Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Fluoromethylated derivatives of carnitine biosynthesis intermediates – synthesis and applications†
Anna M.
Rydzik
a,
Ivanhoe K. H.
Leung
a,
Armin
Thalhammer
a,
Grazyna T.
Kochan‡
b,
Timothy D. W.
Claridge
a and
Christopher J.
Schofield
*a
aDepartment of Chemistry, University of Oxford, Chemistry Research Laboratory, 12 Mansfield Road, Oxford OX1 3TA, UK. E-mail: christopher.schofield@chem.ox.ac.uk
bStructural Genomics Consortium, University of Oxford, Old Road Campus Roosvelt Drive, Headington OX3 7DQ, UK
First published on 21st November 2013
Abstract
A convenient method for the synthesis of fluoromethylated carnitine biosynthesis intermediates, i.e. fluorinated derivatives of γ-butyrobetaine and trimethyllysine, is described. The fluoromethylated probes were useful in both in vitro and cell based assays employing 19F NMR and LC-MS analyses.
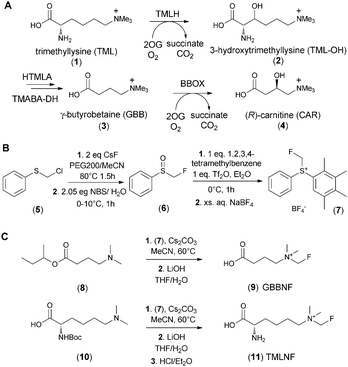
Carnitine, an essential metabolite in humans and other eukaryotes, is vital for the transport of long chain fatty acids into mitochondria.1 In animals carnitine is biosynthesised from trimethyllysine (TML) in four enzyme-catalysed steps.2–4 The first and last steps in the pathway are catalysed by trimethyllysine hydroxylase (TMLH) and γ-butyrobetaine hydroxylase (BBOX), both of which belong to the 2-oxoglutarate (2OG) dependent oxygenase superfamily. TMLH and BBOX catalyse the hydroxylation of Nε-trimethyllysine (TML, (1)) and γ-butyrobetaine (GBB, (3)) respectively (Scheme 1A).5,6 The inhibition of human carnitine biosynthesis reduces fatty acid metabolism and has been targeted for the treatment of cardiovascular diseases.7,8 With respect to efforts aimed at understanding biological roles of 2OG dependent oxygenases in carnitine metabolism,9,10 we are interested in developing reagents that enable assays for carnitine biosynthesis enzyme activities, both in vitro11 and in more biologically representative systems. Given that the monofluoromethyl group is similar in size to the methyl group, we envisioned fluoromethylated GBB (9) and TML (11) analogues as potentially useful tool compounds for monitoring carnitine biosynthesis.
Organofluorine compounds have substantial applications in chemistry and chemical biology; their applications include the modification of physicochemical and conformational properties of small molecules, and use as tracers in radiomedicine12 or labels in NMR studies.13,1419F NMR is emerging as a valuable tool for studying biochemical processes in vitro and in vivo.15 The principle advantage of 19F NMR is that the absence of endogenous fluorinated species in most living organisms eliminates the problem of background interference and signal overlap. However, the use of 19F NMR is often limited by the availability of appropriately labeled fluorinated compounds.
Various approaches for the incorporation of fluorine into small molecules are available, but reagents for electrophilic monofluoromethylation are limited.16 Prakash et al. reported an air and moisture stable electrophilic fluoromethylation reagent (7) that reacts under mild conditions17 with various nucleophiles, including amines.17,18 The reported preparation of (7) required the use of liquefied CH2FCl, which can be inconvenient. We therefore employed a route where chloromethylphenyl sulphide (5) was converted to the corresponding fluoride by reaction with caesium fluoride19 and then directly oxidised with aqueous N-bromosuccinimide to yield fluoromethylsulphinyl benzene (6) – a known intermediate in the synthesis of the fluoromethylating reagent (7) (Scheme 1B). We then used (7) to efficiently prepare N-CH2F modified derivatives of GBB (3) and TML (1) in 2 or 3 steps, respectively, starting from appropriately protected N,N-dimethylated precursors (Scheme 1C).
