 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The causality principle in the reconstruction of sparse NMR spectra†
M.
Mayzel
a,
K.
Kazimierczuk
b and
V. Yu.
Orekhov
*a
aSwedish NMR Centre, University of Gothenburg, Box 465, S-405 30 Göteborg, Sweden. E-mail: vladislav.orekhov@nmr.gu.se
bCentre of New Technologies, University of Warsaw, Banacha 2C, 02-097, Warsaw, Poland
First published on 23rd June 2014
Abstract
Non-uniform sampling offers a dramatic increase in the power and efficiency of magnetic resonance techniques in chemistry, molecular structural biology, and other fields. Here we show that use of the causality property of an NMR signal is a general approach for major reduction of measuring time and quality improvement of the sparsely detected spectra.
The invention of multidimensional magnetic resonance (MR) experiments 40 years ago led to the success of the modern MRI and NMR spectroscopy in medicine, chemistry, molecular structural biology, and other fields. This approach, however, has an important weakness: the detailed site-specific information and ultimate resolution obtained in two and higher dimensional experiments are contingent on the lengthy data collection required for systematic uniform sampling of the large multidimensional space spanned by the indirectly detected spectral dimensions.1 A fundamental solution to this problem stems from an observation that upon appropriate transform, e.g. from the NMR time to frequency domain, the MR signal becomes nearly-black or sparse, i.e. essentially zero in the vast majority of points and thus largely redundant. Darkness of the MR images and NMR spectra is a key for the remarkable success and rapid development of the non-uniform sampling (NUS) methods.1–7 The darker an object is, the less experimental measurements are needed for its recovery.8 The transform that brings data into a dark presentation is called sparsifying transform. In NMR, the Fourier transform connects the complex free induction decay (FID) signal in the time domain and frequency spectrum. A properly phased spectrum consists of the real absorption part used for the analysis and the redundant imaginary dispersion part. Since an absorption signal is much narrower than the dispersion, the latter contributes the most to the total spectrum brightness. The main result of this paper is the notion that for most of the currently used algorithms, e.g. compressed sensing,4,5 SIFT,9 maximum entropy,2,7 MINT,6etc., it is the dispersion part that sets the lower limit for the amount of measured data required for the high quality spectrum reconstruction from the NUS signal. We show that the causality property of the NMR signal can be used to construct a sparsifying transform, which eliminates the spectral dispersion part and, thus, allows spectrum reconstruction with better fidelity and from fewer measurements. In NMR, the causality reflects the fact that the FID signal is only observed after excitation of the spin system, e.g. by a radiofrequency pulse, and is zero before the excitation.
It is well known that the Fourier transform of a causal time signal S(t) leads to a spectrum, whose real and imaginary parts can be produced from each other using the Kramers–Kronig relations also known as the Hilbert transform.10 The Kramers–Kronig relations are illustrated in Fig. 1. Signal SFID(t) (Fig. 1a) and the corresponding spectrum in Fig. 1b are related via the Fourier transform. The spectrum in Fig. 1d is produced from the one in Fig. 1b by zeroing its imaginary part. The inverse Fourier transform of the real spectrum in panel d gives a complex time domain signal (Fig. 1c), whose real and imaginary parts are essentially even and odd parts of the real and imaginary components of the FID (Fig. 1a), respectively. Thus, the signal in Fig. 1c can also be produced by the time reversal and complex conjugate of the FID.
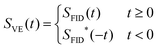 | (1) |
In the following, we call the SVE(t) signal in eqn (1) virtual-echo (VE). The original signal SFID(t) can be obtained from SVE(t) by zeroing the signal for negative time. Direct transition from panel d to panel b in Fig. 1 is done by the Hilbert transform. In practice, the Hilbert transform algorithm takes the detour d → c → a → b (Fig. 1) in order to use the computationally efficient fast Fourier transform.
The spectrum (Fig. 1d) obtained from the VE representation (Fig. 1c) consists of the traditionally looking real part and zero imaginary part. Depending on the signal phase, the real part can contain absorption, dispersion, or a mixture of the both modes. Given a priori, the phase, eqn (1) allows us to obtain the time domain signal corresponding to the pure absorption spectrum and, thus, to construct a sparsifying transform that produces a significantly darker spectrum than the traditional Fourier transform of the original FID.
