Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Site-specific protein labelling and immobilization mediated by microbial transglutaminase†
Samuel K.
Oteng-Pabi
,
Christophe
Pardin
,
Maria
Stoica
and
Jeffrey W.
Keillor
*
Department of Chemistry, University of Ottawa, 10 Marie-Curie, Ottawa, Ontario, Canada K1N 6N5. E-mail: jkeillor@uottawa.ca
First published on 2nd May 2014
Abstract
Microbial transglutaminase (mTG) shows broad substrate specificity that is amenable to in vitro bio-conjugation applications. Herein, test proteins were genetically fused with peptide tags, followed by mTG-mediated propargylation of their reactive Gln residues. The propargylated proteins were subjected to copper-assisted azide-alkyne cycloaddition to demonstrate either fluorescent labelling or immobilization.
The sequencing of the human genome has identified a huge number of putative genes.1 However, the functional role of very few can be inferred from their primary sequences. For most, assigning functional roles will require methods for monitoring the activity and interactions of the gene products. Although a wide range of methods for the chemical modification of proteins have been developed,2,3 these methods usually result in the selective labelling of a given functional group, rather than the site-specific labelling of one residue of a specific protein. However, by labelling proteins in living cells, critical information may be gained regarding their expression, localization and trafficking. For this reason, several methods for the site-specific labelling of specific proteins have emerged and are the subject of recently reviews.4–7 One of the most important of these techniques involves the genetic fusion of a short peptide sequence to a protein of interest (POI), followed by the site-specific modification of this peptide tag, using a co-expressed enzyme.7,8
Several enzymes have been used to this end,7 including farnesyl transferase,9 biotin ligase,10 myristyl transferase,11 formylglycine-generating enzyme,12 sortase,13 lipoic acid ligase14 and tissue transglutaminase (TG2).13 The excellent review by Rashidian et al. summarizes their relative advantages and disadvantages.7 For example, formylglycine-generating enzyme, biotin ligase and lipoic acid ligase all recognize large 13-mer to 15-mer tags, whereas farnesyl transferase, myristyl transferase, sortase and TG2 recognize minimal 4–5 residue tags. Moreover, sortase and TG2 are able to directly incorporate small labels bearing ‘molecular cargo’ such as fluorophores, while farnesyl transferase, myristyl transferase, and lipoic acid ligase all incorporate relatively large second substrates. Furthermore, only TG2 can modify the side-chain of its peptide substrate sequence regardless of where this tag is located in the POI. Finally, farnesyl transferase and TG2 catalyse their reactions efficiently, while other enzymes may be sluggish and/or unstable over the duration of the labelling experiment.
From this cursory analysis, TG2 emerges as having tremendous potential for labelling a POI with minimal perturbation, anywhere in its sequence, by virtue of the versatility and efficiency of this enzyme. In their native role, transglutaminases catalyse the cross-linking of proteins, by mediating a transamidation reaction between the side-chains of peptide-bound Gln and Lys residues (Fig. 1). This activity has been exploited, in our labs16 and others15 through the TG2-mediated incorporation of functionalized amines into a Gln-substrate peptide tag. However, the cellular application of TG2 seems problematic, given its broad specificity for its glutamine-donor sequence, large size (76 kDa) and calcium dependence.
