Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Albumin-micelles via a one-pot technology platform for the delivery of drugs†
Yanyan
Jiang
,
Mingtao
Liang
,
Domenic
Svejkar
,
Gene
Hart-Smith
,
Hongxu
Lu
,
Wei
Scarano
and
Martina H.
Stenzel
*
Centre for Advanced Macromolecular Design (CAMD), School of Chemistry, University of New South Wales, Sydney, NSW 2052, Australia. E-mail: M.Stenzel@unsw.edu.au
First published on 24th April 2014
Abstract
A new micelle delivery platform based on albumin coated nanoparticles is able to selectively deliver the payload to cancerous cells while healthy cells remain less affected. The technology is simple and can be used in a one-pot procedure.
Abraxane©, is an injectable formulation of paclitaxel where the drug is bound to albumin as a delivery vehicle (nab technology). This is one of the few nano-formulations that have made it on the market today. The secret of this technology developed by Desai and co-workers1 lies within the simplicity of the approach while being highly effective during treatment of cancer. The capacity for albumin to adsorb hydrophobic molecules as well as the advantage of discharging the drug via desorption without significant burst effects makes it a unique carrier for a controlled release system.2 Due to pathophysiological conditions in neoplastic tissue, high amounts of albumin accumulate in tumours, and metabolization in malignant cells is enhanced by specific receptor glycoproteins gp60 on the endothelial cell surface. These provide a mediated albumin transport pathway to the subendothelial space and more importantly, even distribution of the drug in tumour site.3 Many formulations so far use a single albumin molecule as carrier with the drug either physically absorbed or chemically bound.3 Processing many albumin molecules into nanoparticles represents a quantum leap in performance since additional accumulation of the drug in the tumour site was caused by the enhanced permeation retention (EPR) effect.3,4
The nab technology produces particles of sizes of well above 100 nm. This may limit its use to certain application since some cancers, such as pancreatic cancers, seem to have an appetite for particles below 50 nm.5 Moreover, the driving force of nanoparticle formation is the presence of hydrophobic drug molecules which act like glue holding the albumin molecules in place. More hydrophilic drug molecules and charged drugs like nucleic acids are therefore not suitable for this technique. In some cases, albumin has been modified to conjugate to DNA,6,7 but these extensive modifications of albumin may have altered the protein structure, which in turn loses its specificity towards gp60.
In order to modify albumin without affecting its bioactivity, one method is to only modify one amino acid with a water-soluble polymer as it has been done in the past 40 years.8 Conjugation of hydrophobic polymer chain is a less explored pathway resulting in biohybrid amphiphiles with a hydrophilic protein head group conjugated to a hydrophobic polymer tail which self-assembles to form micelles in water.9–13 The synthesis of a well-defined biohybrid amphiphile requires precise control over both the protein/polymer ratio and the site at which the protein is modified.9,14
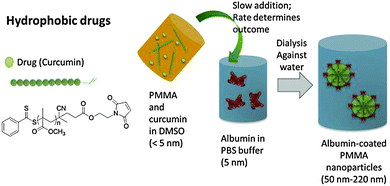
In this Communication, we have demonstrated the successful synthesis of albumin polymer conjugated nanocarriers and also its ability to deliver drugs without losing the specificity of albumin (Fig. 1).15 Maleimide-terminated poly(methyl methacrylate) (PMMA) has been prepared via the reversible addition fragmentation chain transfer (RAFT) polymerization. After drug loading the formation of micelles took place in an aqueous environment. These biohybrid micelles possess a built-in biofunctionality through the presence of the protein based head group compared to PEG based hydrophilic head groups, which is typically used for polymeric micelles.
