Open Access Article
Open Access ArticleAn acyl-SAM analog as an affinity ligand for identifying quorum sensing signal synthases†
Kenji
Kai
*,
Hiroki
Fujii
,
Rui
Ikenaka
,
Mitsugu
Akagawa
and
Hideo
Hayashi
Graduate School of Life and Environmental Sciences, Osaka Prefecture University, 1-1 Gakuen-cho, Naka-ku, Sakai, Osaka 599-8531, Japan. E-mail: kai@biochem.osakafu-u.ac.jp
First published on 16th June 2014
Abstract
N-Acylhomoserine lactones (AHLs) are quorum sensing signals produced by Gram-negative bacteria. We here report the affinity purification of AHL synthases using beads conjugated with an enzyme inhibitor, which was designed based on the catalytic intermediate acyl-SAM.
Bacteria communicate using chemical signals to sense cell density and regulate diverse coordinated behaviors by a process known as quorum sensing (QS).1,2N-Acylhomoserine lactones (AHLs), one class of QS signals in Gram-negative bacteria, are synthesized from the acyl–acyl carrier protein (acyl-ACP) and S-adenosylmethionine (SAM) by LuxI-type synthases (Fig. 1a).3,4 This catalytic reaction is thought to be proceeded by a two-step mechanism involving the intermediate acyl-SAM; the transfer of an acyl group from acyl-ACP to the amino group of SAM and lactonization of the methionine part, concomitant with the release of methylthioadenosine (MTA).3,5,6 AHLs differ in the length and substitution of their respective acyl side chains, which confers them with signal specificity.1,2,7 In certain strains, AHLs possessing the phenylpropanoyl moiety have also been utilized as QS signals.8,9 When the cell density increases and the concentration of AHLs reaches a threshold level, AHLs bind to the cognate LuxR-type receptors, and these complexes then activate the expression of target genes. In many pathogenic bacteria, this process results in pathogenic events such as the formation of biofilms and production of virulence factors.1,2 Therefore, QS modulators have potential uses in pharmaceutical and agrochemical fields by preventing microbial infection in hosts.10–12
 | ||
| Fig. 1 (a) Mechanism of AHL production by LuxI-type AHL synthases. (b) Structures of acyl-SAM analogs (1 and 4) and SAM analogs (2 and 3). | ||
The identification of AHL synthases is a prerequisite for understanding AHL-based QS systems in Gram-negative bacteria, and mostly depends on screening of genome/cDNA libraries or mutants of target bacteria,8,9,13,14 and more recently on genome analyses.15 Although some degenerated primer sets to amplify AHL synthase genes by PCR have been reported, their application has been limited to restricted members of Gram-negative bacteria.16,17 As described above, the catalytic mechanism of AHL synthases is thought to be conserved among Gram-negative bacteria; therefore, chemical probes that can bind to AHL synthases may lead to the development of a ligand-based approach for identifying and isolating the key enzymes. The competitive inhibitors of AHL synthases may be candidates for the affinity ligands. Chung et al. described an AHL synthase inhibitor, named J8-C8, from a chemical library of AHL antagonists and demonstrated that its inhibitory effect against TofI, an AHL synthase of the plant pathogenic bacterium Burkholderia glumae, was synergistic with the reaction byproduct MTA.18 This finding demonstrated, using X-ray crystal analysis, the binding mode of J8-C8 and MTA to the TofI enzyme of the TofI–J8-C8–MTA complex. However, these compounds only exhibited a weak inhibitory effect on the TofI enzyme. Christensen et al. recently reported a strong inhibitor, which inhibited BmaI1, an AHL synthase from Burkholderia mallei, with a Kappi value of 0.7 μM.19 This is the first inhibitor to inhibit AHL synthase with an effective Ki value, and, thus, opens up a new avenue for the chemical inhibition of bacterial QS. However, it is yet to be determined whether this inhibitor is suitable for an affinity ligand for AHL synthases because the mechanism of inhibition is noncompetitive; therefore, the possibility of nonspecific interactions between the inhibitor and proteins other than AHL synthases remains. Therefore, in order to develop a method for the affinity purification of AHL synthase, we firstly need to develop a strong and specific competitive inhibitor of the enzyme.
