Open Access Article
Open Access ArticleConvenient detection of HPV virus in a clinical sample using concurrent rolling circle and junction probe amplifications†
Lei
Yan
*a,
Kailong
Liu
b and
Herman O.
Sintim
*cd
aChangzhou Fangyuan Pharmaceutical Co., Ltd., 108 Hehaixi Road, Xinbei District, Changzhou, Jiangsu 213022, China. E-mail: leiyanzju@gmail.com; Fax: +86 519 86926772; Tel: +86 519 86926771
bChangzhou Care Biotechnology Co., Ltd., 3F Changzhou Biopharmaceutical Incubator, 106 Hehaixi Road, Xinbei District, Changzhou, Jiangsu 213022, China. E-mail: 974898024@qq.com; Fax: +86 519 86926772; Tel: +86 519 86926771
cDepartment of Chemistry and Biochemistry, University of Maryland, College Park, MD 20740, USA. E-mail: hsintim@umd.edu; Fax: +1 301 314 9121; Tel: +1 301 405 0633
dProgram in Oncology, University of Maryland Marlene and Stewart Greenebaum Cancer Center, 22 S. Greene Street, Baltimore, Maryland 21201, USA
First published on 22nd May 2014
Abstract
Herein we show that two isothermal amplification strategies, rolling circle amplification and junction probe strategy, can be used in tandem in the same tube under isothermal conditions to detect HPV16 in clinical cervical swabs. It was discovered that the prior treatment of the clinical sample with a cocktail of restriction endonucleases (REAses) to digest the genomic DNA facilitated the isothermal detection assay.
The detection of nucleic acids using non-PCR (polymerase chain reaction) or non-LCR (ligase chain reaction) based techniques has been intensively pursued for more than a decade.1 Recently the use of nucleases (nicking or restriction endonucleases or exonucleases) for the isothermal detection of RNA or DNA has become popular and many innovative strategies to achieve specific detection of nucleic acid analytes have been reported.2
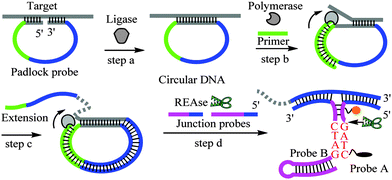
A junction probe (JP) detection platform is an isothermal nucleic acid detection platform and uses restriction endonucleases and probes with limited complementarity that have weak affinity for each other but form Y-junction structures in the presence of a nucleic acid analyte, which binds the probes in a juxtapose manner (template enhanced hybridization processes, TeHyp).2c The JP strategy has been used to detect E. coli via specific ribosome binding, without any sophisticated sample preparation,2f that is, E. coli ribosomal RNA in a crude cell lysate could be detected with JP probes and restriction endonucleases without a prior nucleic acid separating step. The success of JP in detecting nucleic acids in complex biological matrices prompted us to investigate the use of JP to detect clinical samples without any prior purification. A limitation of the JP strategy and many DNA isothermal detection assays is the requirement of a single-stranded DNA template. Herein we reveal that the prior treatment of genomic DNA with combined REAses, SacI and BstXI affords short duplex DNA fragments that could melt into single-stranded regions, without a heating step. These single-stranded regions could be detected via concurrent rolling circle amplification (RCA) and endonuclease-based junction probes in a single tube, without any separating step, all under isothermal conditions. To show the utility of these new nucleic acid detection sequences in clinical diagnostics, we used the endonuclease-RCA-JP strategy to detect human papillomavirus 16 (HPV16) in a clinical swab from a patient.