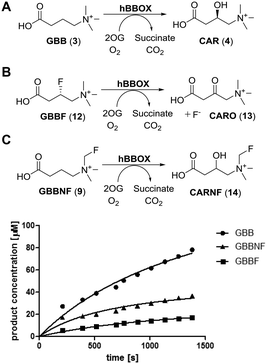
The fluoromethylated GBB analogue (GBBNF, (9)) was evaluated in reaction with the purified recombinant human BBOX (hBBOX) in vitro. GBBNF (9) was found to be a hBBOX substrate as demonstrated by 1H NMR analyses (Fig. S1, ESI†) undergoing hydroxylation at the C3 position to give a fluoromethylated carnitine analogue (CARNF) (14) (Fig. 1C; for NMR assignment of the product see Fig. S2–S4, ESI†), in an analogous manner to hBBOX catalysed GBB hydroxylation (Fig. 1A). We have reported the synthesis of another fluorinated analogue of GBB – (3S)-fluoro-GBB (GBBF, (12)), C-3 hydroxylation of which leads to fluoride release concomitant with ketone (13) formation (Fig. 1B).11 In NMR assays we did not observe fluoride release in reaction of GBBNF (9) with hBBOX, indicating that the fluoromethylated quaternary ammonium group is stable under the assay conditions. Comparison of the initial rates of hydroxylation of GBB (3), GBBNF (9) and GBBF (12) by hBBOX revealed that GBBNF (9) is a better substrate than GBBF (12) (Table 1, Fig. 1). The initial hydroxylation rate of GBBNF (9) is ∼65% of the initial hydroxylation rate of GBB (3), while GBBF (12) is hydroxylated at ∼20% of the initial rate of GBB (3). The KM and kcat values also reveal that the properties of GBBNF (9) as a substrate are closer to those of GBB (3) than GBBF (12) (Table 1). Some 2OG dependent oxygenases catalyse turnover of 2OG independent of substrate transformation (uncoupled 2OG turnover)5 which can manifest with a poor substrate.20 We examined levels of uncoupled turnover when GBB (3) and its fluorinated analogues were used, by comparing rates of succinate formation to that of hydroxylation. The results showed GBBNF (9) to be similar to GBB (3) (ratio of succinate formation to hydroxylation 1.1 for GBB vs. 1.3 for GBBNF), while in the case of GBBF (12), the ratio was ∼2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Table 1 and Fig. S5, ESI†). Note that, as for GBB (3), GBBF (12) displays substantial substrate inhibition (apparently Ki values in micromolar range – see Table 1). Interestingly, the extent of substrate inhibition by GBBNF (9) was much less than for GBB (3) or GBBF (12) (Fig. S6, ESI†). The mechanism of substrate inhibition of hBBOX is unknown, but is of interest as it may be involved in regulating carnitine biosynthesis in cells.
1 (Table 1 and Fig. S5, ESI†). Note that, as for GBB (3), GBBF (12) displays substantial substrate inhibition (apparently Ki values in micromolar range – see Table 1). Interestingly, the extent of substrate inhibition by GBBNF (9) was much less than for GBB (3) or GBBF (12) (Fig. S6, ESI†). The mechanism of substrate inhibition of hBBOX is unknown, but is of interest as it may be involved in regulating carnitine biosynthesis in cells.
| GBB (3) | GBBF (12) | GBBNF (9) | ||
|---|---|---|---|---|
| Rate [μM s−1] | Hydroxylation | 0.123 | 0.027 | 0.083 |
| Succinate formation | 0.162 | 0.063 | 0.097 | |
| Ratio of succinate formation to hydroxylation | 1.1 | 2 | 1.3 | |
| Kinetic parameters | K M [μM] | 4.2 | 19.8 | 16.6 |
| k cat [s−1] | 0.83 | 0.14 | 0.30 | |
| K i [μM] | 24.5 | 135 | — | |
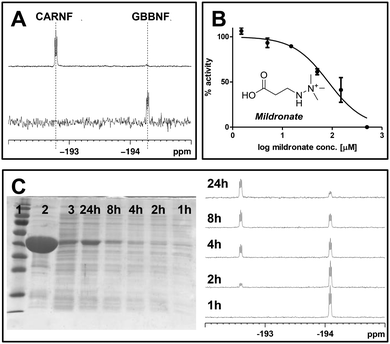
hBBOX catalysed GBBNF (9) hydroxylation can be followed by 19F NMR, because the fluorine shift of the product is distinctively different from a shift of the substrate (Fig. 2A). In these compounds the 19F resonance appears with a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 triplet fine structure, not as a singlet. This structure arises from coupling of the fluorine to the adjacent quadrupolar 14N nucleus (I = 1, 99.6% abundance) and is apparent because the highly symmetrical tetrahedral environment of the 14N centre suppresses rapid quadrupolar relaxation that, in less symmetrical environments, leads to loss of coupling fine structure. 19F NMR can also be employed for IC50 measurements (Fig. 2B). We investigated hBBOX inhibition by Mildronate,7 which is an inhibitor and a competitive substrate for hBBOX.9,10 The obtained IC50 value (82 μM) is similar to that obtained by 1H NMR (34 μM) and fluoride release (65 μM) assays.11 The differences likely reflect the different assay conditions used. Thus, 19F NMR assay employing GBBNF (9) as a substrate is useful for in vitro activity studies. An advantage of the GBBNF (9) based system is its ability to monitor specific hBBOX turnover without the interference of any other non-fluorinated components, making it suitable for biologically relevant assay conditions.