We speculated that acyl-SAM analogs may be good candidates for competitive inhibitors of AHL synthases because many examples of catalytic intermediate analogs were previously reported to be good inhibitors of target enzymes,20–24 and the intermediate acyl-SAM was shown to be specific to the synthesis of AHL in Gram-negative bacteria.3,5,6 Furthermore, to the best of our knowledge, acyl-SAM has not yet been detected in other organisms; thus, the analogs may also be useful for identifying the AHL synthase of an uncultured bacterial symbiont from the host cell lysate.25 In the case of TofI, N-octanoylhomoserine lactone (C8-HSL) was synthesized from octanoyl-ACP and SAM.18 Hence, we firstly designed and synthesized N-octanoyl-S-adenosyl-L-homocysteine (1) (Fig. 1b) as an octanoyl-SAM analog, which lacked the methyl group at the 5′-sulfur atom of the intermediate. The activity of 1 was tested for its ability to inhibit AHL synthase using octanoyl-ACP (10 μM), which was prepared from the ACP of Escherichia coli K-12 by chemical acylation,26,27 with SAM (10 μM) as the substrate and the TofI enzyme (1 μM), which was heterogeneously expressed in E. coli strain BL21(DE3)pLysS.18 The C8-HSL product was quantified by GC–MS.25,28 Analog 1 showed 91% and 51% inhibition at concentrations of 100 and 10 μM, respectively (Table 1). Kinetic analysis indicated that 1 acted as a competitive inhibitor of TofI activity with a Ki value of 4.8 ± 0.7 μM, which was determined by a Dixon plot of 1/V0versus the inhibitor concentration (Fig. S1, ESI†). On the other hand, the SAM analogs, S-adenosylhomocysteine (2) and sinefungin (3) (Fig. 1b), showed moderate or no inhibition of TofI at the concentrations tested (Table 1), although they were reported to have some inhibitory effects on AHL synthase.3 Therefore, it is reasonable to assume that mimics of the reaction intermediate acyl-SAM may be good competitive inhibitors and affinity ligands of AHL synthases.
| Compound | Inhibitiona (%) | K i (μM) | ||
|---|---|---|---|---|
| 100 μM | 10 μM | 1 μM | ||
| a The percentage inhibition was calculated from the ratio of C8-HSL production with and without an inhibitor; n.i. = no inhibition (i.e., <5% inhibition). Data represent the average of triplicate determinations. b K i values were determined by a Dixon plot of V0versus the inhibitor concentration. | ||||
| 1 | 91 | 51 | 4.8 | |
| 2 | 51 | 8 | ||
| 3 | n.i. | n.i. | ||
| 4 | 91 | 43 | 12 | 6.7 |
| 5 | 87 | 40 | 0.73 | |
| 6 | 91 | 52 | 0.22 | |
| 7 | >98 | 83 | 0.11 | |
| 8 | 83 | 21 | 0.88 | |
| 9 | 88 | 48 | ||
| 10 | 94 | 51 | ||
Based on the above results, we next designed and synthesized decarboxy analog 4 (Fig. 1b). This compound could simplify the structure of 1, making it easy to prepare the derivatives of the octanoyl-SAM analog. Analog 4 was able to inhibit the TofI reaction by 91% at 100 μM and 43% at 10 μM (Table 1). The Ki value of 4 was determined to be 6.7 ± 0.7 μM (Fig. S2, ESI†), which was similar to that of 1. These results indicated that the carboxy group of analog 1 was not needed to inhibit TofI.