Human papillomavirus is a DNA virus which can infect humans. There are more than 100 types of HPV. Many humans carry different strains of HPVs and in most cases these common viruses do not cause any diseases. However a few of these viruses can cause genital warts,3 and HPV16 has been implicated in various cancers, including cervical4 and oropharyngeal cancers.5 The number of deaths caused by cervical and oral cancers was estimated to be around 4000 and 8000, respectively, in 2013 in the US alone.6 There is currently no effective therapeutics for these viruses but their early detection in cervical smears is critical for the management of cervical cancers as the physician could then monitor patients who are HPV 16, 18 or 45 positive for abnormal cervical cells, which have a high correlation with cervical cancer progression. Also, from epidemiological perspective the early detection of HPV in patients could save partners of these patients from contacting the virus. Thus far, commercial kits that are used to detect HPV 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68, etc. use polymerase-based methods.7 Although sensitive, these kits require instrumentation that might not be readily accessible in resource-poor areas. Even if the instrumentations and skilled technicians for PCR could be afforded, the issue of aerosol contamination from PCR procedures requires laboratories with regulated air flows, which is both expensive and inconvenient. In our continuing efforts to develop an isothermal PCR-free detection assay for pathogens, such as HPV, we investigated the concurrent use of RCA and JP as the singular use of these technologies (RCA or JP alone) was not sensitive for the clinical detection of HPV. The concurrent use of two or more isothermal nucleic acid amplification technologies is non-trivial because these strategies utilize different enzymes and different probes, which require specific conditions for optimal detection. Sometimes these conditions are not compatible, thereby preventing the concurrent use of these amplification strategies. We therefore began our studies by conducting the two amplification steps (RCA and JP) separately. Firstly, picomolar concentrations of ssDNA, which were used as templates to form a circular template, and a padlock probe (see the ESI† for sequences) were heated to 95 °C, and then cooled to room temperature before a ligase was added to form the circular template for the RCA step (see Scheme 1, step a). After the rolling circle polymerization with Φ29 DNA polymerase8 (Scheme 1, steps b and c), the amplified DNA was purified on sephadex G-50 (GE Healthcare) spin column and the amplified template was then detected with the JP technology (Scheme 1, step d). One of the JP probes contains a fluorescein fluorophore that is quenched by a neighboring dabsyl quencher. Upon cleavage, during the JP step, the fluorescein molecule is separated from the dabsyl quencher, leading to enhanced fluorescence.2c Using these protocols, it was deduced that the signal obtained from 100 pM template, using the tandem RCA-JP protocol was similar to that obtained from 100 nM using only the JP protocol (i.e. without an initial RCA pre-amplification), see Fig. 1. It therefore appears that the RCA was providing a thousand-fold enhancement. Although this first-generation RCA-JP protocol was not ideal, due to the need for a separation step (using sephadex G-50), we proceeded to test the protocol on a clinical sample, hoping to optimize the procedure into a one-tube format, should the first-generation RCA-JP work on the clinical sample.
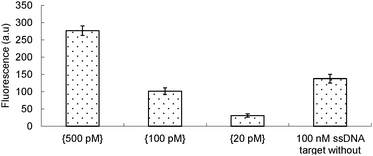 | ||
| Fig. 1 Single-stranded DNA (ssDNA) detection using tandem ligation-RCA-JP protocol. The concentrations of the analyte in the tandem protocol are indicated in the bracket. | ||
Most isothermal detection platforms require single-stranded DNA, but genomic DNA occurs as a double helix. To resolve the confounding issue of the duplex nature of genomic DNA, which makes probe access to single-stranded regions of DNA problematic, we decided to use the restriction enzymes SacI (cognate sequence is GAGCT|C) and BstXI (cognate sequence is CCANNNNN|NTGG) to randomly cleave the genomic DNA. We rationalized that the digestion of the genomic DNA would lead to short duplex fragments, which would then be easier to separate or “melt” under isothermal condition. Saline was added to a cervical swab (a gift from Changzhou Kelai Clinical Laboratory, Inc., Changzhou, Jiangsu, China) and after a brief vortex, the sample was incubated with Tris-HCl buffer (see the ESI† for details). This sample was then used for detection without any sophisticated nucleic acid isolation. Pleasingly, the combined RCA-JP protocol was effective in detecting HPV16 in the clinical sample, which was confirmed to be HPV16 positive using a commercial diagnostic kit (HPV Typing PCR-RDB (reverse dot blot) Kit, Daan Gene Co., Ltd. of Sun Yat-sen University, Guangzhou, Guangdong, China). Of note, the sample that was not treated with the combined REAses, SacI and BstXI, which would digest the genomic DNA into smaller fragments that would readily melt into single-stranded regions (required for the ligation step, see Scheme 1, step a), did not give a signal above the background (see Fig. 2).
