Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Discovery of new G-quadruplex binding chemotypes†
Stephan A.
Ohnmacht
a,
Ehsan
Varavipour
a,
Rupesh
Nanjunda
b,
Ingrida
Pazitna
a,
Gloria
Di Vita
a,
Mekala
Gunaratnam
a,
Arvind
Kumar
b,
Mohamed A.
Ismail
b,
David W.
Boykin
b,
W. David
Wilson
b and
Stephen
Neidle
*a
aSchool of Pharmacy, University College London, London WC1N 1AX, UK. E-mail: s.neidle@ucl.ac.uk; Fax: +44 (0)207 753 5970; Tel: +44 (0)207 753 5969
bDepartment of Chemistry and Center for Biotechnology and Drug Design, Georgia State University, Atlanta, Georgia 30303-3083, USA
First published on 2nd December 2013
Abstract
We report here on the discovery and preliminary evaluation of a novel non-macrocyclic low molecular weight quadruplex-stabilizing chemotype. The lead compounds, based on a furan core, show high G-quadruplex stabilisation and selectivity as well as potent in vitro anti-proliferative activity.
Quadruplexes (G4s) are higher-order nucleic acid arrangements involving a core of π–π stacked guanine-quartets (G-quartets) rather than the Watson–Crick base pairs of double-helical nucleic acids.1 G4-forming sequences are widely prevalent in eukaryotic telomeric sequences as well as being over-represented in other genomes,2 notably promoter and 5′-UTR sequences of genes involved in cellular proliferation.3 The recent demonstration of the presence of G4s in human cells4 has added credence to the concept that G4s can be targets for therapeutic intervention, at the single gene or poly-gene levels.5 Appropriate small molecules can serve to stabilise G4s and the resulting complexes can then act as impediments to telomere maintenance, transcription or translation, depending on the nature of the quadruplex target site.6 These effects have been shown in several target genes of relevance to human cancer such as c-MYC7 and c-KIT.8
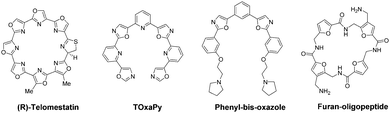
A large number of small molecule chemotypes have been reported as G4-binding ligands.6 The overwhelming majority are heteroaromatic with large flat surfaces, designed to complement the surface characteristics of a terminal G-quartet in a typical quadruplex structure. A second class of ligand is represented by the cyclic polyoxazole natural product telomestatin.9 A number of cyclic and acyclic analogues have been reported, some of which show potent biological activity.10 The acyclic compounds tend to be characterised by a crescent shape. For example, pyridostatin11 and several series of phenyl- and pyridyl-bis-oxazoles12–14 all selectively target G4s (Fig. 1). A more general requirement of most G4-binding ligands is the possession of side-chains terminating in cationic charge.6
Few G4-binding small molecules have proceeded to in vivo evaluation in models of human cancer, and to date only one compound, Quarfloxin, has been evaluated in clinical trials.15 The perceived lack of drug-like characteristics in many G4-binding compounds may have hindered progress to the clinic. We report here on a study to discover novel ligands with MW <400 Da that could be suitable starting-points for future drug discovery efforts.
Thirty-eight representative members of a large chemical library from the anti-parasitic drug discovery programme at Georgia State University16,17 (several hundred compounds), with highly diverse scaffolds and functional groups, were screened using a high-throughput 96-well FRET (Fluorescence Resonance Energy Transfer) assay.18 G4 stabilisation was initially evaluated using dual-labelled F21T (human telomeric 21-mer) and c-KIT2 (a tyrosine kinase oncogene) G4s, as well as a duplex DNA sequence (T-loop). The ten most active compounds were subsequently screened against an expanded panel of fluorescently-labelled promoter G4-forming sequences, with HSP90A, HSP90B (heat shock protein 90 promoter sequences),12 k-RAS21 (in the promoter of the k-RAS oncogene)19 and AR, a G4 recently identified in the promoter of the androgen receptor (involved in prostate cancer development).20
Six acyclic furan- and thiophene-based compounds (Fig. 2: 1–6), representing two distinctive chemotypes were identified with high (>15 °C) ΔTm values. A competition assay using unlabelled calf thymus duplex DNA and compounds 1–6 examined the ability of these compounds to differentiate between duplex and the F21T G4 DNA at high duplex![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G4 ratios. The compounds were also examined in a 96 h short-term sulforhodamine B (SRB) assay, to determine their ability to inhibit cancer cell growth (Tables 1 and 2). All six compounds showed potent G4 stabilising abilities, as judged by the large changes in ΔTm values for the selected G4s. In particular the bis-phenyl-mono-furan compounds 1 and 5 had especially high ΔTm values, broadly comparable to those for established high-affinity G4-binding compounds such as tetra-substituted naphthalene diimides.21 The tri-furan compound 2 is consistently more effective in stabilising the G4s than the polyfuran compound 6. Compound 2 and 6, representatives of a tetrafuran second chemotype, were less selective at high duplex ratios and were inactive in the SRB assay, possibly because of aqueous solubility and cellular uptake issues. Switching from a furan (1, 5) to a thiophene (4), does slightly affect G4 stabilisation and selectivity vs. duplex DNA, though not in vitro potency, which at least in the cell lines examined, is comparable to that of compounds 1 and 5.