We then attempted to improve the TofI inhibitory activity of 4. Based on a structure–activity relationship study of 4 against TofI (see ESI†), we confirmed that the C8-acyl chain including the amide and adenosine moieties of 4 were recognized by TofI and, thus, suggested that the enzyme may have high affinity to the catalytic intermediate octanoyl-SAM. The findings of a previously conducted crystallographic analysis indicated that J8-C8 and MTA bound to TofI, occupying the binding sites of the acyl chain of octanoyl-ACP and the adenosine moiety of SAM, respectively.18 Therefore, it was reasonable to speculate that the binding mode of the respective substructures of octanoyl-SAM to the enzyme was similar to those of J8-C8 and MTA observed in the ternary complex. If this was the case, J8-C8 and MTA in the crystal could be replaced by octanoyl-SAM. However, the crystallographic analysis also revealed that the binding sites of J8-C8 and MTA were separated in TofI and also the presence of the large channel in which the reactions for acylation and lactonization of the methionine part in SAM can occur. Thus, the pocket of the TofI crystal appeared to be slightly larger than the size of octanoyl-SAM. Therefore, we predicted that the length between the sulfur atom and amide in octanoyl-SAM analogs may be an important factor and that elongation of the methylene chain of 4 could improve the inhibitory activity of 4 against TofI.
Thus, we designed and synthesized analogs 5–8 (Fig. 2a) in which the methylene chains between the sulfur atom and amide in 4 were extended to C4–C7, respectively. As expected, the inhibitory activities of these analogs were higher than that of 4, and they almost completely inhibited the production of C8-HSL by TofI at a concentration of 10 μM, while 5, 6, and 7 were effective to a similar degree at 1 μM (Table 1). Analogs 6 and 7 showed high inhibitory activity against TofI with Ki values of 0.22 ± 0.03 and 0.11 ± 0.02 μM, respectively (Fig. S3 and S4, ESI†). A docking simulation provided a reasonable model in which analog 6 bound to the pocket of TofI covering the acyl chain of J8-C8 and the adenosine of MTA (Fig. 2b, Fig. S6a and b, ESI†). However, in a docking model of 7 in TofI, the analog did not match the model of 6, rather the acyl chain of 7 fit to the upper space of the binding site of that of 6 (Fig. S6c, ESI†). The possibility remains that analog 7 also binds to TofI in the same manner as that of 6. Future experiments (e.g., X-ray crystallographic analyses of these inhibitors with TofI) are needed to clarify their binding mode. Because analog 8 showed lower activity than analogs 5–7, the methylene chains between the sulfur atom and amide in 8 were too long to fit well into TofI. Analogs 6 and 7 were conclusively found to be effective inhibitors that satisfied the binding site scaffold of the TofI enzyme. Due to its high hydrophobicity, analog 7 appeared to be difficult to handle during the preparation of affinity beads. Therefore, we selected 6 as an affinity ligand for the isolation of AHL synthases.
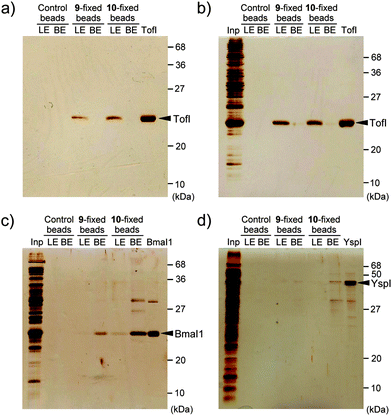
The above docking simulation of analog 6 to TofI also suggested that the 2′- and 3′-hydroxy groups of analog 6 presented almost at the surface of the enzyme (Fig. S6b, ESI†). Therefore, we introduced a propargyl linker to either of the hydroxy groups in an ether form, and the resulting alkynes 9 and 10 that formed were fixed to azide-type magnetic beads (FG beads)29 using click chemistry (Fig. 3). Ligands 9 and 10 also inhibited TofI to the same extent as analog 6 (Table 1). Thus, we confirmed that the propargyl linkers did not affect their inhibitory activity or binding to TofI. The results of pull-down assays of the ligand-fixed beads using purified TofI are shown in Fig. 4a. Control beads (no ligand) did not bind to TofI. In contrast, TofI was bound to 9- and 10-fixed beads and was effectively eluted by 6 solution (250 μM). These results indicated that these ligand-fixed beads possessed the ability to capture the TofI protein. Thus, 9- and 10-fixed beads were used in pull-down assays with the whole cell protein extracts of E. coli strain BL21(DE3)pLysS expressing TofI. The results obtained demonstrated that only TofI was purified from crude samples (Fig. 4b), which indicated that the interaction between TofI and the ligand-fixed beads was very specific. We also confirmed the competitive inhibition of their interactions by the natural ligand SAM (Fig. S7, ESI†). Therefore, these results suggested that 9- and 10-fixed beads were a good affinity matrix for AHL synthases.