 | ||
| Fig. 1 Transglutaminase (TGase)-mediated protein cross-linking. Transamidation between protein-bound Gln and Lys residues leads to the formation of γ-glutamyl-ε-lysyl isopeptide bonds (red). | ||
Recent attention has therefore turned to the use of microbial transglutaminase (mTG) from Streptoverticillium mobaraense, a smaller (38 kDa), calcium-independent enzyme.17 mTG has been used extensively in the food industry to cross-link proteins, through the same Gln/Lys acyl transfer reaction as the mammalian enzymes, as a way of improving texture.18 It has also been used to facilitate the conjugation of biodegradable polymers with the intention of mediating drug delivery,19 and applied to in vitro site-specific protein labelling20 and immobilization.21
Furthermore, two recent discoveries regarding the substrate specificity of mTG have set the stage for its widespread application. Firstly, short peptide sequences have been developed that serve as genetically encodable high-affinity Gln-substrates for mTG (‘Q-tags’). In seminal work, Hitomi and co-workers used phage-displayed peptide libraries to discover sequences that serve as efficient Gln-donor substrates for mTG, while maintaining orthogonality with mammalian transglutaminases.22 Recently, we showed23 that these sequences can be trimmed down to heptamers and genetically fused to test proteins, while retaining their efficient reactivity with mTG. Secondly, we have recently shown that propargylamine serves as a surprisingly efficient amine substrate for TG216 and mTG.24 Through subsequent copper-catalyzed azide-alkyne cycloaddition (CuAAC) we were able to demonstrate the modification of small glutamine peptides16,24 and a native protein.16 The exciting potential of these discoveries can be manifested in a protein-labelling method combining the specificity of mTG Q-tag recognition with the breadth of ‘click’ chemistry, thereby adding a wide range of unnatural functionality to a POI, through selective and site-specific post-translational modification. Herein, we demonstrate two proof-of-principle applications of mTG-mediated protein labelling, in the site-specific modification of a Q-tagged protein for (1) fluorescent labelling and (2) immobilization.
For the fluorescent labelling demonstration, an appropriate test protein was first prepared by sub-cloning onto the C-terminus of Maltose Binding Protein (MBP) a short spacer sequence (GSSGSS), followed by one of three Q-tags23 – 7M48 (WALQRPH), 7M42 (WELQRPY), or 7M38 (YPMQGWF). The Q-tagged MBPs were then expressed and purified according to established protocols25 (see ESI†). These purified proteins were then incubated at 37 °C at a final concentration of 0.02 mg mL−1 in the presence of 1 unit of mTG in a reaction mixture containing 20 mM propargylamine, 200 mM MOPS (pH 7.2) and 1 mM EDTA at a total volume of 500 μL. After 2 hours, 1 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 water
1 water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) tert-butanol solution of 10 mM of dansyl-ethylazide (see ESI† for synthesis), 0.1% copper sulphate and 0.01% sodium ascorbate was added. The ensuing ‘click’ reaction was allowed to proceed overnight at 4 °C. After the reaction, labelled MBP was washed repeatedly over a membrane having a 14-kDa molecular weight cut-off, effectively removing any unbound dansyl-ethylazide.
tert-butanol solution of 10 mM of dansyl-ethylazide (see ESI† for synthesis), 0.1% copper sulphate and 0.01% sodium ascorbate was added. The ensuing ‘click’ reaction was allowed to proceed overnight at 4 °C. After the reaction, labelled MBP was washed repeatedly over a membrane having a 14-kDa molecular weight cut-off, effectively removing any unbound dansyl-ethylazide.
The labelled proteins were then analysed by SDS-PAGE. As shown in Fig. 2, MBP was effectively fluorescently labelled when bearing the Q-tags 7M48 and 7M42. However, when Q-tagged with 7M38, a sequence known to have relatively weaker affinity for mTG,23 little fluorescent labelling was observed, as expected. As a negative control, untagged MBP was also subjected to the same labelling experiment. Although MBP contains nine native Gln residues, no fluorescent labelling was observed. This clearly demonstrates that non-specific labelling of intrinsically unreactive Gln residues is negligible, illustrating the distinctive affinity conferred by select Q-tag sequences.
 | ||
| Fig. 2 mTG-mediated propargylation of MBP bearing high-affinity Q-tags. Following propargylation, test proteins were fluorescently labelled with dansyl-ethylazide. | ||
Furthermore, the three Q-tagged MBPs were then characterized as Gln-donor substrates for mTG, using our continuous enzyme-coupled assay (see ESI† for protocol).23 Comparison of the specificity constants (kcat/KM) for these three proteins shows that the reaction with MBP-7M48 is ten times more efficient than with the other Q-tagged proteins (Table 1). This confirms our previous findings,23 and validates the use of the 7M48 sequence as a high-affinity peptide tag for mTG.