1 triplet fine structure, not as a singlet. This structure arises from coupling of the fluorine to the adjacent quadrupolar 14N nucleus (I = 1, 99.6% abundance) and is apparent because the highly symmetrical tetrahedral environment of the 14N centre suppresses rapid quadrupolar relaxation that, in less symmetrical environments, leads to loss of coupling fine structure. 19F NMR can also be employed for IC50 measurements (Fig. 2B). We investigated hBBOX inhibition by Mildronate,7 which is an inhibitor and a competitive substrate for hBBOX.9,10 The obtained IC50 value (82 μM) is similar to that obtained by 1H NMR (34 μM) and fluoride release (65 μM) assays.11 The differences likely reflect the different assay conditions used. Thus, 19F NMR assay employing GBBNF (9) as a substrate is useful for in vitro activity studies. An advantage of the GBBNF (9) based system is its ability to monitor specific hBBOX turnover without the interference of any other non-fluorinated components, making it suitable for biologically relevant assay conditions.
To test the turnover of fluorinated analogues in cells we used E. coli BL21 cells producing a prokaryotic BBOX homologue from Pseudomonas sp. AK1 (psBBOX). We observed that GBBNF (9) is converted to CARNF (14) in crude cell lysates, provided that 2OG was added to the reaction mixture, and that the turnover level is dependent on 2OG concentration (Fig. S7, ESI†). The extent of GBBNF (9) turnover is dependent on the amount of psBBOX present in the extracts (Fig. 2C and Fig. S8, ESI†). Thus, 19F NMR can be used to estimate psBBOX activities in cell lysates.
In addition to the use as a label for 19F NMR studies, fluorine is a convenient marker for small molecules in MS based studies. We therefore investigated the metabolism of TMLNF (11) in human embrionic kidney cells (HEK 293T cells) using LC-MS. HEK 293T cells were grown either with or without TML (1) or TMLNF (11) added to the growth media. Cells were harvested, lysed and analysed for the carnitine related metabolites (Fig. 3), using appropriate standards (Fig. S10, ESI†). In none of the samples could TML (1) or TMLNF (11) be detected. In the sample treated with TML (1), elevated levels of GBB (3) were observed, indicating that TML (1) penetrates cell membranes and is converted to GBB (3). Cells treated with TMLNF (11) contained lower amounts of GBB (3) than controls and substantial levels of GBBNF (9), which can only be formed from TMLNF (11), demonstrating that TMLNF (11) is carried through the first enzyme catalysed step of carnitine biosynthesis. All of the samples contained similar levels of carnitine (within error), but no CARNF (14) was observed in the TMLNF (11) treated sample; this result may reflect the differences in affinities of GBB (3) and GBBNF (11) to hBBOX, as observed by in vitro kinetic data (Table 1).
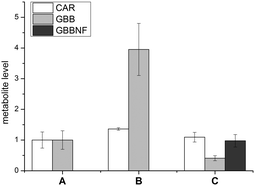 | ||
| Fig. 3 Levels of CAR (4), GBB (3) and GBBNF (9) in HEK 293T cells: A – control, B – treated with TML (1), C – treated with TMLNF (11). Error bars represent standard deviation of n = 3 samples. | ||
In conclusion we have described an efficient procedure for the synthesis of the electrophilic monofluoromethylation reagent17 (7) that enables convenient preparation of fluoromethylated quaternary ammonium derivatives, e.g. trimethyllysine and carnitine related compounds. The utility of these compounds in enzymatic assays was demonstrated employing 19F NMR with recombinant hBBOX and crude cell lysates. LC-MS studies enabled tracking of fluorinated intermediates in human cells. Quaternary ammonium derivatives have widespread pharmaceutical and industrial applications e.g. as antimicrobial agents21 or phase transfer catalysts in organic chemistry.22 They are also a ubiquitous class of metabolites, present in all living organisms.23 In humans and animals N-methylated lysine, arginine and nucleic acid compounds play crucial roles in epigenetic regulation. We hope that the development of appropriate fluorinated small molecules will enable work aimed at understanding the role of methylation in epigenetics.