For the attachment of hydrophobic polymer chains to albumin, one of the prerequisite is that the polymer chains must have high reactive end group fidelity for the conjugation. PMMA was primarily chosen as the suitable hydrophobic block to be attached to albumin, but the process should be suitable for any hydrophobic block including degradable polymers. For proof of concept, PMMA was prepared with a RAFT agent that contains the functionality of furan protected maleimide, but a myriad of other reactive groups for protein conjugation are possible.16 The main challenge is the slow addition of the albumin solution in PBS buffer to the polymer solution in DMSO. Increasing amounts of water in the DMSO solution can lead to the precipitation of polymer while the presence of DMSO causes the protein to precipitate. The rate of polymer–protein conjugation needs to be faster than the change in solvent quality to obtain a stable system. The system was optimised by the slow addition of albumin containing aqueous solution to the DMSO solution over several hours with the help of a syringe pump, followed by dialysis to remove DMSO. The size of albumin coated PMMA particles obtained were found to be dependent on the molecular weight of the hydrophobic polymers and larger PMMA chains lead to larger nanoparticles (ESI† and Table 1). Therefore by simply changing the polymer length from Mn = 5000 g mol−1 to 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 g mol−1 the particle size can be adjusted between 68 to 210 nm.
500 g mol−1 the particle size can be adjusted between 68 to 210 nm.
| Molecular weight/g mol−1 | Curcumin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) polymer molar ratio polymer molar ratio |
Volume of DMSO/mL | Volume of PBS buffer/mL | D h/nma | |
|---|---|---|---|---|---|
| a After dialysis against water. | |||||
| 1 | 42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
— | 2 | 8 | 213 ± 37 |
| 2 | 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— | 2 | 8 | 164 ± 22 |
| 3 | 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— | 2 | 8 | 122 ± 15 |
| 4 | 8500 | — | 2 | 8 | 99 ± 12 |
| 5 | 5400 | — | 2 | 8 | 68 ± 11 |
| 6 | 5400 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 5 |
3 | 7 | 104 ± 25 |
| 7 | 5400 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
3 | 7 | 59 ± 18 |
| 8 | 5400 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 15 |
3 | 7 | 50 ± 12 |
| 9 | 5400 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 15 |
2 | 8 | 112 ± 20 |
| 10 | 5400 | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 15 |
4 | 6 | 119 ± 23 |
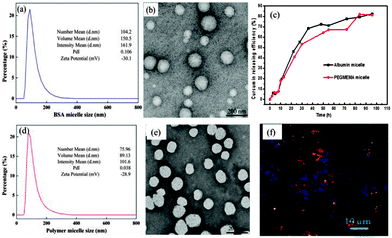
The formation of polymer–protein conjugates and the simultaneous self-assembly into nanoparticles was not only confirmed using DLS and TEM, but also with MALDI-TOF and GPC using THF and water as the mobile phase. MALDI-TOF analysis gives direct evidence for the presence of the protein drug conjugate at around 71![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g mol−1 (ESI,† Fig. S3 and S4), although it needs to be considered that ionization of the polymer–protein conjugate compared to the free protein is difficult and the height of the signals is not representative of the actual amounts. The spectrum shown in ESI,† Fig. S4 also shows the presence of BSA dimers, which have potentially polymers attached. It is commonly assumed that dimerization is the result of coupling of Cys-34, which is possibly the main attachment point for polymer conjugation, but other reports highlight that dimerization is not necessarily prevented when Cys-34 has been reacted.17 Further investigations are therefore necessary to understand the co-existence of dimers and polymer-conjugates. In addition, the conjugation efficiency could be measured using GPC (ESI,† Fig. S5 and S6). Resuspension of the freeze dried final product in THF led to the extraction of the unreacted PMMA into the THF. In contrast, water will only separate free BSA while the conjugate remains insoluble. Based on the results from the GPC measurements, equimolar amount of albumin and PMMA led to a conjugation efficiency of about 58%. (ESI,† Fig. S6) Results from the THF GPC, which suggests a conjugation efficiency of 87% (ESI,† Fig. S5), were neglected since the incomplete removal of unreacted PMMA from the particle is very likely. A control experiment was carried out that enlisted PMMA without any reactive endgroup and BSA. Mixing of both compounds using similar conditions led to the formation of a cloudy solution with a broad particle size distribution. Analysis of the reaction mixture by washing it with THF to extract unreacted PMMA revealed the easy separation of PMMA and BSA (ESI,† Fig. S7).
