Open Access Article
Open Access ArticleIdentification of bromophenol thiohydantoin as an inhibitor of DisA, a c-di-AMP synthase, from a 1000 compound library, using the coralyne assay†
Yue
Zheng
,
Jie
Zhou
,
David A.
Sayre
and
Herman O.
Sintim
*
Department of Chemistry and Biochemistry, University of Maryland, College Park, Maryland 20742, USA. E-mail: hsintim@umd.edu; Fax: +1 3013149121; Tel: +1 3014050633
First published on 6th August 2014
Abstract
c-di-AMP is an important bacterial second messenger found in Gram-positive and mycobacteria. c-di-AMP regulates myriads of processes in bacteria as well as immune response in higher organisms so interest in small molecules that would attenuate the activity of c-di-AMP metabolism enzymes is high. Herein, we report the first small molecule inhibitor of a c-di-AMP synthase, DisA, using a coralyne-based assay.
The ability to sense the environment and communicate with co-inhabitants of an ecosystem is critical for the survival of any organism. Consequently intricate sensing and regulatory mechanisms have evolved in organisms to regulate physiological processes, in response to a changing environment. In 2008, Hopfner showed in a seminal paper that DisA, a Bacillus subtilis protein that controls sporulation checkpoint when DNA double strand break occurs, has a nucleotide-binding domain that binds to ATP to make a novel cyclic dinucleotide, c-di-AMP.1 It turned out that the presence of c-di-AMP signaled the presence of DNA structures that would interfere with proper chromosome segregation. Hopfner also predicted that many bacteria harbored proteins that contained the diadenylate cyclase (DAC) domain and hence c-di-AMP could have a wider role in bacteria, beyond reporting DNA strand breaks.1 The importance of Hopfner’s discovery became apparent when soon thereafter many reports by other laboratories confirmed that c-di-AMP is indeed produced in other bacteria, some of which are of clinical relevance.2,3 c-di-AMP has now been shown to control a dazzling array of processes in bacteria (see Fig. 1), including cell wall formation,4,5 cell size regulation,6 heat stress,7 virulence,8 ion transport,9 resistance to acid5,10etc. Additionally, when c-di-AMP is introduced into the cytoplasm of eukaryotic cells, for example when intracellular pathogens secret the nucleotide or during endocytosis of bacteria, it binds to STING to elicit type I inteferon response.2,11
The modulation of bacterial cell wall synthesis by c-di-AMP is particularly interesting because a high percentage of antibiotics, in clinical use, target components of the bacterial cell wall, suggesting that small molecules that could perturb the intracellular concentrations of c-di-AMP could synergize with antibiotics such as the β-lactams or vancomycins to kill pathogenic bacteria.12 An obvious way to reduce intracellular concentration of c-di-AMP would be to inhibit c-di-AMP synthases. High-throughput assays that could be used to monitor the production of c-di-AMP by DAC enzymes would facilitate the discovery of c-di-AMP signaling inhibitors.
Recently, we described a surprisingly simple detection of c-di-AMP using commercially available coralyne and halide quenchers.13 In this assay, the fluorescence of coralyne is quenched by a bromide or iodide anion but when coralyne becomes entrapped by a supramolecular aggregate of c-di-AMP, iodide can no longer quench the fluorescence and hence enhancement of fluorescence occurs only when c-di-AMP is present (see Fig. 2). In our prior disclosure, the synthesis of c-di-AMP by DisA was monitored as an end-point assay, that is aliquots of the enzymatic reaction were taken at different time points and analyzed using coralyne and KI. For high-throughput assay, this format was obviously not ideal so we wondered if we could add the coralyne and KI to the DisA reaction and monitor the progress of c-di-AMP formation in “real-time”. Pleasingly, the presence of coralyne or KI did not affect the enzymatic proficiency of DisA to a great extent and so we could use the coralyne assay to screen for inhibition using a library of 1000 compounds (obtained from TimTec kinase set, and Sigma, see ESI† for compounds ID). The Sigma compounds were randomly selected by us from the Sigma website but our selection was biased by compounds that were either drug-like or known to be kinase inhibitors.