G4 ratios. The compounds were also examined in a 96 h short-term sulforhodamine B (SRB) assay, to determine their ability to inhibit cancer cell growth (Tables 1 and 2). All six compounds showed potent G4 stabilising abilities, as judged by the large changes in ΔTm values for the selected G4s. In particular the bis-phenyl-mono-furan compounds 1 and 5 had especially high ΔTm values, broadly comparable to those for established high-affinity G4-binding compounds such as tetra-substituted naphthalene diimides.21 The tri-furan compound 2 is consistently more effective in stabilising the G4s than the polyfuran compound 6. Compound 2 and 6, representatives of a tetrafuran second chemotype, were less selective at high duplex ratios and were inactive in the SRB assay, possibly because of aqueous solubility and cellular uptake issues. Switching from a furan (1, 5) to a thiophene (4), does slightly affect G4 stabilisation and selectivity vs. duplex DNA, though not in vitro potency, which at least in the cell lines examined, is comparable to that of compounds 1 and 5.
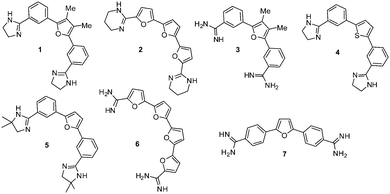 | ||
| Fig. 2 Structures of the lead compounds 1–6 identified in this study, together with a control compound 7. | ||
| Compound | Mol. Wt | F21T | c-KIT2 | HSP-90A | HSP-90B | k-RAS21R | AR | T-loop | G4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT 1 CT 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 1 |
G4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT 1 CT 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
G4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT 1 CT 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 100 |
G4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CT 1 CT 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 300 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 384.5 | 22.3 | 16.8 | 27.7 | 23.7 | 19.6 | 15.4 | <2 | 100 | 100 | 100 | 100 |
| 2 | 364.4 | 20.4 | 16.3 | 26.0 | 22.0 | 18.5 | 13.8 | <2 | 100 | 100 | 92.7 | 63.3 |
| 3 | 330.5 | 17.6 | 17.8 | 21.2 | 18.1 | 15.1 | 11.9 | 2.2 | 100 | 100 | 65.9 | 43.9 |
| 4 | 372.5 | 18.0 | 13.9 | 22.5 | 19.1 | 15.9 | 10.2 | <2 | 100 | 100 | 60.5 | 39.9 |
| 5 | 412.5 | 22.6 | 18.5 | 26.6 | 22.7 | 18.3 | 14.7 | <2 | 100 | 100 | 100 | n/a |
| 6 | 350.3 | 18.6 | n/a | 20.6 | 16.2 | 13.4 | 10.1 | <2 | 100 | 100 | 53.1 | 35.4 |
| 7 | 302.5 | 14.4 | 12.2 | 19.0 | 15.7 | 16.6 | 9.9 | 3.4 | 100 | 100 | 24.5 | 2.5 |
| 8 | 830.6 | 26.6 | 22.0 | 33.1 | 28.6 | n/a | 15.9 | 4.9 | 100 | 100 | n/a | 27.2 |
| A549 | MCF7 | ALT | Mia-PaCa2 | Panc1 | RCC4 | 786-O | WI38 | |
|---|---|---|---|---|---|---|---|---|
| 1 | 0.3 | 0.9 | 1.0 | 0.3 | 0.5 | 4.9 | 3.0 | 1.3 |
| 2 | >25 | >25 | >25 | >25 | >25 | >25 | >25 | >25 |
| 3 | 0.8 | 0.9 | 6.6 | 2.0 | 1.7 | >25 | >25 | >25 |
| 4 | 0.4 | 1.4 | 1.9 | 0.7 | 1.0 | 3.3 | 5.6 | 2.0 |
| 5 | 0.5 | 1.2 | 1.4 | 0.5 | 1.0 | 3.9 | 4.8 | 2.0 |
| 6 | 4.2 | 15.6 | 18.8 | 7.0 | >25 | >25 | 12.8 | >25 |
| 7 | 3.7 | 11.7 | >25 | 8.90 | 10.9 | >25 | >25 | >25 |
| 8 | 0.019 | 0.070 | 0.063 | 0.011 | 0.003 | 0.560 | 0.320 | 0.230 |
Overall, F21T and the two HSP90 G4s have been most stabilised by compounds 1–6. Comparison with the behaviour of a tetra-substituted naphthalenediimide compound previously examined by us,20 shows that 1–6 exhibit only moderate ΔTm values with the AR G4, which are generally lower than with other G4s. Compounds 1–6 produced slightly reduced but still significant stabilisation with the c-KIT2 and k-RAS21 G4s, suggesting that these compounds have the ability to act simultaneously on multiple G4 targets (G4 poly-targeting). The stabilisation of a duplex DNA sequence (T-loop) was not significantly affected by any of the compounds at the biologically relevant concentration employed here (1 μM). Compounds 1 and 5 in particular are highly selective for G4 versus duplex DNA, as found in a series of competition assays, where the G4 stabilisation ability of both compounds is undiminished by adding calf thymus DNA in excess, at ratios up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100/300. A control compound, 7, an established duplex DNA minor groove binder, also showed significant G4 stabilisation, albeit with greater effects on the duplex DNA used (with a sub-optimal sequence for this compound).