Obtaining NMR spectrum from a time-domain signal is a typical example of the mathematical inverse problem. When all data points in the signal are present, the solution of the problem is trivial and is given by the Discrete Fourier Transform (DFT). In the case of NUS, most of the data in the time-domain signal are missing and the unconstrained inverse problem has an infinite number of solutions (i.e. spectra). A unique and “correct” spectrum is obtained by introducing additional assumptions such as minimal power, maximum entropy, maximal sparseness, etc. The VE presentation is equally applicable to traditional fully sampled and NUS signals. When the former is processed using DFT, FID and VE presentations lead to the equivalent spectra as illustrated in Fig. 1. However, when reconstructing spectra from the NUS signal and in some other cases,11 use of the Kramers–Kronig relations, namely path a → c → d in Fig. 1, represents a significant advantage over the traditional processing, which is a → b → d.
Fig. 2 demonstrates the benefits of the VE signal for two modern spectra recovering algorithms used for the NUS signal: spectroscopy by Integration of Frequency and Time Domain (SIFT)9 and Compressed Sensing by Iterative Reweighted Least Squares (CS-IRLS).4,12 Similar results for the alternative CS algorithm, Iterative Soft Thresholding (CS-IST),4,13,14 are presented in Fig. S3 (ESI†). Both CS algorithms and SIFT can be applied without modifications to either the traditional FID or VE signal. With SIFT making use of the prior knowledge about positions of dark regions in a spectrum and CS searching for the darkest among all possible spectra consistent with the measured data, both methods are expected to benefit from the darker representation of the spectrum provided by VE.
For a given number of NUS measurements, quality of the SIFT reconstruction improves, when the larger fraction of the spectrum area is free from signals and contains only the baseline noise. In our calculations, the signal-free area is defined by a mask, which excludes rectangles of defined size around all peaks in the spectrum. This corresponds, for example, to a set-up in relaxation and kinetics studies,15 where the peak positions are known and only their intensities or integrals need to be defined. Fig. 2a and b show reconstructions of a 2D 1H–15N HSQC spectrum of human alpha-synuclein obtained using only 15% of the data from the full experiment.
By avoiding broad dispersion peaks, the VE signal ensures that a larger fraction of the spectrum is “dark” and thus SIFT produces a much better spectrum (Fig. 2b and Fig. S4, ESI†) and more accurate peak intensities in comparison to the reconstruction from the original FID (Fig. 2e and Fig. S5, ESI†). Fig. 2e (inset) illustrates that prior information about the signal phase does not have to be exact. For the SIFT example, the peak intensities in the VE reconstruction obtained for the uncorrected up to 15° phase are still better reproduced than those measured in the spectrum calculated for the traditional FID representation. A similar behaviour is also observed for the CS algorithms. For most of the multidimensional experiments, zero order phases for the indirect spectral dimensions are known and thus can be corrected in the time domain to values close to zero prior to the spectrum reconstruction.
Similarly to SIFT, CS also assumes that the major part of a spectrum is dark. However, no assumption is made about the exact location of the dark regions, which creates an apparently unsolvable combinatorial problem. Yet, it has been recently reformulated as a relatively simple task of spectral lp-norm (0 < p ≤ 1) minimization:16
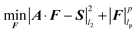 | (2) |
| |F|lp = (|F1|p + |F2|p + ⋯ + |FN|p)1/p | (3) |
In the present paper p = 1 is used for the IST algorithm13 and lp-norm with p iteratively approaching 0 for the IRLS algorithm.4,17 The use of the CS method in NMR spectroscopy has been commented recently by many authors,4,5,18,19 with important conclusions on the limited applicability to non-random sampling20 and superior performance of non-convex lp-norms (p < 1).19,21
Here we apply the CS IRLS algorithm4 to reconstruct a 3D HNCO spectrum sampled at the level of 0.7%, without VE (Fig. 2c) and with VE in both indirect dimensions (Fig. 2d). It can be seen that VE improves the reconstruction significantly by providing better line shapes, more accurate peak intensities (Fig. 2f), and revealing low intensity signals. Fig. S3 (ESI†) shows a notable improvement for the 2D 1H–15N HSQC spectrum of intrinsically disordered protein alpha-synuclein processed with CS-IST.