000 g mol−1 (ESI,† Fig. S3 and S4), although it needs to be considered that ionization of the polymer–protein conjugate compared to the free protein is difficult and the height of the signals is not representative of the actual amounts. The spectrum shown in ESI,† Fig. S4 also shows the presence of BSA dimers, which have potentially polymers attached. It is commonly assumed that dimerization is the result of coupling of Cys-34, which is possibly the main attachment point for polymer conjugation, but other reports highlight that dimerization is not necessarily prevented when Cys-34 has been reacted.17 Further investigations are therefore necessary to understand the co-existence of dimers and polymer-conjugates. In addition, the conjugation efficiency could be measured using GPC (ESI,† Fig. S5 and S6). Resuspension of the freeze dried final product in THF led to the extraction of the unreacted PMMA into the THF. In contrast, water will only separate free BSA while the conjugate remains insoluble. Based on the results from the GPC measurements, equimolar amount of albumin and PMMA led to a conjugation efficiency of about 58%. (ESI,† Fig. S6) Results from the THF GPC, which suggests a conjugation efficiency of 87% (ESI,† Fig. S5), were neglected since the incomplete removal of unreacted PMMA from the particle is very likely. A control experiment was carried out that enlisted PMMA without any reactive endgroup and BSA. Mixing of both compounds using similar conditions led to the formation of a cloudy solution with a broad particle size distribution. Analysis of the reaction mixture by washing it with THF to extract unreacted PMMA revealed the easy separation of PMMA and BSA (ESI,† Fig. S7).
It should be noted here that the resulting core–shell nanoparticles are not micelles in the traditional sense, but rather nano-precipitated aggregates with an albumin surface layer. The detailed internal structure will be subject to further investigations.
Subsequently, curcumin was employed as the model-drug, not only because of its bright yellow colour which facilitates the analysis, but also because of its anti-cancer properties.18 Curcumin has recently been delivered using the nab-technology, but the particles were well above 200 nm.19 Incorporating curcumin in the synthesis of albumin–polymer conjugates results in three processes that take place simultaneously: polymer–protein conjugation, self-assembly into micelles and drug encapsulation into the PMMA compartments of the nanoparticle. For the following experiment, PMMA with a molecular weight of Mn = 5400 g mol−1 was employed. Initial investigations were devoted to the effect of the amount of curcumin used during micelle formation process. Molar ratios between curcumin and PMMA of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 to 1
5 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 were employed for the investigation (Table 1). The DLS analysis of the only slightly opaque (ESI,† Fig. S8) curcumin loaded nanoparticles reveals a desirable aggregation sizes below 100 nm with a clearly-defined, narrow peak (ESI,† Fig. S9). A clear relationship between the amount of curcumin used and the final particle size is shown in Table 1 and Fig. S4 (ESI†). Higher amounts of curcumin lead to larger particles. Included in this study was the optimum ratio between DMSO and water. The drug-loaded nanoparticle formation is a sensitive interplay between different parameters. Drug encapsulation, polymer–protein conjugation and self-assembly need to occur prior to precipitation in the DMSO–water mixture that can neither dissolve albumin nor PMMA. A volume ratio of 30/70% (v/v) was deemed the most suitable resulting in narrow particle size distributions (ESI,† Fig. S10) while the overall hydrodynamic diameter was small compared to the other mixtures (Table 1). Higher DMSO content led to premature precipitation of albumin while PMMA cannot be dissolved in high water-contents.
15 were employed for the investigation (Table 1). The DLS analysis of the only slightly opaque (ESI,† Fig. S8) curcumin loaded nanoparticles reveals a desirable aggregation sizes below 100 nm with a clearly-defined, narrow peak (ESI,† Fig. S9). A clear relationship between the amount of curcumin used and the final particle size is shown in Table 1 and Fig. S4 (ESI†). Higher amounts of curcumin lead to larger particles. Included in this study was the optimum ratio between DMSO and water. The drug-loaded nanoparticle formation is a sensitive interplay between different parameters. Drug encapsulation, polymer–protein conjugation and self-assembly need to occur prior to precipitation in the DMSO–water mixture that can neither dissolve albumin nor PMMA. A volume ratio of 30/70% (v/v) was deemed the most suitable resulting in narrow particle size distributions (ESI,† Fig. S10) while the overall hydrodynamic diameter was small compared to the other mixtures (Table 1). Higher DMSO content led to premature precipitation of albumin while PMMA cannot be dissolved in high water-contents.
