the thermal denaturation temperatures (Tm's) of 9-mer mixed-sequence model duplexes by 10–11 °C, while leucine-conjugated monomer Y results in slightly less thermostable duplexes (ΔTm = +7.0 to +10.5 °C). ONs containing lysine-conjugated monomer Z display the highest RNA affinity in this series (ΔTm = +11.0 to +14.0 °C). In fact, Z is as affinity-enhancing as C5-aminopropynyl LNA-U (monomer N).7 Similar trends are observed towards DNA targets, although slightly less pronounced stabilization is observed (Table S2, ESI†).
| ON | Sequence | ΔTm/mod | ||||
|---|---|---|---|---|---|---|
| B = Lb | B = Nb | B = X | B = Y | B = Z | ||
| a ΔTm/mod = change in Tm per modification relative to unmodified reference duplex (R1:D2 or D1:R2 both Tm = 28.0 °C); R1: 5′-GUG AUA UGC; R2: 3′-CAC UAU ACG; Tm's determined as the maximum of the first derivative of the melting curve (A260vs. T) recorded in medium salt phosphate buffer ([Na+] = 110 mM, [Cl−] = 100 mM, pH 7.0 (NaH2PO4/Na2HPO4)), using 1.0 μM of each strand. Monomer L = LNA-T. Monomer N = C5-(3-aminopropyn-1-yl)-LNA-U. b Previously reported.7 | ||||||
| B1 | 5′-GTG ABA TGC | +8.5 | +13.0 | +10.5 | +9.5 | +12.5 |
| B2 | 3′-CAC BAT ACG | +5.5 | +10.0 | +10.5 | +10.5 | +11.0 |
| B3 | 3′-CAC TAB ACG | +8.5 | +12.5 | +10.0 | +7.0 | +14.0 |
| B4 | 3′-CAC BAB ACG | +7.5 | +11.0 | +10.8 | +9.3 | +13.0 |
Thermodynamic parameters for duplex formation were determined from denaturation curves through curve fitting (Table S3, ESI†).11 Changes in free energy (ΔG) follow similar trends as Tm's. Thus, ONs containing monomers X/Y/Z show greater affinity toward RNA than DNA targets (RNA: ΔG298 = −8 to −23 kJ mol−1; DNA: ΔG298 = −5 to −17 kJ mol−1), and greater target affinity than ONs modified with LNA-T. The additional stabilization provided by the C5-amino acid functionalized LNA monomers is a result of more favorable enthalpy. This is likely due to extended π-conjugation of the nucleobase and improved stacking within the duplex,6b as well as electrostatic screening of the negatively charged duplex by protonated amino acid residues.6c–g The latter is a reasonable assumption considering that the pKa for α-amino groups of free amino acids and the ε-amino group of lysine is >9.16 and 10.67, respectively.12
To study in greater detail if protonation of the amino acid residues of monomers X/Y/Z contributes to duplex stabilization, thermal denaturation profiles were recorded in buffers of decreasing ionic strength (110, 40, and 10 mM sodium phosphate buffer).6a As expected, lower absolute Tm's are observed at low ionic strengths due to decreased electrostatic shielding of the two negatively charged duplex strands (Tables S4 and S5, ESI†). Interestingly, ONs modified with monomers X/Y/Z show greater relative affinity increases toward RNA and DNA targets at lower ionic strengths as evidenced by the trend in ΔTm's (Fig. 1 and Fig. S2, ESI†). For example, the ΔTm for the duplex between lysine-modified Z1 and complementary RNA is 12.5 °C and 18.0 °C at [Na+] = 110 mM and 10 mM, respectively (Table S4, ESI†). These effects are much weaker for LNA-modified ONs (L1–L4), which strongly suggests that the C5-amino acid moieties of monomers X/Y/Z indeed are protonated and contribute to duplex stability.6a