 | ||
| Fig. 2 Coralyne detection of c-di-AMP formation.13 DAC converts ATP to c-di-AMP. Once c-di-AMP is formed, it can form complexes with coralyne. Iodide in the solution quenches coralyne fluorescence in the absence of c-di-AMP. c-di-AMP protects coralyne from quenchers and c-di-AMP–coralyne complex’s fluorescence can be detected. | ||
The majority of the tested compounds, 99%, did not affect the synthesis of c-di-AMP by DisA to any meaningful extent (see ESI† and Fig. 3). However, about 11 out of the 1000 compounds reduced the fluorescence of the coralyne assay, implying that these compounds either inhibited the DisA enzyme or competed with the synthesized c-di-AMP for binding to coralyne or both. Conceivably the compounds could form a π-complex with coralyne leading to a “dark” aggregate. This would be especially true for compounds that contain heavy halogens, which are known to be fluorescence quenchers.14 We therefore investigated the extent to which the fluorescence of coralyne was reduced by the 11 apparent “hits” in the presence and absence of c-di-AMP. We also used HPLC to monitor the synthesis of c-di-AMP by DisA in the presence of the 11 apparent hits, see ESI.† Out of the 11 apparent hits, only one ST056083 (5-[(3,5-dibromo-2-hydroxyphenyl)methylene]-2-thioxo-1,3-diazolidin-4-one,15 referred to as bromophenol-TH in this manuscript, see Fig. 3 for structure) could inhibit c-di-AMP synthesis, judged by HPLC analysis (see ESI†). It therefore appears that the coralyne assay has false positives due to the direct quenching of coralyne’s fluorescence by some small molecules. But this problem is not unique to the coralyne assay and in fact would exist in all turn-off assays as the potential for library members to affect the fluorescence of an assay would always exist. An excellent paper by Inglese has documented the problem of false positives that arise from fluorogenic library members.16 Nonetheless, there are hardly any false-negatives. That is if the fluorescence of coralyne in the presence of a non-fluorescent inhibitor is similar to when no small molecule was added, control, then it implies that small molecule is not an inhibitor. To provide support for this statement randomly selected 25 compounds that were negative for the coralyne assay were retested using HPLC assay and none showed inhibition, see ESI,† Fig. S4. Thus, the assay is still very useful because it reduced a large library size of 1000 to 11, which could then be investigated in detail using traditional low-throughput assays, such as HPLC and radioactive labelling.
 | ||
| Fig. 3 (a) Structures of bromophenol-TH and molecules in the 1000 compound library that have similar structures. (b) Real-time coralyne-based fluorescent detection of DisA reaction.13 | ||
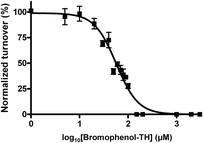
Bromophenol-TH contains both a halogenated phenol and thiohydantoin groups, however it appears that the mere presence of these groups is not good enough for DisA inhibition. For example, ST003014 and ST003440 both contain thiohydantoin and haloarene/phenolic moiety yet they did not inhibit c-di-AMP synthesis by DisA (see Fig. 3). Intrinsic tryptophan fluorescence can be used to monitor ligand binding to a protein if the binding event results in a change in the environment of the tryptophan. Pleasingly, the fluorescence of a tryptophan residue in DisA changed upon the addition of bromophenol-TH and so we could determine the apparent Kd for the bromophenol-TH–DisA complex from tryptophan fluorescence titration (see Fig. 4 and Table 1).17 Interestingly, ST028249, which also contains bromophenolic fragment and a rhodanine moiety (which is similar to a thiohydantoin group), binds to DisA more tightly than bromophenol-TH (Kapparentd of 11 vs. 21 μM, Table 118) yet it is unable to inhibit c-di-AMP synthesis. Using α-32P-ATP, we determined that the IC50 for inhibiting 5 μM of DisA by bromophenol-TH, when 50 μM of ATP is used as substrate is 56 μM (see Fig. 5). When the ATP concentration was increased by 10-fold to 500 μM, the IC50 only changed by a factor of 2 to 100 μM (see ESI,† Fig. S6). To eliminate the possibility that the observed inhibition is due to non-specific aggregation, we also investigated the inhibition of DisA by bromophenol-TH in the presence of a surfactant, Triton X-100. Pleasingly, bromopenol-TH could still inhibit DisA in the presence of Triton X-100 (see ESI,† Fig. S7). Interestingly, bromophenol-TH does not bind to the same site as ATP because the addition of up to 1 mM ATP did not abrogate the binding of bromophenol-TH to DisA (see Fig. 6 and ESI,† Fig. S8). Bromophenol-TH contains a Michael acceptor and could potentially react with nucleophiles. To investigate if conjugate addition by nucleophiles would readily occur, we performed incubation with 1 mM cysteine and followed the reaction progress using UV absorbance. The UV intensity, from 250 nm to 450 nm, did not change after cysteine addition and remained unchanged for 1 h (see ESI,† Fig. S9), implying that nucleophilic addition to bromophenol is not facile under the assay condition. Future structure–activity relationship studies are planned to optimize bromophenol-TH into a more potent inhibitor.
