 Open Access Article
Open Access ArticleUse of thiolated oligonucleotides as anti-fouling diluents in electrochemical peptide-based sensors†
Adam
McQuistan
a,
Anita J.
Zaitouna
a,
Elena
Echeverria
b and
Rebecca Y.
Lai
*a
aDepartment of Chemistry, Hamilton Hall, University of Nebraska-Lincoln, Lincoln, NE 68588-0304, USA. E-mail: rlai2@unl.edu; Fax: +1 402 472 9402; Tel: +1 402 472 5340
bDepartment of Physics and Astronomy, Theodore Jorgensen Hall, University of Nebraska-Lincoln, Lincoln, NE 68588-0299, USA
First published on 17th March 2014
Abstract
We incorporated short thiolated oligonucleotides as passivating diluents in the fabrication of electrochemical peptide-based (E-PB) sensors, with the goal of creating a negatively charged layer capable of resisting non-specific adsorption of matrix contaminants. The E-PB HIV sensors fabricated using these diluents were found to be more specific and selective, while retaining attributes similar to the sensor fabricated without these diluents. Overall, these results highlight the advantages of using oligonucleotides as anti-fouling diluents in self-assembled monolayer-based sensors.
In the past few decades, remarkable advances have been made in medicine, specifically, in the understanding of pathogenesis of various diseases at the molecular level. Complexities in the molecular aspect of diseases have led to the development of new disease diagnostic devices and sensors. A wide range of optical, electrochemical and piezoelectric biosensors have since been reported.1,2 Among them, electrochemical biosensors have proven to be useful in disease diagnosis and monitoring, with desirable features that include rapid response, high sensitivity, specificity and selectivity.2 To date, numerous electrochemical biosensors have been developed for detection of biomedically relevant targets, including nucleic acids, proteins, and small molecules.2,3
Among the currently available electrochemical biosensors designed for detection of antibodies, the electrochemical peptide-based (E-PB) sensors have shown to be capable of real-time and reagentless detection of anti-HIV antibodies.4 For this class of sensors, the peptide recognition probe is often modified with an 11-carbon alkanethiol linker at the n-terminus and a methylene blue (MB) redox label at the c-terminus.4 In the absence of the target antibody (IgG-target), the peptide probe is flexible, electron transfer between the electrode and the MB label is efficient, resulting in high MB current. In the presence of the large target antibody, probe flexibility is significantly dampened; the limited surface mobility is reflected by the reduction in the MB current (Scheme 1).4 In this study, the recognition element is a 21-residue peptide epitope (DRY-MB) from the HIV-1 p24 antigen (Fig. S1, ESI†).5 The p24 capsid protein is known to be highly immunogenic, the serum concentration of anti-p24 antibodies ranges from few nanomolar to tens of nanomolar.6 The epitope used here is different from the previously employed p24 epitope, but both are known to be highly antigenic.4–6
 | ||
| Scheme 1 Design and signalling mechanism of the E-PB sensor fabricated with a DNA anti-fouling diluent (T2, T4 or T6). | ||
Similar to the electrochemical DNA and aptamer-based sensors, E-PB sensors are sensitive, inherently "reagentless", and easy to operate. However, independent of the signal transduction mechanism, sensor specificity and selectivity are often dependent on the specific probe–target interaction, as well as the type of surface passivation employed.4 Thus, the use of anti-fouling diluents to prevent non-specific adsorption of random targets and matrix contaminants has been implemented in the fabrication of a wide range of sensors, in particular, self-assembled monolayer (SAM)-based sensors designed for use with surface-plasmon resonance spectroscopy. Thiolated oligo-ethyleneglycol (OEG) is one of the most commonly used sensor passivating diluents, owing to its ability to resist surface fouling.7 While thiolated OEG has shown to be effective in preventing non-specific adsorption of proteins on the sensor surface, there are other neutral or charged diluent molecules that serve the same function. Uniformly charged DNA, however, has not been explored for use as an anti-fouling diluent. Thus, the goal of this study is to compare E-PB sensors fabricated with and without DNA-based anti-fouling diluents. The use of short DNA as diluents could prove to be instrumental in improving sensor specificity and selectivity. Furthermore, this sensor passivation strategy is versatile and can potentially be used with all E-PB sensors.