Finally, to address the utility of 9- and 10-fixed beads in the purification of other AHL synthases, we carried out pull-down assays of BmaI1, a Burkholderia mallei AHL synthase, and YspI, a Yersinia pestis AHL synthase,19 using the beads. As a result of the pull-down assay of the whole cell protein extracts of E. coli strain Tuner(DE3) expressing BmaI1, we obtained BmaI1 as a major band from the samples obtained by ligand and boiling elutions of ligand-fixed beads (Fig. 4c). The binding of BmaI1 to the ligand-fixed beads should be stronger than that of Tof I because BmaI1 was difficult to elute with 6 solution, but was effectively eluted by boiling the beads in the SDS sample buffer. 10-fixed beads captured BmaI1 more effectively than 9-fixed beads, suggesting that the preparation of several conjugation-type beads is needed to successfully isolate AHL synthases by pull-down assays. In contrast, YspI could not be effectively purified by pull-down assays from the E. coli cell lysate, in which only weak binding of YspI to the ligand-fixed beads was observed (Fig. 4d). This was attributed to YspI mainly producing N-(3-oxooctanoyl)homoserine lactone (3-oxo-C8-HSL) and N-(3-oxohexanoyl)homoserine lactone (3-oxo-C6-HSL),30 while BmaI1 produced C8-HSL.19 Therefore, we confirmed the specificity of these beads for C8-HSL-type AHL synthases.
In conclusion, we developed strong AHL synthase inhibitors, which were designed based on the catalytic intermediate acyl-SAM, for affinity ligands of AHL synthases. We also demonstrated the usefulness of acyl-SAM analog-fixed beads in pull-down assays to isolate AHL synthases. This is the first study to show that QS signal synthases could be purified by a ligand-based affinity protocol. Furthermore, acyl-SAM analogs are expected to be the lead compounds for antivirulence agents because they showed high specific binding to AHL synthases. The identification of AHL synthases and their genes is currently based on gene/mutant screening or genome analysis. In addition to these approaches, acyl-SAM analog-based affinity ligands will provide a new method to identify unknown AHL synthases in Gram-negative bacteria.
We gratefully acknowledge Prof. Ingyu Hwang (Seoul National University) and Prof. E. Peter Greenberg (University of Washington) for providing plasmids, Dr Toshiyuki Harada (Sumitomo Chemical) for his assistance in constructing docking models, Prof. Naoharu Watanabe and Dr Hiroyuki Takemoto (Shizuoka University) for HRMS measurements, and Prof. Jun Hiratake (Kyoto University) for his valuable discussions. This work was supported by JSPS KAKENHI Grant Number: 23658101, 26450140.
Notes and references
- C. M. Waters and B. L. Bassler, Annu. Rev. Cell Dev. Biol., 2005, 21, 319 CrossRef CAS PubMed.
- W. Galloway, J. Hodgkinson, S. Bowden, M. Welch and D. Spring, Chem. Rev., 2011, 111, 28 CrossRef CAS PubMed.
- M. R. Parsek, D. L. Val, B. L. Hanzelka, J. E. Cronan and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 4360 CrossRef CAS.
- W. T. Watson, T. D. Minogue, D. L. Val, S. B. von Bodman and M. E. A. Churchill, Mol. Cell, 2002, 9, 685 CrossRef CAS.
- A. Raychaudhuri, A. Jerga and P. Tipton, Biochemistry, 2005, 44, 2974 CrossRef CAS PubMed.
- A. Raychaudhuri, A. Tullock and P. Tipton, Biochemistry, 2008, 47, 2893 CrossRef CAS PubMed.
- A. Yajima, Tetrahedron Lett., 2014, 55, 2273 CrossRef PubMed.
- A. L. Schaefer, E. P. Greenberg, C. M. Oliver, Y. Oda, J. J. Huang, G. Bittan-Banin, C. M. Peres, S. Schmidt, K. Juhaszova, J. R. Sufrin and C. S. Harwood, Nature, 2008, 454, 595 CrossRef CAS PubMed.
