Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A high-throughput polymer microarray approach for identifying defined substrates for mesenchymal stem cells†
Cairnan R. E.
Duffy‡
a,
Rong
Zhang‡
b,
Siew-Eng
How
c,
Annamaria
Lilienkampf
d,
Guilhem
Tourniaire
d,
Wei
Hu
b,
Christopher C.
West
e,
Paul
de Sousa
*a and
Mark
Bradley
*d
aCentre for Regenerative Medicine, University of Edinburgh, Chancellor's Building, 49 Little France Crescent, Edinburgh, EH16 4SB, UK. E-mail: paul.desousa@ed.ac.uk
bSchool of Materials Science & Engineering, Changzhou University, Changzhou 213164, Jiangsu, China
cSchool of Science & Technology, University Malaysia Sabah, Jalan UMS, 88400 Kota Kinabalu, Sabah, Malaysia
dSchool of Chemistry, EaStCHEM, University of Edinburgh, Joseph Black Building, West Mains Road, Edinburgh, EH9 3JJ, UK. E-mail: mark.bradley@ed.ac.uk
eMRC Centre for Regenerative Medicine, University of Edinburgh, SCRM Building, 5 Little France Drive, Edinburgh, EH16 4UU, UK
First published on 10th July 2014
Abstract
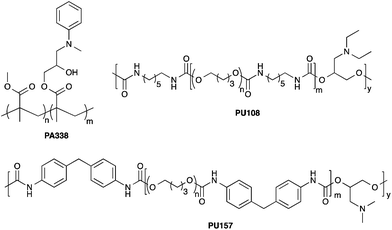
Mesenchymal stem cells (MSCs) hold great promise in regenerative medicine due to their wide multilineage potential as well as their ability to suppress/modulate the immune response. Maintaining these cells in vitro and expanding them on a clinically relevant scale remains a challenge that needs to be addressed to realise their full potential. Current culture methods for MSCs typically rely on animal sourced substrates and often result in a heterogeneous population of cells with varying degrees of differentiation capacity. Here, a high-throughput platform was used to identify synthetic substrates for MSC culture that not only facilitated growth but also maintained the MSC phenotype. Two polymers, PU157 (synthesised from poly(butyleneglycol) and 4,4′-methylenediphenyldiisocyanate with 3-(dimethylamino)-1,2-propanediol as a chain extender) and PA338 (N-methylaniline modified poly(methylmethacrylate-co-glycidylmethacrylate)) were able to maintain the growth and phenotype of human embryonic derived mesenchymal progenitors (hES-MPs) and adipose derived MSCs (ADMSCs) for five and ten passages, respectively. Cell phenotype and multipotency were confirmed by flow cytometry analysis of ten MSC markers and differentiation analysis. These new polymer substrates provide a chemically defined synthetic surface for efficient, long-term MSC culture.
Introduction
Mesenchymal stem cells (MSCs) were first isolated from a bone marrow aspirate by Friedenstein as a ‘rare population’ of fibroblastic cells termed ‘colony forming units’.1 Subsequently, MSCs have been isolated from various sources including umbilical cord blood,2 Wharton's jelly,3 adipose tissue,4 and other vascularised tissues. This wide variety of sources arises from their in vivo niche adjacent to the vasculature.5 MSCs exhibit multilineage potential and can differentiate into osteogenic, adipogenic and chondrogenic lineages6 as well as to other cell types such as smooth muscle7 and neurons.8 The remarkable multilineage differentiation capacity and access has made MSCs the focus of numerous applications in tissue engineering.9 Furthermore, MSCs have been shown to possess the ability to modulate the immune system and have been co-transplanted as a treatment for graft versus host disease.10 A number of studies are currently underway where the immune modulatory property of MSCs is being applied in the treatment of autoimmune diseases such as type 1 diabetes,11 multiple sclerosis,12 and arthritis.13Given the clinical importance of MSCs, their culture is crucial for any downstream application. MSCs grown on tissue culture plastic have been shown to lose both osteogenic and adipogenic potential when compared to gelatin.14,15 On the other hand, the use of animal derived substrates such as gelatin, collagen or fibronectin is problematic due to the presence of a multitude of undefined factors, batch variation, and the potential for pathogen transfer. In order to capitalise on the full clinical potential of MSCs, there is undoubtedly a pressing need for defined synthetic substrates for long-term culture. Since MSC populations contain cells of varying degrees of multipotency,16 the ideal substrate would also only enrich the most multipotent cell type.