Having established that a REAse digestion-ligation-RCA-JP protocol can be used to detect HPV16 in a clinical swab under isothermal conditions, we sought to make this protocol more convenient by combining all of these steps in a single tube, without any separation step and under isothermal conditions (without the heating to 95 °C and cooling to room temperature steps). A single tube concurrent reaction that encompasses the above steps will involve the co-existence of five enzymes (three different REAses, a ligase and a polymerase). Using 10 pM of a purified single strand DNA template, 5′-GGCATTTGTTGGGGTAACCAACTATTTGT-3′, a fragment of the HPV16 virus, we first established that we could use the five enzymes in a single tube to detect the target analyte over background noise (see the ESI† for details). With this success, we proceeded to detect HPV in the HPV16-positive cervical swab with our new REAse-ligation-RCA-JP sequence in a single tube under isothermal conditions. Pleasingly, the signals from the HPV16-positive sample and a plasmid containing a fragment of the HPV sequence were appreciably different from those of background, HPV-negative9 and HPV55-positive samples (see Fig. 3).
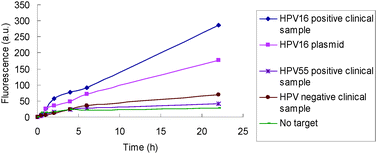 | ||
| Fig. 3 Detection of HPV16 in a clinical sample using RCA-JP in a single tube, without a “separation” step. | ||
In conclusion, we have demonstrated that it is possible to combine various endonucleases, a ligase and a polymerase to achieve the detection of pathogenic genes in a real life sample, cervical swab, under isothermal conditions without PCR cycles. We anticipate further development of this concurrent protocol to achieve shorter assay time and more convenient and/or cheaper visualization formats. For example, the incorporation of strategies that allow for colorimetric or chemiluminescence readouts would make the described nucleic acid protocol even more convenient, and we are currently pursuing these avenues.
Notes and references
- (a) K. Knez, D. Spasic, K. P. F. Janssen and J. Lammertyn, Analyst, 2014, 139, 353 RSC; (b) B. T. Roembke, S. Nakayama and H. O. Sintim, Methods, 2013, 64, 185 CrossRef CAS PubMed; (c) H. Zhang, F. Li, B. Dever, X. Li and X. Le, Chem. Rev., 2012, 113, 2812 CrossRef PubMed; (d) D. M. Kolpashchikov, Chem. Rev., 2010, 110, 4709 CrossRef CAS PubMed; (e) J. Zhou, Y. Zheng, S. Nakayama, B. Roembke, A. Gamson and H. O. Sintim, Mol. BioSyst., 2014, 10, 970 RSC.