Prior to further investigations in regards to the drug loading efficiency, drug release and in vitro investigations a control experiment was set-up to be able to compare the performance of the albumin micelle with a tradition polymer micelle with a poly[oligo(ethylene glycol) methylether methacrylate] (POEGMEMA) surface layer. To simulate the negative charge of the albumin micelle (ξ = −30.1 mV), methacrylic acid (MAA) was co-polymerized with OEGMEMA using RAFT polymerization. The macroRAFT agent was then chain extended with MMA to yield P(OEGMEMA104-co-MAA5)-b-PMMA76, yielding micelles with the same hydrophobic compartment as the albumin-micelles, but a PEG-based shell with a similar negative charge (ξ = −28.9 mV). Two systems – one with albumin shell, the other with a PEG based shell – were compared to identify the effect of albumin. The drug encapsulation efficiency of albumin-loaded micelle was depicted in Fig. 1 was determined using UV/Vis analysis and calculated to be 72% (ESI,† Fig. S11). The curcumin in the POEGMEMA based micelle was loaded using similar concentrations, but with a more traditional solvent exchange technique (ESI†). The drug encapsulation efficiency was determined to be 79%. The physical parameter of both albumin and POEGMEMA based micelles are summarized in Fig. 2 highlighting their similarities. In addition, the rate of drug release was found to be similar as well (Fig. 2(c)). This is not unexpected considering that the retention of the drug is influenced partly by the compatibility with the polymer matrix, in both cases here PMMA.20
It is hypothesised that the replacement of the more tradition PEG shell of the micelle by albumin has more advantages, not just the biocompatibility factor as albumin is already abundant in the body. The uptake efficiency of both micelles was tested against different cell lines and was determined visually by confocal fluorescent microscopy (Fig. 2(f) and ESI,† Fig. S12). The fluorescent labelled micelles were found to be localized inside the endosomes within a time window of a few minutes. The efficient delivery of the anti-cancer drugs into the cell can often be correlated to amount of cell death, which is expressed by the IC50 value. To investigate the cytotoxicity of these drug loaded micelles the ovarian cancer cell line (A2870) and the pancreatic cancer cell line (AsPC-1) were used. A non-cancerous cell line, Chinese hamster ovary (CHO) was also tested to evaluate the selectivity of the albumin-carrier towards tumour cells.
In vitro studies (ESI,† Fig. S13–S15) show that free curcumin is toxic to all cell lines including healthy Chinese hamster ovary (CHO) cells (Table 2, entry 1). It needs to be noted here that the empty albumin-PMMA micelle is non-toxic. Drugs loaded into PEGMEMA based micelles increases the toxicity simply due to its improved drug uptake. This improved effect is due to the change in uptake pathway where the drugs are now entering the cell via an endocytic pathway instead of diffusion. Therefore compared to free drug, the PEG based micelles shows a higher toxicity towards cancer cells, but also shows high toxicity towards healthy CHO cells (Table 2, entry 2). Remarkably, the delivery of the drug into the two cancerous cell lines using albumin-coated micelles seems to be enhanced, which leads to higher toxicities. In contrast, the healthy CHO cells seemed to be less affected when incubated with the albumin micelle (Table 2, entry 3). This clearly highlights the selectivity of the albumin micelles towards cancerous cells. It has been highlighted earlier that the accumulation of albumin and albumin-bound drugs in the tumour is facilitated by SPARC (secreted protein, acidic and rich in cysteine). It was reported that SPARC is an albumin bound protein which may play an important role in the increased tumour accumulation of albumin-bound drugs.21 Interestingly, SPARC is overexpressed in several aggressive cancers but is absent in the corresponding normal tissues.
| IC50 (μM) A 2870 (cancerous) | IC50 (μM) AsPC-1 (cancerous) | IC50 (μM) CHO (healthy) | |
|---|---|---|---|
| 1 | 15.4 | 11.5 | 10.07 |
| 2 | 8.38 | 12.89 | 6.6 |
| 3 | 2.09 | 4.33 | 11 |
In conclusion, initial results show that the use of albumin-micelles is a simple and effective approach to deliver anti-cancer drugs. As depicted in Fig. 1, the nanoparticles can often be prepared in one pot while the size can be adjusted by the molecular weight of the polymer. The advantage of this system is not only the simplicity in synthesis, but also the specificity of albumin towards cancerous cells. This technique can therefore be described as inexpensive while being efficient to select cancer cells and considered to be able to deliver different types of drugs which will be further investigated in future. One aspect that has not yet been touch upon is the activity of the albumin after conjugation and the effect of the molecular weight the protein, which will be subject to a more detailed upcoming study.