We thank the Wellcome Trust, Biotechnology and Biological Sciences Research Council, European Union, Dulverton Trust (A.M.R.) and Cancer Research UK (A.T.) for financial support.
Notes and references
- R. J. Wanders, P. Vreken, M. E. den Boer, F. A. Wijburg, A. H. van Gennip and L. Ijlst, J. Inherited Metab. Dis., 1999, 22, 442–487 CrossRef CAS.
- K. Strijbis, F. M. Vaz and B. Distel, IUBMB Life, 2010, 62, 357–362 CAS.
- C. Hoppel, Am. J. Kidney Dis., 2003, 41, S4–S12 CrossRef CAS.
- C. J. Rebouche and H. Seim, Annu. Rev. Nutr., 1998, 18, 39–61 CrossRef CAS PubMed.
- R. P. Hausinger, Crit. Rev. Biochem. Mol. Biol., 2004, 39, 21–68 CrossRef CAS PubMed.
- C. Loenarz and C. J. Schofield, Nat. Chem. Biol., 2008, 4, 152–156 CrossRef CAS PubMed.
- M. Dambrova, E. Liepinsh and I. Kalvinsh, Trends Cardiovasc. Med., 2002, 12, 275–279 CrossRef CAS.
- Y. Hayashi, T. Kirimoto, N. Asaka, M. Nakano, K. Tajima, H. Miyake and N. Matsuura, Eur. J. Pharmacol., 2000, 395, 217–224 CrossRef CAS.
- I. K. H. Leung, T. J. Krojer, G. T. Kochan, L. Henry, F. Von Delft, T. D. W. Claridge, U. Oppermann, M. A. McDonough and C. J. Schofield, Chem. Biol., 2010, 17, 1316–1324 CrossRef CAS PubMed.
- L. Henry, I. K. H. Leung, T. D. W. Claridge and C. J. Schofield, Bioorg. Med. Chem. Lett., 2012, 22, 4975–4978 CrossRef CAS PubMed.
- A. M. Rydzik, I. K. H. Leung, G. T. Kochan, A. Thalhammer, U. Oppermann, T. D. W. Claridge and C. J. Schofield, ChemBioChem, 2012, 13, 1559–1563 CrossRef CAS PubMed.
- S. Purser, P. R. Moore, S. Swallow and V. Gouverneur, Chem. Soc. Rev., 2008, 37, 320–330 RSC.
- A. Vulpetti and C. Dalvit, Drug Discovery Today, 2012, 17, 890–897 CrossRef CAS PubMed.
- C. Dalvit, Prog. Nucl. Magn. Reson. Spectrosc., 2007, 51, 243–271 CrossRef CAS PubMed.
- H. Chen, S. Viel, F. Ziarelli and L. Peng, Chem. Soc. Rev., 2013, 42, 7971–7982 RSC.
- J. Hu, W. Zhang and F. Wang, Chem. Commun., 2009, 7465–7478 RSC.
- G. K. S. Prakash, I. Ledneczki, S. Chacko and G. A. Olah, Org. Lett., 2008, 10, 557–560 CrossRef CAS PubMed.
- G. K. S. Prakash and S. Chacko, Curr. Opin. Drug Discovery Dev., 2008, 11, 793–802 CAS.
- D. P. Matthews, R. A. Persichetti and J. R. McCarthy, Org. Prep. Proced. Int., 1994, 26, 605–608 CrossRef CAS.
- E. Flashman, S. L. Davies, K. K. Yeoh and C. J. Schofield, Biochem. J., 2010, 427, 135–142 CrossRef CAS PubMed.
- M. Tischer, G. Pradel, K. Ohlsen and U. Holzgrabe, ChemMedChem, 2012, 7, 22–31 CrossRef CAS PubMed.
- T. Ooi and K. Maruoka, Angew. Chem., Int. Ed., 2007, 46, 4222–4266 CrossRef CAS PubMed.
- U. Anthoni, C. Christophersen, L. Hougaard and P. H. Nielsen, Comp. Biochem. Physiol., Part B: Biochem. Mol. Biol., 1991, 99, 1–18 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis procedures, assay conditions, and NMR assignments. See DOI: 10.1039/c3cc47581f |
| ‡ Current address: Navarrabiomed-Fundacion Miguel Servet, C/Irunlarrea 3, Complejo Hospitalario de Navarra, 31008 Pamplona, Navarra, Spain. |
| This journal is © The Royal Society of Chemistry 2014 |