Polymer microarrays allow the high-throughput interrogation of defined surfaces for various biomedical applications.17,18 For example, polymer microarrays have enabled the identification of polymers that capture protozoan pathogens,19 resist bacterial adhesion,20 or activate platelets.21 In stem cell research, polymer microarrays have been used to identify polymers that can support human embryonic stem cells (hESCs) in undefined22 and defined media.23 Thermo-responsive hydrogels have also been identified that can maintain MSCs in undefined, and hESCs in defined, media in the absence of enzymatic or chemical passaging.24,25 Besides substrates that promote adhesion and growth, polymer microarrays enable the screening for substrates that can maintain or enhance the MSC phenotype, i.e., cells can be evaluated for multiple MSC markers in a high-throughput manner.
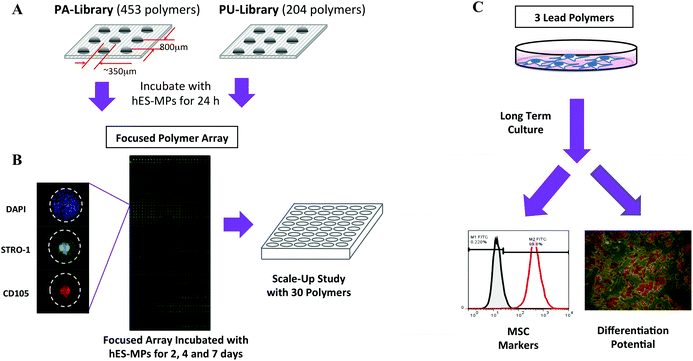
In this study, the high-throughput screening strategy outlined in Fig. 1 was used to identify polymers as MCS substrates. This approach yielded polymers that showed good cellular adhesion and proliferation for both human embryonic derived mesenchymal progenitors (hES-MPs) and adipose derived mesenchymal stem cells (ADMSCs), while maintaining the expression of several MSC markers, as well as their differentiation potential in long-term culture. In the screening process, hES-MPs were utilised as a stable and readily available surrogate for adult derived MSCs.
Materials and methods
Materials
Reagents for cell culture were purchased from Invitrogen unless otherwise stated. Other chemicals were from Sigma Aldrich.Cell culture
All cells were maintained in a humidified incubator at 37 °C with 5% CO2. ADMSCs were derived from the stromal vascular fraction of lipoaspirate4 and grown in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% FCS, bFGF (4 ng mL−1), L-glutamine (100 units mL−1) and pen/strep (100 units mL−1) with media changes every 2–3 days. Ethical approval for the collection of adipose tissue and subsequent research was granted by the South East Scotland Research Ethics Committee (Ref 10/S1103/45 h). hES-MPs (hES-MP™002.5, Cellectis bioresearch) were cultured under the same conditions. For all the experiments cells were recovered by trypsinisation (TrypLE™ Express) for 5 min at 37 °C.Microarray screening
A suspension of hES-MPs (4 mL) was added to the microarray slides (initial screen n = 2, the ‘focused arrays’ n = 3) at 1.2 × 104 cells cm−2, placed in a 4-well rectangular chamber. After incubation, the slides were washed with PBS and the cells fixed with 4% formaldehyde in PBS for 10 min, stained with DAPI (0.1 μg mL−1 in PBS) for 10 min, and washed 3 times with PBS. In the initial screen, polymers with ≥5 cells per feature were selected. The ‘focused arrays’ were assessed for cell count (DAPI), cell viability (CellTracker Green) and immunostained for STRO-1 and CD105.Immunocytochemistry
Cells on the arrays were stained using standard immunocytochemistry protocols. For the ‘focused arrays’, polymer coated coverslips and the gelatin and tissue culture plastic controls, the cells were blocked for 1 h with 10% FCS in PBS. They were then incubated overnight at 4 °C with primary antibodies against STRO-1 (mouse anti-human, Millipore), CD105 (goat anti-human, R&D systems) both at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution, and for the scale-up experiment also with CD271 (rabbit anti-human, Millipore) at 1
100 dilution, and for the scale-up experiment also with CD271 (rabbit anti-human, Millipore) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 dilution. The cells were incubated for 4 h with the appropriate AlexaFluor®-conjugated secondary antibodies; STRO-1 (donkey anti-mouse 488) at 1
50 dilution. The cells were incubated for 4 h with the appropriate AlexaFluor®-conjugated secondary antibodies; STRO-1 (donkey anti-mouse 488) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200, CD105 (donkey anti-goat 555) at 1
200, CD105 (donkey anti-goat 555) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 and CD271 (chicken anti-rabbit) at 1
400 and CD271 (chicken anti-rabbit) at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 dilution. Excess antibody was removed by washing 3 times with PBS. Finally, cells were stained with DAPI (0.1 μg mL−1 in PBS) and washed 3 times with PBS.