100/300. A control compound, 7, an established duplex DNA minor groove binder, also showed significant G4 stabilisation, albeit with greater effects on the duplex DNA used (with a sub-optimal sequence for this compound).
Circular dichroism (CD) was employed to qualitatively evaluate the binding mode of the lead compounds and to examine induced structural transitions in the telomeric G4. The CD spectra (Fig. 3 and ESI†) show that compounds 1 and 3–6 produce very small induced CD signals in a human telomeric quadruplex sequence. Such weak induced CD signals are characteristic of quadruplex end-stacking compounds and the small differences in the CD signal patterns for different compounds indicate minor differences in the stacking geometries of the ligands at the terminal G-quartets.
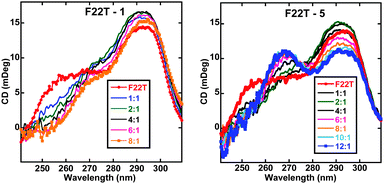 | ||
Fig. 3 Circular dichroism (CD) spectra for compounds 1 and 5 at differing ligand![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) G4 ratios. F22T is a 22-mer analogue of the F21T sequence. G4 ratios. F22T is a 22-mer analogue of the F21T sequence. | ||
All the compounds exhibiting weak induced CD signals that show a binding preference for an anti-parallel type quadruplex conformation as observed by decreases in the CD signal around 260 nm. Compound 5 however behaves differently, inducing a much larger conformational transition in the telomeric quadruplex upon complex formation. Upon titrating 5, the CD intensity around 290 nm decreases with a subsequent increase in the CD signal intensity around 260 nm. This is most likely due to an induced conformational transition from a hybrid to a more parallel-type G4 form.
Analysis of the equilibrium conformation for compound 1 was undertaken with classical molecular mechanics (MM2 and AMBER) force fields and by ab initio (STO-3G) calculations. The MM2 analysis suggested that overall arrangement involved coplanar rings. This somewhat implausible conclusion was at variance with results from the AMBER force field and the ab initio calculations, which concurred in suggesting a twisted conformation due to the repulsive effects of the two furanoid methyl groups on the attached phenyl rings. The barrier between planar and twisted conformations is likely to be low so a qualitative analysis of plausible G4-bound conformations for compounds 1 and 2 has been undertaken. The NMR structure of an anti-parallel human telomeric G4 complexed with a telomestatin derivative has been used as a starting-point.22 Compounds 1 and 2 have similarities to the overall shape and curvature of telomestatin (Fig. 4), albeit in an acyclic manner. The NMR structure shows that the telomestatin derivative is non-planar and its out-of-plane distortions complement the non-planarity of the G-quartet to which it is bound. The qualitative low-energy structures proposed for compounds 1 and 2 incorporate such distortions, which still enable effective π–π stacking onto the terminal G-quartet of this G4, as well as some additional stabilisation from a thymine. The modelling show that the meta position of the dihydroimidazole substituents in 1 and the tetrahydropyrimidine substituents in 2, is crucial for shape similarity to telomestatin. The para substitution in the control compound 7 enables it to effectively bind to the minor groove of duplex DNA, which is not possible for any of the six compounds arising from the screen.