- N. A. Ahlgren, C. S. Harwood, A. L. Schaefer, E. Giraud and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 7183 CrossRef PubMed.
- M. E. Mattmann and H. E. Blackwell, J. Org. Chem., 2010, 75, 6737 CrossRef CAS PubMed.
- G. D. Geske, J. C. O'Neill and H. E. Blackwell, Chem. Soc. Rev., 2008, 37, 1432 RSC.
- W. R. J. D. Galloway, J. T. Hodgkinson, S. Bovvden, M. Welch and D. R. Spring, Trends Microbiol., 2012, 20, 449 CrossRef CAS PubMed.
- M. Mattiuzzo, I. Bertani, S. Ferluga, L. Cabrio, J. Bigirimana, C. Guarnaccia, S. Pongor, H. Maraite and V. Venturi, Environ. Microbiol., 2011, 13, 145 CrossRef CAS PubMed.
- J. Zan, E. M. Cicirelli, N. M. Mohamed, H. Sibhatu, S. Kroll, O. Choi, C. L. Uhlson, C. L. Wysoczinski, R. C. Murphy, M. E. A. Churchill, R. T. Hill and C. Fuqua, Mol. Microbiol., 2012, 85, 916 CrossRef CAS PubMed.
- J. Gao, A. Ma, X. Zhuang and G. Zhuang, Appl. Environ. Microbiol., 2014, 80, 951 CrossRef CAS PubMed.
- K. Jangid, R. Kong, M. S. Patole and Y. S. Shouche, BMC Microbiol., 2007, 7 DOI:10.1186/1471-2180-7-93.
- Y. Huang, Y. Zeng and Z. Yu, Curr. Microbiol., 2013, 67, 183 CrossRef CAS PubMed.
- J. Chung, E. Goo, S. Yu, O. Choi, J. Lee, J. Kim, H. Kim, J. Igarashi, H. Suga, J. S. Moon, I. Hwang and S. Rhee, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12089 CrossRef CAS PubMed.
- Q. H. Christensen, T. L. Grove, S. J. Booker and E. P. Greenberg, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 13815 CrossRef CAS PubMed.
- J. V. Schloss and W. W. Cleland, Biochemistry, 1982, 21, 4420 CrossRef CAS.
- H. Oikawa, K. Yagi, S. Ohashi, K. Watanabe, T. Mie, A. Ichihara, M. Honma and K. Kobayashi, Biosci., Biotechnol., Biochem., 2000, 64, 2368 CrossRef CAS.
- M. S. Maurice and S. L. Bearne, Biochemistry, 2000, 39, 13324 CrossRef PubMed.
- M. P. Storz, C. K. Maurer, C. Zimmer, N. Wagner, C. Brengel, J. C. de Jong, S. Lucas, M. Müsken, S. Häussler, A. Steinbach and R. W. Hartmann, J. Am. Chem. Soc., 2012, 134, 16143 CrossRef CAS PubMed.
- A. M. Haapalainen, K. Thomas, P. C. Tyler, G. B. Evans, S. C. Almo and V. L. Schramm, Structure, 2013, 21, 963 CrossRef CAS PubMed.
- K. Kai, K. Furuyabu, A. Tani and H. Hayashi, ChemBioChem, 2012, 13, 1776 CrossRef CAS PubMed.
- C. O. Rock and J. E. Cronan, Methods Enzymol., 1981, 71, 341 CAS.
- J. E. Cronan and A. L. Klages, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 5440 CrossRef CAS.
- K. Kai, A. Tani and H. Hayashi, Bioorg. Med. Chem., 2010, 18, 3776 CrossRef CAS PubMed.
- S. Sakamoto, Y. Kabe, M. Hatakeyama, Y. Yamaguchi and H. Handa, Chem. Rec., 2009, 9, 66 CrossRef CAS PubMed.
- J. P. Kirwan, T. A. Gould, H. P. Schweizer, S. W. Bearden, R. C. Murphy and M. E. A. Churchill, J. Bacteriol., 2006, 188, 784 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4cc03094j |
| This journal is © The Royal Society of Chemistry 2014 |