100 dilution. Excess antibody was removed by washing 3 times with PBS. Finally, cells were stained with DAPI (0.1 μg mL−1 in PBS) and washed 3 times with PBS.
Coating of polymers on coverslips
Glass coverslips (Menzel-Gläser, Germany), 10 mm (scale-up experiments) and 32 mm diameter (long term culture), were washed with tetrahydrofuran (THF) and spin-coated with 1% (w/v) polymer solutions in THF or NMP for 2 s at 2000 rpm. The coverslips were dried overnight in a vacuum oven (200 mbar) at 40 °C. The coated coverslips were sterilised with UV light for 20 min prior to cell culture.Scale-up culture and analysis on 30 polymers
48-Well plates were coated with 0.5% agarose solution (100 μL per well) and dried overnight at 50 °C. Polymer coated coverslips (n = 4) were placed into the wells and sterilised by UV irradiation for 20 min and washed twice with PBS, the tissue culture plastic controls consisted of 48-well plates, the gelatin controls were 48-well plates coated with a 500 μL 0.1% gelatin solution (in PBS). This was followed by seeding with hES-MPs at 1 × 104 cells per well. Media was changed every 2 days. At day 7, cells were washed with PBS, fixed with 4% formaldehyde, stained with DAPI (1 μg mL−1 in PBS) and triple immunostained for STRO-1, CD105 and CD271. Each well was imaged 9 times on four channels: 360 nm (DAPI), 488 nm (STRO-1), 555 nm (CD105) and 649 nm (CD271). Stained nuclei were counted and the fluorescence intensity measured for each image to give cells mm−2 and intensity mm−2 (the intensity was normalised to the cell number). The background fluorescence for each polymer was calculated using a replicate stained only with the secondary antibody and deducted from sample fluorescence to give a corrected fluorescence intensity value for each polymer.Long-term passaging
hES-MPs and ADMSCs were seeded at 3 × 104 cells per well onto 32 mm diameter polymer coated coverslips (n = 3), which were placed into agarose coated 6-well plates (0.5% agarose, 500 μL per well). For gelatin controls, 6-well tissue culture plates were treated with 0.1% gelatin in PBS (3 mL per well) for 5 min, and the excess solution removed prior to the application of cells and media. Media was changed every 2–3 days and cells were passaged every 5–6 days at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 splitting ratio. At passage 5 (hES-MPs and ADMSCs) and passage 10 (ADMSCs) cells where stained for CD105, STRO-1, CD271, CD90, CD73, CD34, CD14, CD146, CD140b, and HLA-DR, and analysed by flow cytometry.
6 splitting ratio. At passage 5 (hES-MPs and ADMSCs) and passage 10 (ADMSCs) cells where stained for CD105, STRO-1, CD271, CD90, CD73, CD34, CD14, CD146, CD140b, and HLA-DR, and analysed by flow cytometry.