To assess the effect of different DNA diluents, we designed and fabricated four sensors using the same peptide probe. Three of them were passivated with 6-mercapto-1-hexanol (C6-OH) and a DNA diluent, whereas the last one was passivated with C6-OH only. The three DNA diluents used in this study contain 2, 4, or 6 thymine (T) bases connected to a 6-carbon alkanethiol linker (Fig. S2, ESI†). T was chosen since it is relatively flexible and is not known to interact with matrix contaminants that can interfere with the analysis.8 We systematically evaluated performance of the sensors fabricated with one of the three diluents (T2, T4 or T6) using alternating current (AC) voltammetry, with the goal of understanding the effect of DNA diluents of different chain length on sensor specificity and selectivity.
When interrogated using AC voltammetry, we observed a relatively small yet well-defined MB reduction peak in the absence of the target for all four systems, suggesting successful immobilization of the peptide probe (Fig. S3, ESI†).4 The surface probe coverage was determined using a previously developed method and the values are shown in Table 1.9 The probe coverage was lower for the T2, T4, and T6 sensors when compared to the T0 sensor (i.e., the sensor passivated with C6-OH only), presumably because of the displacement of peptide probes by the DNA diluent. All three sensors fabricated using a DNA diluent had similar probe coverage, despite differences in the DNA chain length. As can be seen, the sensors responded well to 65 nM IgG-target; the observed % signal suppression (%SS) ranged from 22 to 40%, verifying the sensors' ability to recognize the target antibody. It is worth noting that all four sensors responded rather rapidly independent of the addition of a DNA anti-fouling diluent, most of the signal change (∼80%) was seen in less than 20 min after target addition. Incorporation of a DNA diluent did not appear to have a negative impact on the binding kinetics.
| Peptide probe coverage (1010 molecules per cm2) | DNA diluent coverage (1013 molecules per cm2) | |
|---|---|---|
| T0 sensor | 15.6 ± 7.7 | 0.0 |
| T2 sensor | 3.2 ± 0.5 | 9.5 ± 1.1 |
| T4 sensor | 4.6 ± 0.6 | 6.3 ± 1.1 |
| T6 sensor | 4.7 ± 1.2 | 2.7 ± 1.4 |
Since the amount of DNA diluent being incorporated into the monolayer can affect the overall sensor performance, the surface coverage of the diluent has to be carefully analyzed and monitored. Here we used X-ray photoelectron spectroscopy (XPS) to analyze the amount of DNA diluent on the sensor surface.10 Specifically, we analyzed the size of the phosphorous 2p peak with respect to the number of phosphate groups per thiolated DNA strand (Fig. S4, ESI†).10 Our XPS results show a strong inverse correlation between the diluent chain length and surface coverage. For example, more T2 DNA strands were incorporated in the monolayer when compared to the larger diluents (i.e., T4 and T6 diluents) (Table 1 and Table S1, ESI†). These results are not unexpected since steric hindrance from the peptide probe could affect immobilization of the larger DNA diluents. Despite the difference in DNA diluent coverage, all three sensors showed improved specificity when compared to the T0 sensor (Fig. 1). For the T0 sensor, ∼35 %SS was evident when challenged with 70 nM IgG-random, whereas ∼5 %SS was observed with the rest of the sensors. Addition of the DNA diluent, however, did not significantly affect the sensors' response to IgG-target. Slightly lower %SS was observed for both T2 and T6 sensors in the presence of 70 nM IgG-target, but the T4 sensor showed similar %SS as the T0 sensor. It is worth noting that a discrimination factor (F) (F = %SSIgG-target/%SSIgG-random) of ∼12 was determined for the T4 sensor, a value much higher than that obtained for the T0 sensor (Fig. S5, ESI†). For the T6 sensor, the lower DNA diluent concentration could be the reason behind the lower F value. The low F value seen with the T2 sensor could be, in part, due to the quality of the T2 DNA diluent; purification of this probe was rather challenging. Independent of the DNA chain length, all three sensors showed at least a 7-fold reduction in nonspecific binding of IgG-random when compared to the T0 sensor. These findings corroborate with our hypothesis that the negatively charged backbone of DNA helps in alleviating surface fouling. However, while XPS can be used to determine surface coverage, it does not provide information on how the thiolated DNA strands are distributed on the sensor surface. Based on our previous studies on SAM-based sensors, clustering of thiolated molecules is not uncommon, especially for multi-component SAM systems like the ones used in this study.11 In this case, if the anti-fouling diluent molecules are not uniformly distributed on the sensor surface, their ability to resist non-specific adsorption of random targets could be hindered. We are currently working on ways to prevent cluster formation on sensor surfaces; if successful, specificity of these sensors could be further improved.