| MBP-Q-tag | k cat/KM (μM−1 s−1) |
|---|---|
| MBP-7M38 | (1.3 ± 0.3) × 10−2 |
| MBP-7M42 | (9.7 ± 0.6) × 10−2 |
| MBP-7M48 | (6.8 ± 0.1) × 10−1 |
In our second proof-of-principle experiment, we sought to immobilize a test protein. Protein immobilization is used extensively throughout molecular biology26 and for a variety of commercial purposes,27 while the site-specific immobilization of enzymes is of critical importance in bio-catalysis. For the purposes of our demonstration, we sub-cloned a GSSGSS spacer and our 7M48 Q-tag onto the C-terminus of the highly photostable monomeric red fluorescent protein mRuby2, derived from eqFP611.28 The resulting mRuby2-7M48 test protein was then expressed and purified (see ESI†) and diluted to 0.01 mg mL−1 prior to mTG-mediated propargylation, as described above. After washing, the propargylated test protein was incubated with 10 mg of magnetized azide-functionalized nanoparticles (TurboBeads®), in the presence of 0.1% copper sulphate and 0.01% sodium ascorbate, in 1 mL of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 water
1 water![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) tert-butanol solution. After reaction overnight at 4 °C, the functionalized beads were washed and characterized.
tert-butanol solution. After reaction overnight at 4 °C, the functionalized beads were washed and characterized.
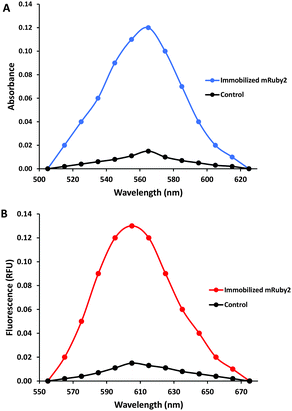
Spectral analysis in a microtiter plate reader was used to verify the immobilization of propargylated mRuby2-7M48. As shown in Fig. 3, the labelled beads showed significant absorbance and fluorescence, characteristic of mRuby2. When mRuby2-7M48 was treated in the same way, but in the absence of mTG (as a negative control), the beads showed minimal absorbance and fluorescence, which was found to diminish with repeated washing, suggesting it is due to non-specific binding of mRuby2. Comparison of the spectra unambiguously confirms the efficacy of the mTG-mediated immobilization. Additional qualitative evidence in support of this conclusion was obtained by fluorescence microscopy (see ESI†).
These experiments clearly demonstrate the effectiveness of mTG for the efficient, site-specific propargylation of the 7M48 Q-tag sequence genetically fused to a POI. Subsequent ‘click’ chemistry can then be used to functionalize the POI with a broad range of azide derivatives. The negligible background reactivity of the Gln residue, the site-selectivity of the mTG-mediated propargylation of a select Q-tag and the rich diversity of the subsequent bioorthogonal CuAAC ‘click’ reaction29 all illustrate the potential for the extension of this method to the labelling of specific proteins in cell culture. We are currently exploring the scope of this approach, as a method complementary to others we have developed,30 for the labelling of specific proteins in living cells.
This work was funded through a Discovery Grant to JWK from the Natural Sciences and Engineering Research Council of Canada (NSERC).
Notes and references
- F. Liang, I. Holt, G. Pertea, S. Karamycheva, S. L. Salzberg and J. Quackenbush, Nat. Genet., 2000, 25, 239–240 CrossRef CAS PubMed.
- T. Nakazawa, Curr. Proteomics, 2006, 3, 33–54 CrossRef CAS.
- M. Iwakura, D. Nakamura, T. Takenawa and Y. Mitsuichi, Protein Eng., 2001, 14, 583–589 CrossRef CAS PubMed.
- M. S. T. Gonçalves, Chem. Rev., 2009, 109, 190–212 CrossRef PubMed.
- N. Soh, Sensors, 2008, 8, 1004–1024 CrossRef CAS PubMed.
- Y. Takaoka, A. Ojida and I. Hamachi, Angew. Chem., Int. Ed., 2013, 52, 4088–4106 CrossRef CAS PubMed.
- M. Rashidian, J. K. Dozier and M. D. Distefano, Bioconjugate Chem., 2013, 24, 1277–1294 CrossRef CAS PubMed.
- M. Sunbul and J. Yin, Org. Biomol. Chem., 2009, 7, 3361–3371 CAS.
- B. P. Duckworth, Z. Zhang, A. Hosokawa and M. D. Distefano, ChemBioChem, 2007, 8, 98–105 CrossRef CAS PubMed.