The effect can be explained using the basic CS theorem, binding the number of properly reconstructed spectral points, which is essentially a measure of spectrum darkness, with the sampling level.16 With the VE, fewer points contribute to each peak in the spectrum and thus relatively low sampling level is sufficient to fulfil the condition for the successful CS reconstruction. It should be emphasized that the striking advantage of the VE demonstrated in Fig. 2 and Fig. S3–S5 (ESI†) is mostly due to the very low sampling level. Without the VE, high quality reconstructions by CS and SIFT are also possible, but require at least twice as many sampling points for the presented spectra (inset in Fig. 2f and Fig. S4, ESI†).
As pointed out by Donoho et al.,8 there is an unambiguous relationship between the darkness of the NMR spectrum and the quality of the spectral reconstruction by the maximum entropy or minimum l1-norm minimisation. It is therefore likely that most of the related methods including FM-reconstruction,22 MINT,6 hmsIST,14 QME,7etc. will also benefit from the VE signal.
We show that the causality property of the NMR signal can be exploited to dramatically enhance the performance of the CS, SIFT and probably many other algorithms commonly used for the reconstruction of NUS spectra. Our findings open a way for significant reduction in measurement time and improvement of the quality of NUS spectra and thus should increase the power and appeal of multidimensional NMR spectroscopy in multitude of its existing and future applications. The method is particularly useful for short living systems, time resolved measurements, and high-dimensional experiments on intrinsically disordered proteins.
The work was supported by the Swedish Research Council (research grant 2011-5994); Swedish National Infrastructure for Computing (grant SNIC 001/12-271); Polish National Centre of Science (grant DEC-2012/07/E/ST4/01386); Polish Ministry of Science and Higher Education (grant IP2011 023171); and Foundation for Polish Science, TEAM programme. We thank Dina Katabi and Haitham Hassanieh (Dept Electr Eng & Comput. Sci., Massachusetts Institute of Technology) for an inspiring discussion and Anna Zawadzka-Kazimierczuk (Biological and Chemical Research Centre, University of Warsaw established from EU Regional Development Fund) for the HSQC spectrum of alpha-synuclein; The EU FP7 Bio-NMR project (contract 261863); The Knut and Alice Wallenberg foundation project NMR for Life.
Notes and references
-
M. Billeter and V. Y. Orekhov, in Novel Sampling Approaches in Higher Dimensional NMR, ed. M. Billeter and V. Y. Orekhov, Springer, Heidelberg Dordrecht London New York, 2012, vol. 316, pp. ix–xiv Search PubMed
.
- J. C. J. Barna, E. D. Laue, M. R. Mayger, J. Skilling and S. J. P. Worrall, Biochem. Soc. Trans., 1986, 14, 1262–1263 Search PubMed
; S. G. Hyberts, K. Takeuchi and G. Wagner, J. Am. Chem. Soc., 2010, 132, 2145–2147 CrossRef CAS PubMed
.
- V. Y. Orekhov, I. Ibraghimov and M. Billeter, J. Biomol. NMR, 2003, 27, 165–173 CrossRef CAS
; B. E. Coggins, R. A. Venters and P. Zhou, J. Am. Chem. Soc., 2004, 126, 1000–1001 CrossRef PubMed
; R. Bruschweiler and F. L. Zhang, J. Chem. Phys., 2004, 120, 5253–5260 CrossRef PubMed
; V. Tugarinov, L. E. Kay, I. Ibraghimov and V. Y. Orekhov, J. Am. Chem. Soc., 2005, 127, 2767–2775 CrossRef PubMed
; D. Marion, J. Biomol. NMR, 2006, 36, 45–54 CrossRef PubMed
; V. Jaravine, I. Ibraghimov and V. Y. Orekhov, Nat. Methods, 2006, 3, 605–607 CrossRef PubMed
; M. Lustig, D. Donoho and J. M. Pauly, Magn. Reson. Med., 2007, 58, 1182–1195 CrossRef PubMed
; S. Hiller, R. G. Garces, T. J. Malia, V. Y. Orekhov, M. Colombini and G. Wagner, Science, 2008, 321, 1206–1210 CrossRef PubMed
; D. Sakakibara, A. Sasaki, T. Ikeya, J. Hamatsu, T. Hanashima, M. Mishima, M. Yoshimasu, N. Hayashi, T. Mikawa, M. Wälchli, B. O. Smith, M. Shirakawa, P. Güntert and Y. Ito, Nature, 2009, 457, 102–105 CrossRef PubMed
.