 | ||
| Fig. 1 %SS recorded in the presence of 70 nM IgG-random and 70 nM IgG-target in Phys2 for all four sensors. | ||
In addition to having fast binding kinetics, all four sensors responded well to IgG-target, even at very low target concentrations. Shown in Fig. 2A are the dose response curves for the sensors. Although the dissociation constant (Kd) was found to be different for each sensor, incorporation of the DNA diluent did not have a major effect on sensor sensitivity. A limit of detection of 1 nM was determined for all four sensors. The dynamic range was similar among the sensors, but the signal attenuation was lower for both T2 and T4 sensors. This effect was not observed with the T6 sensor, which behaved entirely like the T0 sensor. This could be a result of the steric hindrance attributed by the incorporated DNA, in which the T2 and T6 sensors had the highest and lowest DNA diluent coverage, respectively. Further investigation is needed to elucidate the effect of DNA diluents on signal attenuation, these aspects are currently under investigation in the lab.
Good selectivity is another feature of the folding- and dynamics-based electrochemical sensing platform; the previously developed E-PB sensor for detection of HIV antibodies was characterized in 50% urine proxy.4 In this study, to determine sensor selectivity, we interrogated all four sensors in 10% synthetic human saliva. The noninvasive nature of salivary testing has made it an attractive alternative to blood and urine testing in disease diagnosis.12 The T0 sensor showed a reduction in MB current (14 ± 1 %SS) when used with the saliva, whereas substantially lower signal change was observed with sensors fabricated with DNA diluents (Fig. 3). Addition of 70 nM IgG-target resulted in 34(±2), 16(±3), 29(±2), and 30(±2) %SS for the T0, T2, T4, and T6 sensors, respectively. These results clearly show the advantages of using DNA diluents in the fabrication of E-PB sensors. Other than the T2 sensor, the signal attenuation in the presence of IgG-target was relatively similar for all three sensors. For both T4 and T6 sensors, target recognition was not affected by the presence of matrix contaminants. These results further highlight the ability of these diluents in enhancing sensor selectivity.
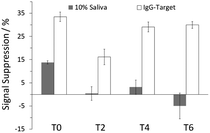 | ||
| Fig. 3 %SS observed in the presence of 10% saliva and after the addition of 70 nM IgG-target for all four sensors. | ||
Here we have demonstrated the advantages of incorporating short thiolated DNA as anti-fouling diluents in the fabrication of E-PB sensors. Among the sensors fabricated with a DNA diluent, the T4 sensor shows promise for real world applications. It has the best combination of properties, which includes good signal attenuation, specificity and selectivity. With proper optimization, DNA diluents can potentially be used to prevent surface fouling in sensor devices. This approach is also highly versatile; these diluents can be used with most SAM-based biosensing platforms, independent of the signal transduction mechanism.
The authors acknowledge the UNL UCARE program and National Science Foundation (GAANN #P200A100041, CHE-0955439, MRSEC DMR-0820521) for financial support. The authors would like to thank Dr P. A. Dowben for his help with the XPS data analysis.