- (a) Y. Weizmann, M. K. Beissenhirtz, Z. C. R. Nowarski, M. Kotler and I. Willner, Angew. Chem., Int. Ed., 2006, 45, 7384 CrossRef CAS PubMed; (b) W. Xu, X. Xue, T. Li, H. Zeng and X. Liu, Angew. Chem., Int. Ed., 2009, 48, 6849 CrossRef CAS PubMed; (c) S. Nakayama, L. Yan and H. O. Sintim, J. Am. Chem. Soc., 2008, 130, 12560 CrossRef CAS PubMed; (d) X. Zuo, F. Xia, Y. Xiao and K. W. Plaxco, J. Am. Chem. Soc., 2010, 132, 1816 CrossRef CAS PubMed; (e) S. Liu, C. Wang, C. Zhang, Y. Wang and B. Tang, Anal. Chem., 2013, 85, 2282 CrossRef CAS PubMed; (f) L. Yan, S. Nakayama, S. Yitbarek, I. Greenfield and H. O. Sintim, Chem. Commun., 2011, 47, 200 RSC; (g) Z. Shen, S. Nakayama, S. Semancik and H. O. Sintim, Chem. Commun., 2012, 48, 7580 RSC; (h) F. Xuan, X. Luo and I. M. Hsing, Anal. Chem., 2012, 84, 5216 CrossRef CAS PubMed; (i) L. Yan, S. Nakayama and H. O. Sintim, Bioorg. Med. Chem., 2013, 21, 6181 CrossRef CAS PubMed; (j) Q. Wang, L. Yang, X. Yang, K. Wang, L. He, J. Zhu and T. Su, Chem. Commun., 2012, 48, 2982 RSC; (k) Q. Xu, A. Cao, L. Zhang and C. Zhang, Anal. Chem., 2012, 84, 10845 CrossRef CAS PubMed; (l) H. Zhou, S. Xie, S. Zhang, G. Shen, R. Yu and Z. Wu, Chem. Commun., 2013, 49, 2448 RSC; (m) Y. V. Gerasimova, S. Peck and D. M. Kolpashchikov, Chem. Commun., 2010, 46, 8761 RSC; (n) J. Li, X. Qi, Y. Du, H. Fu, G. Chen and H. Yang, Biosens. Bioelectron., 2012, 36, 142 CrossRef CAS PubMed; (o) R. Miaranda-Castro, D. Marchal, B. Limoges and F. Mavré, Chem. Commun., 2012, 48, 8772 RSC; (p) S. Liu, C. Zhang, J. Ming, C. Wang, T. Liu and F. Li, Chem. Commun., 2013, 49, 7947 RSC; (q) S. Bi, J. Zhang and S. Zhang, Chem. Commun., 2010, 46, 5509 RSC; (r) Y. Weizmann, Z. Cheglakov, V. Pavlov and I. Willner, Angew. Chem., Int. Ed., 2006, 45, 2238 CrossRef CAS PubMed.
- C. J. Lacey, C. M. Lowndes and K. V. Shah, Vaccine, 2006, 24(suppl 3), S35 CrossRef PubMed.
- F. X. Bosch, M. M. Manos, N. Muñoz, M. Sherman, A. M. Jansen, J. Peto, M. H. Schiffman, V. Moreno, R. Kurman and K. V. Shah, J. Natl. Cancer Inst., 1996, 87, 796 CrossRef PubMed.
- P. M. Weinberger, Z. Yu, B. G. Haffty, D. Kowalski, M. Harigopal, J. Brandsma, C. Sasaki, J. Joe, R. L. Camp, D. L. Rimm and A. Psyrri, J. Clin. Oncol., 2006, 24, 736 CrossRef CAS PubMed.
- R. Siegel, D. Naishadham and A. Jemal, Ca-Cancer J. Clin., 2013, 63, 11 CrossRef PubMed.
- A. Arney and K. M. Bennett, Lab. Med., 2010, 41, 523 CrossRef.
- E. M. Harcourt and E. T. Kool, Nucleic Acids Res., 2012, 40, e65 CrossRef CAS PubMed.
- This sample was a cervical swab that was negative for HPV6, 11, 16, 18, 31, 33, 39, 43, 45, 51, 52, 53, 56, 58, 59, 66, 68 and CP8304.
Footnote |
| † Electronic supplementary information (ESI) available: DNA sequences and details of buffer compositions and reactions. See DOI: 10.1039/c4cc02532f |
| This journal is © The Royal Society of Chemistry 2014 |