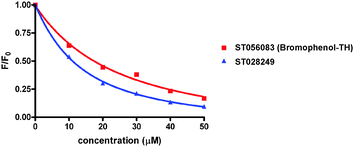 | ||
| Fig. 4 Tryptophan fluorescence titration. Addition of small molecules to protein causes reduction of protein’s tryptophan fluorescence. Normalized fluorescence of ST056083 (bromophenol-TH) and ST028249 binding to DisA at 340 nm, which is fluorescence [F] divided by the fluorescence measured without ligand [F0].17 | ||
| Compound | Bromophenol (μM) | ST028249 (μM) | 2-Thiohydantoin (mM) |
|---|---|---|---|
| K apparentd | 21 | 11 | >1 |
| Compound | 3,5-Dibromosalicylic acid (μM) | 3,5-Dibromosalicylaldehyde (μM) |
|---|---|---|
| K apparentd | 125 | 67 |
To guide future SAR and optimization studies, we determined the binding parameters for different fragments of bromophenol-TH, 3,5-dibromosalicylic acid, 3,5-dibromosalicylaldehyde and 2-thiohydantoin (see Fig. 7). It appears that the bromophenol fragment is more important for binding to DisA than the thiohydantoin moiety (compare Kapparentd of 67 μM for 3,5-dibromosalicylaldehyde with >1 mM for 2-thiohydantoin, Table 1).
In conclusion, we have demonstrated that the coralyne assay for c-di-AMP detection is a powerful assay to do high-throughput screening for discovering novel c-di-AMP synthesis inhibitors. Interestingly, although we initially selected TimTec’s kinase inhibitor library set for screening, hoping to uncover an ATP-competitive inhibitor, the hit that we found does not compete with ATP for DisA binding. Hopefully this proof-of-concept paper would encourage others, especially those with access to large compound libraries, to utilize this cheap and commercially available reagent, coralyne, to discover nucleotide signaling inhibitors. Beyond the demonstrations that this is an enabling assay for inhibitor discovery, we also report the first small molecule inhibitor of c-di-AMP synthesis. The inhibition of a diadenylate cyclase by a thiohydantoin adds to the impressive array of enzymes that this privilege compound structure inhibits.19 Considering that c-di-AMP plays a crucial role in bacterial lifestyle, analogs of bromophenol-TH or other scaffolds, which could be discovered via the assay used in this paper, have great promise to be used as stand-alone antibacterial agents or be used in tandem with traditional antibiotics. We anticipate an explosion in c-di-AMP research and the design of small molecules that inhibit c-di-AMP.
NSF (CHE 1307218) and Camille Dreyfus foundation supported this work. We thank Prof. Karl-Peter Hopfner and Dr Angelika Gründling for plasmids encoding DisA.
Notes and references
- G. Witte, S. Hartung, K. Buettner and K.-P. Hopfner, Mol. Cell, 2008, 30, 167 CrossRef CAS PubMed.
- J. J. Woodward, A. T. Iavarone and D. A. Portnoy, Science, 2010, 328, 1703 CrossRef CAS PubMed.
- (a) R. M. Corrigan and A. Gründling, Nat. Rev. Microbiol., 2013, 11, 513 CrossRef CAS PubMed; (b) J. W. Nelson, N. Sudarsan, K. Furukawa, Z. Weinberg, J. X. Wang and R. R. Breaker, Nat. Chem. Biol., 2013, 9, 834 CrossRef CAS PubMed; (c) C. Pozzi, E. M. Waters, J. K. Rudkin, C. R. Schaeffer, A. J. Lohan, P. Tong, B. J. Loftus, G. B. Pier, P. D. Fey, R. C. Massey and J. P. O'Gara, PLoS Pathog., 2012, 8, e1002626 CAS; (d) Y. Oppenheimer-Shaanan, E. Wexselblatt, J. Katzhendler, E. Yavin and S. Ben-Yehuda, EMBO Rep., 2011, 12, 594 CrossRef CAS PubMed; (e) F. M. Mehne, K. Gunka, H. Eilers, C. Herzberg, V. Kaever and J. Stülke, J. Biol. Chem., 2013, 288, 2004 CrossRef CAS PubMed; (f) V. Dengler, N. McCallum, P. Kiefer, P. Christen, A. Patrignani, J. A. Vorholt, B. Berger-Baechi and M. M. Senn, PLoS One, 2013, 8, e73512 CAS; (g) K. H. Cho and S. O. Kang, PLoS One, 2013, 8, e69425 CAS; (h) K. Tadmor, Y. Pozniak, T. Burg Golani, L. Lobel, M. Brenner, N. Sigal and A. A. Herskovits, Front. Cell. Infect. Microbiol., 2014, 4, 16 CrossRef PubMed; (i) K. Manikandan, V. Sabareesh, N. Singh, K. Saigal, U. Mechold and K. M. Sinha, PLoS One, 2014, 9, e86096 Search PubMed; (j) T. Yamamoto, H. Hara, K. Tsuchiya, S. Sakai, R. Fang, M. Matsuura, T. Nomura, F. Sato, M. Mitsuyama and I. Kawamura, Infect. Immun., 2012, 80, 2323 CrossRef CAS PubMed.