- K. Kazimierczuk and V. Y. Orekhov, Angew. Chem., Int. Ed., 2011, 50, 5556–5559 CrossRef CAS PubMed
.
- D. J. Holland, M. J. Bostock, L. F. Gladden and D. Nietlispach, Angew. Chem., Int. Ed., 2011, 50, 6548–6551 CrossRef CAS PubMed
.
- S. Paramasivam, C. L. Suiter, G. Hou, S. Sun, M. Palmer, J. C. Hoch, D. Rovnyak and T. Polenova, J. Phys. Chem. B, 2012, 116, 7416–7427 CrossRef CAS PubMed
.
- J. Hamatsu, D. O'Donovan, T. Tanaka, T. Shirai, Y. Hourai, T. Mikawa, T. Ikeya, M. Mishima, W. Boucher, B. O. Smith, E. D. Laue, M. Shirakawa and Y. Ito, J. Am. Chem. Soc., 2013, 135, 1688–1691 CrossRef CAS PubMed
.
- D. L. Donoho, I. M. Johnstone, J. C. Hoch and A. S. Stern, J. R. Stat. Soc. Ser. B, 1992, 54, 41–81 Search PubMed
.
- Y. Matsuki, M. T. Eddy and J. Herzfeld, J. Am. Chem. Soc., 2009, 131, 4648–4656 CrossRef CAS PubMed
.
-
S. H. Hall and H. L. Heck, IEEE, Wiley, Hoboken, N.J., 2009 CAS
; E. Bartholdi and R. R. Ernst, J. Magn. Reson., 1973, 11, 9–19 CAS
.
- A. Gibbs and G. A. Morris, J. Magn. Reson., 1991, 91, 77–83 CAS
.
- E. J. Candes, M. B. Wakin and S. P. Boyd, J. Fourier Anal. Appl., 2008, 14, 877 CrossRef
.
- A. Papoulis, IEEE Trans. Circuits Syst., 1975, 22, 735–742 CrossRef
.
- S. G. Hyberts, A. G. Milbradt, A. B. Wagner, H. Arthanari and G. Wagner, J. Biomol. NMR, 2012, 52, 315–327 CrossRef CAS PubMed
.
- D. M. Korzhnev, I. V. Ibraghimov, M. Billeter and V. Y. Orekhov, J. Biomol. NMR, 2001, 21, 263–268 CrossRef CAS
; Y. Matsuki, T. Konuma, T. Fujiwara and K. Sugase, J. Phys. Chem. B, 2011, 115, 13740–13745 CrossRef PubMed
; P. Selenko, D. P. Frueh, S. J. Elsaesser, W. Haas, S. P. Gygi and G. Wagner, Nat. Struct. Mol. Biol., 2008, 15, 321–329 Search PubMed
; M. Mayzel, J. Rosenlow, L. Isaksson and V. Y. Orekhov, J. Biomol. NMR, 2014, 58, 129–139 CrossRef PubMed
.
- E. J. Candes and M. B. Wakin, IEEE Signal Process. Mag., 2008, 25, 21–30 CrossRef
.
- A. E. Yagle, http://webeecsumichedu/~aey/sparsehtml, 2008.
- I. Drori, Eurasip J. Adv. Signal Process., 2007, 20248 CrossRef PubMed
.
- X. B. Qu, D. Guo, X. Cao, S. H. Cai and Z. Chen, Sensors, 2011, 11, 8888–8909 CrossRef CAS PubMed
.
- A. S. Stern, D. L. Donoho and J. C. Hoch, J. Magn. Reson., 2007, 188, 295–300 CrossRef CAS PubMed
.
- K. Kazimierczuk and V. Y. Orekhov, J. Magn. Reson., 2012, 223, 1–10 CrossRef CAS PubMed
.
- S. G. Hyberts, D. P. Frueh, H. Arthanari and G. Wagner, J. Biomol. NMR, 2009, 45, 283–294 CrossRef CAS PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4cc03047h |
| This journal is © The Royal Society of Chemistry 2014 |