- I. Chen, M. Howarth, W. Lin and A. Y. Ting, Nat. Methods, 2005, 2, 99–104 CrossRef CAS PubMed.
- W. P. Heal, S. R. Wickramasinghe, R. J. Leatherbarrow and E. W. Tate, Org. Biomol. Chem., 2008, 6, 2308–2315 CAS.
- P. Wu, W. Shui, B. L. Carlson, N. Hu, D. Rabuka, J. Lee and C. R. Bertozzi, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 3000–3005 CrossRef CAS PubMed.
- M. W.-L. Popp and H. L. Ploegh, Angew. Chem., Int. Ed., 2011, 50, 5024–5032 CrossRef CAS PubMed.
- C. Uttamapinant, K. A. White, H. Baruah, S. Thompson, M. Fernandez-Suarez, S. Puthenveetil and A. Y. Ting, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 10914–10919 CrossRef CAS PubMed.
- C.-W. Lin and A. Y. Ting, J. Am. Chem. Soc., 2006, 128, 4542–4543 CrossRef CAS PubMed.
- C. Gnaccarini, W. Ben-Tahar, A. Mulani, I. Roy, W. D. Lubell, J. N. Pelletier and J. W. Keillor, Org. Biomol. Chem., 2012, 10, 5258–5265 CAS.
- T. Kashiwagi, J. Biol. Chem., 2002, 277, 44252–44260 CrossRef CAS PubMed.
- K. Yokoyama, N. Nio and Y. Kikuchi, Appl. Microbiol. Biotechnol., 2004, 64, 447–454 CrossRef CAS PubMed.
- A. Besheer, T. C. Hertel, J. Kressler, K. Mäder and M. Pietzsch, J. Pharm. Sci., 2009, 98, 4420–4428 CrossRef CAS PubMed.
- N. Kamiya and H. Abe, Methods Mol. Biol., 2011, 751, 81–94 CAS.
- J.-H. Lee, C. Song, D.-H. Kim, I.-H. Park, S.-G. Lee, Y.-S. Lee and B.-G. Kim, Biotechnol. Bioeng., 2013, 110, 353–362 CrossRef CAS PubMed.
- Y. Sugimura, K. Yokoyama, N. Nio, M. Maki and K. Hitomi, Arch. Biochem. Biophys., 2008, 477, 379–383 CrossRef CAS PubMed.
- S. K. Oteng-Pabi and J. W. Keillor, Anal. Biochem., 2013, 441, 169–173 CrossRef CAS PubMed.
- M. T. Gundersen, J. W. Keillor and J. N. Pelletier, Appl. Microbiol. Biotechnol., 2013, 1–12 Search PubMed.
- P. Riggs, Expression and Purification of Maltose-Binding Protein Fusions, John Wiley & Sons, Inc., Hoboken, NJ, USA, 2001 Search PubMed.
- Y.-G. Shi, L. Qian, N. Zhang, C.-R. Han, Y. Liu, Y.-F. Zhang and Y.-Q. Ma, Molecules, 2011, 16, 10046–10058 CrossRef CAS PubMed.
- R. K. Singh, M. K. Tiwari, R. Singh and J.-K. Lee, Int. J. Mol. Sci., 2013, 14, 1232–1277 CrossRef CAS PubMed.
- A. J. Lam, F. St-Pierre, Y. Gong, J. D. Marshall, P. J. Cranfill, M. A. Baird, M. R. McKeown, J. Wiedenmann, M. W. Davidson, M. J. Schnitzer, R. Y. Tsien and M. Z. Lin, Nat. Methods, 2012, 9, 1005–1012 CrossRef CAS PubMed.
- L. Carroll, H. L. Evans, E. O. Aboagye and A. C. Spivey, Org. Biomol. Chem., 2013, 11, 5772–5781 CAS.
- J. Guy, R. Castonguay, N. B. Campos-Reales Pineda, V. Jacquier, K. Caron, S. W. Michnick and J. W. Keillor, Mol. BioSyst., 2010, 6, 976–987 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental information for expression and purification of mTG, cloning and expression of Q-tagged proteins, synthesis of dansylethylazide and fluorescence microscopy of mRuby2-conjugated nanoparticles. See DOI: 10.1039/c4cc00994k |
| This journal is © The Royal Society of Chemistry 2014 |