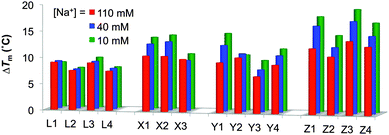 | ||
| Fig. 1 Thermostabilities of duplexes between B1–B4-series and RNA at different ionic strengths. ΔTm = change in Tm's relative to unmodified reference duplex D1:R2 (Tm,110mM = 28.0 °C, Tm,40mM = 21.0 °C, Tm,10mM = 11.5 °C); R1:D2 (Tm,110mM = 28.0 °C, Tm,40mM = 22.0 °C, Tm,10mM = 12.0 °C). Buffer conditions: [Na+] = 110 mM and [Cl−] = 100 mM; [Na+] = 40 mM and [Cl−] = 30 mM; [Na+] = 10 mM. pH 7.0 (NaH2PO4/Na2HPO4) in all cases. See also Table S4 (ESI†). | ||
The specificity of ONs modified with C5-amino acid functionalized LNA-U monomers X/Y/Z was evaluated by measuring the Tm's of duplexes between modified ONs and RNA or DNA containing a mismatched nucleotide either directly across from the central modification (B1-series) or 2′-deoxyadenosine (B4-series). Just like LNA-T (L1) or C5-aminopropynyl modified (N1), X1/Y1/Z1 show comparable or improved discrimination of mismatched RNA and DNA relative to D1 (Table 2 and Table S6, ESI†). Discrimination of the challenging T/U:G mismatch is particularly effective. Doubly modified X4/Y4/Z4 also discriminate mismatched RNA targets well, whereas discrimination of mismatched DNA targets is far less efficient (Table S7, ESI†). The latter may be a result of stabilizing non-specific electrostatic interactions occurring in DNA duplexes.
| ON | Sequence | RNA: 3′-CAC UMU ACG | |||
|---|---|---|---|---|---|
| T m (°C) | ΔTm (°C) | ||||
| M = A | M = C | M = G | M = U | ||
| a ΔTm = change in Tm relative to fully matched B1:R2 duplex (M = A) shown in bold. b Previously reported.7 | |||||
| D1 | 5′-GTG ATA TGC | 28.0 | <−18.0 | −5.5 | <−18.0 |
| L1 | 5′-GTG ALA TGC | 36.5 | −20.0 | −9.0 | −19.5 |
| N1 | 5′-GTG ANA TGC | 41.0 | −18.5 | −11.5 | −22.5 |
| X1 | 5′-GTG AXA TGC | 38.5 | −17.0 | −11.5 | −19.0 |
| Y1 | 5′-GTG AYA TGC | 37.5 | −16.0 | −9.0 | −19.0 |
| Z1 | 5′-GTG AZA TGC | 40.5 | −17.0 | −11.5 | −19.0 |
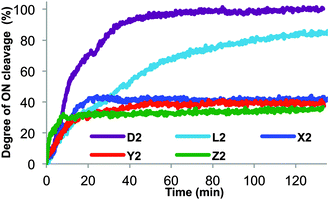
ONs with C5-amino acid functionalized LNA-U monomers positioned close to the 3′-terminus (X2/Y2/Z2) were evaluated for nuclease stability using snake venom phosphodiesterase (SVPDE), a 3′-exonuclease (Fig. 2). As expected, unmodified DNA (D2) is completely cleaved within 40 min. Incorporation of a single LNA-T (L2) still results in complete digestion, although at a significantly slower rate. Satisfyingly, X2/Y2/Z2 were completely stable to SVPDE once degradation of the 2–3 terminal 2′-deoxyribonucleotides had ceased, as evidenced by the plateau in their degradation profiles.
Encouraged by these results, we designed a fully phosphorothioated 3-14-3 gapmer antisense ON (ASO) targeting firefly luciferase mRNA (Table S8, ESI†). Very preliminary experiments using murine 3T3-L1 cells indicate that ASOs modified with two C5-lysine LNA-U Z monomers, one in each LNA wing, display similar knockdown efficiency (∼50% at 80 nM), with no observable cell death, as the corresponding ASO with LNA-T when cationic transfection agents are used (results not shown). Additional studies are necessary to assess the full potential of C5-amino acid functionalized LNA-U monomers as ASO modifications, including their promise for enhanced and/or gymnotic ASO delivery.