Notes and references
- (a) X. Fan, I. A. White, S. Oi. Shopova, H. Zhu, J. D. Suter and Y. Sun, Anal. Chim. Acta, 2008, 610, 8 CrossRef PubMed; (b) J. Ngeh-Ngwainbi, A. A. Suleiman and G. G. Guilbault, Biosens. Bioselectron., 1990, 5, 13 CrossRef CAS; (c) D.-L. Ma, H.-Z. He, K.-H. Leung, H.-J. Zhong, D. S.-H. Chan and C.-H. Leung, Chem. Soc. Rev., 2013, 42, 3427 RSC; (d) H.-Z. He, D. S.-H. Chan, C.-H. Leung and D.-L. Ma, Nucleic Acids Res., 2013, 41, 4345 CrossRef CAS PubMed.
- (a) N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747 RSC; (b) M. Vestergaard, K. Kerman and E. Tamiya, Sensors, 2007, 7, 3442 CrossRef CAS; (c) T. G. Drummond, M. G. Hill and J. K. Barton, Nat. Biotechnol., 2003, 21, 1192 CrossRef CAS PubMed.
- (a) K. Kerman and H.-B. Kraatz, Analyst, 2009, 134, 2400 RSC; (b) S. Zhao, W. Yang and R. Y. Lai, Biosens. Bioelectron., 2011, 26, 2442 CrossRef CAS PubMed; (c) B. R. Baker, R. Y. Lai, M. S. Wood, E. H. Doctor, A. J. Heeger and K. W. Plaxco, J. Am. Chem. Soc., 2006, 128, 3138 CrossRef CAS PubMed.
- (a) J. Y. Gerasimov and R. Y. Lai, Chem. Commun., 2010, 46, 395 RSC; (b) J. Y. Gerasimov and R. Y. Lai, Chem. Commun., 2011, 47, 8688 RSC.
- G. Tonarelli, J. Lottersberger, J. L. Salvetti, S. Jacchieri, R. A. Silva-Lucca and L. M. Beltramin, Lett. Pept. Sci., 2000, 7, 217 CAS.
- M. S. Cohen, C. L. Gay, M. P. Busch and F. M. Hecht, J. Infect. Dis., 2010, 202, S270 CrossRef PubMed.
- (a) K. Uchida, H. Otsuka, M. Kaneko, K. Kataoka and Y. Nagasaki, Anal. Chem., 2005, 77, 1075 CrossRef CAS PubMed; (b) E. Hifumi, N. Kubota, Y. Niimi, K. Shimizu, N. Egashira and Y. Uda, Anal. Sci., 2002, 18, 863 CrossRef CAS PubMed; (c) H. B. Lu, C. T. Campbell and D. G. Castner, Langmuir, 2000, 16, 1711 CrossRef CAS.
- (a) Y. Wu and R. Y. Lai, Chem. Commun., 2013, 49, 3422 RSC; (b) Z. Yu and R. Y. Lai, Chem. Commun., 2012, 48, 10523 RSC.
- J. J. Sumner, K. S. Weber, L. A. Hockett and S. E. Creager, J. Phys. Chem. B, 2000, 104, 7449 CrossRef CAS.
- (a) C.-Y. Lee, P. Gong, G. M. Harbers, D. W. Grainger, D. G. Castner and L. J. Gamble, Anal. Chem., 2006, 78, 3326 CrossRef PubMed; (b) Z.-L. Zhang, D.-W. Pang and R.-Y. Zhang, Bioconjugate Chem., 2002, 13, 104 CrossRef CAS.
- S. J. P. Cañete, Z. Zhang, L. Kong, V. L. Schlegel, B. A. Plantz, P. A. Dowben and R. Y. Lai, Chem. Commun., 2011, 47, 11918 RSC.
- W. Schramm, B. A. Gustavo, P. C. Torres and A. Burgess-Cassler, Clin. Vaccine Immunol., 1999, 6, 577 CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Reagents and experimental procedures. Structure and sequence information of the peptide and DNA probes. AC voltammograms of the sensors in the absence and presence of IgG-target. XPS spectra of the T0 and T4 sensors. Discrimination factor plot for all four sensors. DNA diluent coverage based on XPS results. See DOI: 10.1039/c4cc01290a |
| This journal is © The Royal Society of Chemistry 2014 |