Notes and references
- N. Desai, Drug Del. Report, 2007, pp. 37–41 Search PubMed.
- V. T. G. Chuang, U. Kragh-Hansen and M. Otagiri, Pharm. Res., 2002, 19, 569–577 CrossRef.
- B. Elsadek and F. Kratz, J. Controlled Release, 2012, 157, 4–28 CrossRef CAS PubMed.
- F. Kratz, J. Controlled Release, 2008, 132, 171–183 CrossRef CAS PubMed.
- H. Cabral, Y. Matsumoto, K. Mizuno, Q. Chen, M. Murakami, M. Kimura, Y. Terada, M. R. Kano, K. Miyazono, M. Uesaka, N. Nishiyama and K. Kataoka, Nat. Nanotechnol., 2011, 6, 815–823 CrossRef CAS PubMed.
- K. Eisele, R. A. Gropeanu, C. M. Zehendner, A. Rouhanipour, A. Ramanathan, G. Mihov, K. Koynov, C. R. W. Kuhlmann, S. G. Vasudevan, H. J. Luhmann and T. Weil, Biomaterials, 2010, 31, 8789–8801 CrossRef CAS PubMed.
- D. Fischer, T. Bieber, S. Brüsselbach, H.-P. Elsässer and T. Kissel, Int. J. Pharm., 2001, 225, 97–111 CrossRef CAS.
- K. L. Heredia and H. D. Maynard, Org. Biomol. Chem., 2007, 5, 45–53 CAS.
- A. J. Dirks, R. J. M. Nolte and J. J. L. M. Cornelissen, Adv. Mater., 2008, 20, 3953–3957 CrossRef CAS.
- E. Garanger and S. Lecommandoux, Angew. Chem., Int. Ed., 2012, 51, 3060–3062 CrossRef CAS PubMed.
- H.-N. Kim, J. Lee, H.-Y. Kim and Y.-R. Kim, Chem. Commun., 2009, 7104–7106 RSC.
- H. Y. Cho, M. A. Kadir, B.-S. Kim, H. S. Han, S. Nagasundarapandian, Y.-R. Kim, S. B. Ko, S.-G. Lee and H.-j. Paik, Macromolecules, 2011, 44, 4672–4680 CrossRef CAS.
- B. Le Droumaguet and K. Velonia, Angew. Chem., Int. Ed., 2008, 47, 6263–6266 CrossRef CAS PubMed.
- J. Ge, E. Neofytou, J. Lei, R. E. Beygui and R. N. Zare, Small, 2012, 8, 3573–3578 CrossRef CAS PubMed.
- M. H. Stenzel, W. Scarano, D. Svejkar and M. Liang, Nanoparticles for drug delivery, PCT/AU2014/000346, 2014 Search PubMed.
- R. M. Broyer, G. N. Grover and H. D. Maynard, Chem. Commun., 2011, 47, 2212–2226 RSC.
- A. Brahma, C. Mandal and D. Bhattacharyya, Biochim. Biophys. Acta, 2005, 1751, 159–169 CrossRef CAS PubMed.
- F. H. Sarkar and Y. Li, Cancer Treat. Rev., 2009, 35, 597–607 CrossRef CAS PubMed.
- A. Jithan, K. Madhavi, M. Madhavi and K. Prabhakar, Int. J. Pharm. Invest., 2011, 1, 119–125 CrossRef CAS PubMed.
- Y. Kim, E. D. Liemmawa, M. H. Pourgholami, D. L. Morris and M. H. Stenzel, Macromolecules, 2012, 45, 5451–5462 CrossRef CAS.
- N. Desai, V. Trieu, B. Damascelli and P. Soon-Shiong, Transl. Oncol., 2009, 2, 59–64 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: All experimental procedures as well as all raw data and the investigations in the optimization of the process. See DOI: 10.1039/c4cc00616j |
| This journal is © The Royal Society of Chemistry 2014 |