In summary, C5-amino acid functionalized LNA uridine phosphoramidites represent a novel approach to chemical diversification of LNA. ONs containing monomers X/Y/Z display significantly higher affinity toward complementary RNA than canonical LNA ONs due to extended conjugation of the nucleobase, as well as stabilizing electrostatic interactions. The excellent specificity of LNA modified ONs towards singly mismatched RNA is maintained. Additionally, monomers X/Y/Z are inert towards enzymatic digestion by 3′-endonuclease (SVPDE), which sets them apart from other high-affinity C5-LNA-U monomers.7 The promising biophysical properties and cationic character render C5-amino acid functionalized LNA positively poised for antisense applications.
We appreciate financial support from the National Institute of General Medical Sciences, National Institutes of Health (award number GM088697), and NIH Grant #P20 RR016454 from the INBRE Program of the National Center of Research Resources and P20 GM103408 (National Institute of General Medical Sciences). We thank the EBI Murdock Mass Spectrometry Center and Lee Deobald (Univ. Idaho) and Michael Østergaard (Isis Pharmaceuticals) for MS analysis, Alex Blumenfeld (Univ. Idaho) for NMR analysis, and Deep Pokharel and Rod Hill (Univ. Idaho) for advice on cell culture studies.
Notes and references
- For reviews see: (a) S. M. Freier and K.-H. Altmann, Nucleic Acids Res., 1997, 25, 4429 CrossRef CAS PubMed; (b) C. F. Bennett and E. E. Swayze, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 259 CrossRef CAS PubMed; (c) G. F. Deleavey and M. J. Damha, Chem. Biol., 2012, 19, 937 CrossRef CAS PubMed.
- For reviews see: (a) S. Obika, S. M. A. Rahman, A. Fujisaka, Y. Kawada, T. Baba and T. Imanishi, Heterocycles, 2010, 81, 1347 CrossRef CAS PubMed; (b) C. Zhou and J. Chattopadhyaya, Chem. Rev., 2012, 112, 3808 CrossRef CAS PubMed; (c) P. P. Seth and E. E. Swayze, in Natural Products in Medicinal Chemistry, ed. S. Hanessian, Wiley-VCH, Weinheim, 1st edn, 2014, pp. 403–439 Search PubMed.
- (a) S. K. Singh, P. Nielsen, A. A. Koshkin and J. Wengel, Chem. Commun., 1998, 455 RSC; (b) S. Obika, D. Nanbu, Y. Hari, J.-I. Andoh, K.-I. Morio, T. Doi and T. Imanishi, Tetrahedron Lett., 1998, 39, 5401 CrossRef CAS; (c) H. Kaur, B. R. Babu and S. Maiti, Chem. Rev., 2007, 107, 4672 CrossRef CAS PubMed.
- For recent examples see: (a) Y. Hari, T. Morikawa, T. Osawa and S. Obika, Org. Lett., 2013, 15, 3702 CrossRef CAS PubMed; (b) M. T. Migawa, T. P. Prakash, G. Vasquez, P. P. Seth and E. E. Swayze, Org. Lett., 2013, 15, 4316 CrossRef CAS PubMed; (c) S. Hanessian, J. Wagger, B. L. Merner, R. D. Giacometti, M. E. Østergaard, E. E. Swayze and P. P. Seth, J. Org. Chem., 2013, 78, 9064 CrossRef CAS PubMed; (d) A. R. Shrestha, Y. Kotobuki, Y. Hari and S. Obika, Chem. Commun., 2014, 50, 575 RSC.