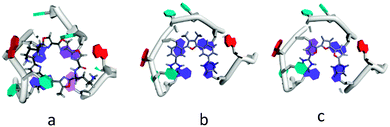 | ||
| Fig. 4 Plots showing the terminal G-quartet of anti-parallel G4 complexes, with guanines shown in dark blue, thymines in cyan and adenines in red. Ligands are shown in stick representation. (a) The telomestatin derivative complex,22 (b) results of qualitative modelling with compound 1 and (c) with compound 2. | ||
Several of the compounds showed low μM anti-proliferative activity (Table 2) in a cancer cell line panel [A549 (lung cancer), MCF7 (breast cancer), RCC4 and 786-O (renal cancer), Panc1 and Mia-PaCa2 (pancreatic cancer), ALT (transformed WI38 lung fibroblast cells characterised by Alternative Maintenance of Telomeres) and WI38 (non-transformed lung fibroblast cells)].
The two poly-furan compounds 2 and 6 have low anti-proliferative activity, even though both have G4-stabilising activity comparable to the other four compounds in the group. This may be due to cell uptake and nuclear localisation problems as well as limited aqueous solubility; the lack of observed precipitation during the SRB assay supports the former suggestions. Compounds from the mono-furan and mono-thiophene series on the other hand show activity in several cell lines in the low-μM range. The lung cancer cell line A549 is slightly more sensitive to these compounds, although we do not currently have a molecular explanation for this.
We cannot exclude the possibility of non-G4 targets being involved in these cellular effects: experiments are currently underway to examine links with G4 affinity for these compounds. G4 selectivity appears to be limited even though for the two lead compounds at least, duplex DNA is less likely to be a target. This suggests that they may be acting as poly-targeting agents, affecting a number of genes and oncogenes involved in cellular proliferation and also that the compounds do not have identical cellular targets. It is notable that compounds 3 and 6 have high selectivity for several of the cancer lines compared to the normal WI38 line and have some activity in the ALT line, which maintains telomere length by non-telomerase mechanisms. This suggests that telomere maintenance rather than telomerase per se, is being targeted.
We report here that screening putative ligands using a HTS-FRET assay against a panel of G4s with a duplex control sequence, has resulted in the discovery of meta-substituted bisphenyl-monofurans as a novel G4 stabilizing chemotype. A similar chemotype, with a urea group replacing the furan ring, has been reported23 as having high G4 affinity. These compounds are structurally-simple, conformationally flexible and chemically readily accessible with MW <400 Da. They have G4 stabilisation ability comparable to those previously observed with polycyclic heteroaromatic compounds21 (cf. compound 8 (4,9-bis((3-(4-methyl-piperazin-1-yl)propyl)amino)-2,7-bis(3-morpholinopropyl)benzo-[lmn][3,8]phenanthroline-1,3,6,8(2H,7H)-tetraone) in Table 1), but with low duplex DNA affinity. They inhibit cancer cell growth at low μM/high nM levels, suggesting that these or related compounds may have potential as drug-like poly-quadruplex targeting agents.
Synthesis and biophysical studies at Georgia State University were supported by National Institutes of Health NIAID Grant AI-064200 (WDW, DWB). Work at UCL was supported by a MRC Confidence in Concept grant (SN).
References
- S. Burge, G. N. Parkinson, P. Hazel, A. K. Todd and S. Neidle, Nucleic Acids Res., 2006, 34, 5402 CrossRef CAS PubMed.
- J. Huppert and S. Balasubramanian, Nucleic Acids Res., 2005, 33, 2908 CrossRef CAS PubMed; A. Todd, M. Johnston and S. Neidle, Nucleic Acids Res., 2005, 33, 2901 CrossRef PubMed.
- J. L. Huppert and S. Balasubramanian, Nucleic Acids Res., 2007, 35, 406 CrossRef CAS PubMed; J. L. Huppert, A. Bugaut, S. Kumari and S. Balasubramanian, Nucleic Acids Res., 2008, 36, 6260 CrossRef PubMed; S. Balasubramanian, L. H. Hurley and S. Neidle, Nat. Rev. Drug Discovery, 2011, 10, 261 CrossRef PubMed.
- G. Biffi, D. Tannahill, J. McCafferty and S. Balasubramanian, Nat. Chem., 2013, 5, 182–186 CrossRef CAS PubMed.
- V. Sekaran, J. Soares and M. B. Jarstfer, J. Med. Chem., 2013 DOI:10.1021/jm400528t.