- M. Kaplan Zeevi, N. S. Shafir, S. Shaham, S. Friedman, N. Sigal, R. Nir Paz, I. G. Boneca and A. A. Herskovits, J. Bacteriol., 2013, 195, 5250 CrossRef PubMed.
- C. E. Witte, A. T. Whiteley, T. P. Burke, J.-D. Sauer, D. A. Portnoy and J. J. Woodward, mBio, 2013, 4, e00282–e00313 CrossRef CAS PubMed.
- R. M. Corrigan, J. C. Abbott, H. Burhenne, V. Kaever and A. Gruendling, PLoS Pathog., 2011, 7, e1002217 CAS.
- W. M. Smith, P. Thi Huong, L. Lei, J. Dou, A. H. Soomro, S. A. Beatson, G. A. Dykes and M. S. Turner, Appl. Environ. Microbiol., 2012, 78, 7753 CrossRef CAS PubMed.
- (a) M. Ye, J. J. Zhang, X. Fang, G. B. Lawlis, B. Troxell, Y. Zhou, M. Gomelsky, Y. Lou and X. F. Yang, Infect. Immun., 2014, 82, 1840 CrossRef CAS PubMed; (b) B. Du, W. Ji, H. An, Y. Shi, Q. Huang, Y. Cheng, Q. Fu, H. Wang, Y. Yan and J. Sun, Microbiol. Res., 2014, 169, 749 CAS.
- Y. Bai, J. Yang, T. M. Zarrella, Y. Zhang, D. W. Metzger and G. Bai, J. Bacteriol., 2014, 196, 614 CrossRef CAS PubMed.
- F. Rao, R. Y. See, D. Zhang, D. C. Toh, Q. Ji and Z.-X. Liang, J. Biol. Chem., 2010, 285, 473 CrossRef CAS PubMed.
- (a) K. Parvatiyar, Z. Zhang, R. M. Teles, S. Ouyang, Y. Jiang, S. S. Iyer, S. A. Zaver, M. Schenk, S. Zeng, W. Zhong, Z.-J. Liu, R. L. Modlin, Y.-j. Liu and G. Cheng, Nat. Immunol., 2012, 13, 1155 CrossRef CAS PubMed; (b) K. A. Archer, J. Durack and D. A. Portnoy, PLoS Pathog., 2014, 10, e1003861 Search PubMed; (c) J. R. Barker, B. J. Koestler, V. K. Carpenter, D. L. Burdette, C. M. Waters, R. E. Vance and R. H. Valdivia, mBio, 2013, 4, e00018 CrossRef CAS PubMed; (d) G. Yi, V. P. Brendel, C. Shu, P. Li, S. Palanathan and C. C. Kao, PLoS One, 2013, 8, e77846 CAS; (e) T. Kamegaya, K. Kuroda and Y. Hayakawa, Nagoya J. Med. Sci., 2011, 73, 49 CAS.
- D. W. Green, Expert Opin. Ther. Targets, 2002, 6, 1 CrossRef CAS PubMed.
- J. Zhou, D. A. Sayre, Y. Zheng, H. Szmacinski and H. O. Sintim, Anal. Chem., 2014, 86, 2412 CrossRef CAS PubMed.
- M. Mac, A. Danel, K. Kizior, P. Nowak, A. Karocki and B. Tokarczyk, Phys. Chem. Chem. Phys., 2003, 5, 988 RSC.
- After identifying this compound as a hit, 50 mg was purchased from TimTec, characterized (see ESI†) to confirm that indeed the active compound is the structure given in the purchased library.
- A. Simeonov, A. Jadhav, C. J. Thomas, Y. Wang, R. Huang, N. T. Southall, P. Shinn, J. Smith, C. P. Austin, D. S. Auld and J. Inglese, J. Med. Chem., 2008, 51, 2363 CrossRef CAS PubMed.
-
K
apparentd was determined by assuming a 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding. This allows for direct comparison of the compounds even if the binding constant is apparent.
1 binding. This allows for direct comparison of the compounds even if the binding constant is apparent. - C. Bodenreider, D. Beer, T. H. Keller, S. Sonntag, D. Wen, L. Yap, Y. H. Yau, S. G. Shochat, D. Huang, T. Zhou, A. Caflisch, X. C. Su, K. Ozawa, G. Otting, S. G. Vasudevan, J. Lescar and S. P. Lim, Anal. Biochem., 2009, 395, 195 CrossRef CAS PubMed.
- T. Mendgen, C. Steuer and C. D. Klein, J. Med. Chem., 2012, 55, 743 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ID number of compounds that were screened, HPLC analysis of DisA synthesis of c-di-AMP in presence of selected compounds and protein expression. See DOI: 10.1039/c4cc02916j |
| This journal is © The Royal Society of Chemistry 2014 |