- For reviews see: (a) I. Luyten and P. Herdewijn, Eur. J. Med. Chem., 1998, 33, 515 CrossRef CAS; (b) M. Ahmadian and D. E. Bergstrom, in Modified Nucleosides in Biochemistry, Biotechnology and Medicine, ed. P. Herdewijn, Wiley-VCH, Weinheim, 1st edn, 2008, pp. 251–276 Search PubMed.
- For representative examples see: (a) H. Hashimoto, M. G. Nelson and C. Switzer, J. Am. Chem. Soc., 1993, 115, 7128 CrossRef CAS; (b) R. W. Wagner, M. D. Matteucci, J. G. Lewis, A. J. Gutierrez, C. Moulds and B. C. Froehler, Science, 1993, 260, 1510 CAS; (c) H. Nara, A. Ono and A. Matsuda, Bioconjugate Chem., 1995, 6, 54 CrossRef CAS; (d) M. Ahmadian, P. M. Zhang and D. E. Bergstrom, Nucleic Acids Res., 1998, 26, 3127 CrossRef CAS PubMed; (e) L. E. Heystek, H. Q. Zhou, P. Dande and B. Gold, J. Am. Chem. Soc., 1998, 120, 12165 CrossRef CAS; (f) V. Roig and U. Asseline, J. Am. Chem. Soc., 2003, 125, 4416 CrossRef CAS PubMed; (g) J. Booth, T. Brown, S. J. Vadhia, O. Lack, W. J. Cummins, J. O. Trent and A. N. Lane, Biochemistry, 2005, 44, 4710 CrossRef CAS PubMed; (h) T. Kottysch, C. Ahlborn, F. Brotzel and C. Richert, Chem. – Eur. J., 2004, 10, 4017 CrossRef CAS PubMed.
- P. Kumar, M. Østergaard, B. Bharal, B. A. Anderson, D. C. Guenther, M. Kaura, D. J. Raible, P. K. Sharma and P. J. Hrdlicka, J. Org. Chem., 2014, 79, 5047 CrossRef CAS PubMed.
- F. Debart, S. Abes, G. Deglane, H. M. Moulton, P. Clair, M. J. Gait, J.-J. Vasseur and B. Lebleu, Curr. Top. Med. Chem., 2007, 7, 727 CrossRef CAS.
- For recent examples see: (a) G. Deglane, S. Abes, T. Michel, P. Prevot, E. Vives, F. Debart, I. Barvik, B. Lebleu and J.-J. Vasseur, ChemBioChem, 2006, 7, 684 CrossRef CAS PubMed; (b) M. Park, D. Canzio and T. C. Bruice, Bioorg. Med. Chem. Lett., 2008, 18, 2377 CrossRef CAS PubMed; (c) K. T. Gagnon, J. K. Watts, H. M. Pendergraff, C. Montaillier, D. Thai, P. Potier and D. R. Corey, J. Am. Chem. Soc., 2011, 133, 8404 CrossRef CAS PubMed.
- (a) M. W. Johannsen, L. Crispino, M. C. Wamber, N. Kalra and J. Wengel, Org. Biomol. Chem., 2011, 9, 243 RSC; (b) J. Lietard, D. Ittig and C. J. Leumann, Bioorg. Med. Chem., 2011, 19, 5869 CrossRef CAS PubMed; (c) K. Mori, T. Kodama, T. Baba and S. Obika, Org. Biomol. Chem., 2011, 9, 5272 RSC.
- J. L. Mergny and L. Lacroix, Oligonucleotides, 2003, 13, 515 CrossRef CAS PubMed.
- Handbook of Chemistry and Physics, ed. D. R. Lide, CRC Press, Boca Raton, 87th edn, 2006, p. 7-1 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthetic protocols, NMR spectra and MS data for new nucleosides and ONs; thermal denaturation curves; additional Tm and thermodynamic data; sequences of AONs. See DOI: 10.1039/c4cc03623a |
| ‡ For an alternative synthesis of compound 2, see ESI.† |
| § Coupling between Fmoc-protected amino acids and 2 was unsuccessful. |
| This journal is © The Royal Society of Chemistry 2014 |