- D. Monchaud and M.-P. Teulade-Fichou, Org. Biomol. Chem., 2008, 6, 627 Search PubMed; T. M. Ou, Y. J. Lu, J. H. Tan, Z. S. Huang, K. Y. Wong and L. Q. Gu, ChemMedChem, 2008, 3, 690 CrossRef CAS PubMed; D. Z. Yang and K. Okamoto, Future Med. Chem., 2010, 2, 619 CrossRef.
- J. Dai, M. Carver, L. H. Hurley and D. Yang, J. Am. Chem. Soc., 2011, 133, 17673 CrossRef CAS PubMed.
- S. Rankin, A. P. Reszka, J. Huppert, M. Zloh, G. N. Parkinson, A. K. Todd, S. Ladame, S. Balasubramanian and S. Neidle, J. Am. Chem. Soc., 2005, 127, 10584 CrossRef CAS PubMed; H. Fernando, A. P. Reszka, J. Huppert, S. Ladame, S. Rankin, A. R. Venkitaraman, S. Neidle and S. Balasubramanian, Biochemistry, 2006, 45, 7854 CrossRef PubMed.
- M. C. Nielsen and T. Ulven, Curr. Med. Chem., 2010, 17, 3438 CrossRef CAS.
- J. Linder, T. P. Garner, H. E. Williams, M. S. Searle and C. J. Moody, J. Am. Chem. Soc., 2011, 133, 1044 CrossRef CAS PubMed.
- S. Müller, D. A. Sanders, M. Di Antonio, S. Matsis, J.-F. Riou, R. Rodriguez and S. Balasubramanian, Org. Biomol. Chem., 2012, 10, 6537 Search PubMed.
- S. A. Ohnmacht, M. Micco, V. Petrucci, A. K. Todd, A. P. Reszka, M. Gunaratnam, M. A. Carvalho, M. Zloh and S. Neidle, Bioorg. Med. Chem. Lett., 2012, 22, 5930 CrossRef CAS PubMed.
- S. A. Ohnmacht, C. Ciancimino, G. Vignaroli, M. Gunaratnam and S. Neidle, Bioorg. Med. Chem. Lett., 2013, 23, 5351 CrossRef CAS PubMed.
- F. Hamon, E. Large, A. Guédin-Beaurepaire, M. Rouchon-Dagois, A. Sidibe, D. Monchaud, J.-L. Mergny, J.-F. Riou, C.-H. Nyugen and M.-P. Teulade-Fichou, Angew. Chem., Int. Ed., 2011, 50, 8745 CrossRef CAS PubMed.
- D. Drygin, A. Siddiqui-Jain, S. O'Brien, M. Schwaebe, A. Lin, J. Bliesath, C. B. Ho, C. Proffitt, K. Trent, J. P. Whitten, J. K. Lim, D. Von Hoff, K. Anderes and W. G. Rice, Cancer Res., 2009, 69, 7653 CrossRef CAS PubMed.
- B. Nguyen, C. Tardy, C. Bailly, P. Colson, C. Houssier, A. Kumar, D. W. Boykin and W. D. Wilson, Biopolymers, 2002, 63, 281 CrossRef CAS PubMed.
- R. Nanjunda, C. Musetti, A. Kumar, M. A. Ismail, A. A. Farahat, S. Wang, C. Sissi, M. Palumbo, D. W. Boykin and W. D. Wilson, Curr. Pharm. Des., 2012, 18, 1934 CrossRef CAS.
- See B. Guyen, C. M. Schultes, P. Hazel, J. Mann and S. Neidle, Org. Biomol. Chem., 2004, 2, 981 CAS and ESI† for further details.
- S. Cogoi and L. E. Xodo, Nucleic Acids Res., 2006, 34, 2536 CrossRef CAS PubMed.
- T. Mitchell, A. Ramos-Montoya, M. Di Antonio, P. Murat, S. Ohnmacht, M. Micco, S. Jurmeister, L. Fryer, S. Balasubramanian, S. Neidle and D. E. Neal, Biochemistry, 2013, 26, 1429 CrossRef PubMed.
- M. Micco, G. W. Collie, A. G. Dale, S. A. Ohnmacht, I. Pazitna, M. Gunaratnam, A. P. Reszka and S. Neidle, J. Med. Chem., 2013, 56, 2959 CrossRef CAS PubMed.
- W. J. Chung, B. Heddi, M. Tera, K. Iida, K. Nagasawa and A. T. Phan, J. Am. Chem. Soc., 2013, 135, 13495 CrossRef CAS PubMed.
- A. Benz, V. Singh, T. U. Mayer and J. S. Hartig, ChemBioChem, 2011, 12, 1422 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3cc48616h |
| This journal is © The Royal Society of Chemistry 2014 |