Flow cytometry
For flow cytometry analysis, cells from each replicate (n = 3) were trypsinised. After removing media and washing with PBS, TrypLE™ Express (500 μL) was added to each well for 5 min and subsequently neutralised with media (2 mL). The cells were spun down (5 min, 1000 rpm) and resuspended in 600 μL of FACS-PBS (PBS with 0.1% of BSA and NaN3). To 100 μL of cell suspension 2 μL of the designated fluorescent-conjugated antibody was added; CD105–FITC (Biolegend), STRO-1–APC (Biolegend), CD73–PE (BD Bioscience), HLA-DR–FITC (BD bioscience), CD14–PE (BD Biosciences), CD34–PE (Biolegend) CD140b–APC (BD Biosciences), CD271–PE (BD Bioscience), CD146–FITC (eBiosciences) and CD90–FITC (Biolegend). After incubation for 20 min at 4 °C, 2 mL of FACS-PBS was added to each tube and the tubes centrifuged for 7 min (475 rpm). The PBS was gently decanted ensuring the pellet was not dislodged and the cells resuspended in 250 μL of FACS-PBS and analysed on a FACSCalibur flow cytometer. Flow cytometry histograms were generated using FlowJo.Osteogenic and adipogenic differentiation
After 5 passages on PU157, PU108, PA338, tissue culture plastic and gelatin, ADMSCs were plated at 1.9 × 104 cells per well (n = 3) in standard tissue culture 12-well plates for osteogenic differentiation, and 3.8 × 104 cells per well (n = 3) for adipogenic differentiation. After 24 h the media was changed to StemPro osteogenic or adipogenic differentiation media and the cells were cultured for 28 days with media changes every 3–4 days. After washing with PBS, the cells were fixed in 4% formaldehyde and stained with DAPI (1 μg mL−1). Osteo-induced cells were stained with 2% Alizarin red S solution (pH 4.2) and adipo-induced cells with Oil Red O solution (3 parts of 0.5% Oil Red O in isopropanol to 2 parts of water). Adipocyte differentiation was quantified by comparing the number of DAPI stained nuclei with the number of differentiated cells. Osteogenic differentiation was evaluated by amount of Alizarin red S positive pixels in osteogenic differentiation images with imageJ by setting the image/adjust/colour threshold option. The analyse option (analyse particles) calculated the percentage of pixels above threshold. The best and poorest Alizarin red S stained wells were used as reference points.26,27Image capture
Image capture and analyses from the polymer microarrays and the coated cover slips (on 48-well plates), tissue culture plastic and gelatin wells, was carried out using a Nikon Eclipse 50i microscope with the Pathfinder software (IMSTAR, France). The differentiated cells and the stem cells in the long term cultures on coated cover slips were imaged with a Zeiss Observer microscope.Results and discussion
Screening on polymer microarrays
Two polymer libraries, a ‘PA-library’ (consisting of 453 copolymers synthesised from acrylate, acrylamide and vinyl monomers) and a ‘PU-library’ (204 polyurethanes), were investigated using polymer microarray technology. The libraries were constructed to contain a diverse range of monomers (ESI Fig. S1–S3†) with the aim to produce cell compatible surfaces with a range of physico-chemical properties, such as variation in overall charge, lipophilicity and wettability.The two libraries were printed onto agarose coated glass slides with each polymer printed in quadruplicate (Fig. 1A).28 In the initial screen hES-MPs were incubated for 24 h on the microarrays to enable the identification of polymers that facilitated cell binding, as assessed by counting DAPI stained nuclei on each polymer feature. From this initial screen, 171 polymer candidates were identified (ESI Fig. S4†).
Based on cell binding from the initial screen, a new ‘focused array’ (171 polymers, n = 6) was fabricated (Fig. 1B). The new arrays were incubated with hES-MPs for 2, 4 or 7 days, and analysed for cell binding on each polymer. In order to compensate for any variation in feature size between different polymers, the features were measured and the cell counts given as cells mm−2. Certain polymers showed an increase in cell numbers with increasing culture time indicating that the substrates facilitated both binding and growth (ESI Fig. S5†). In addition, cell viability was assessed at day 7 (ESI Fig S6†).
The phenotype of the hES-MPs was assessed at each time point by staining the arrays for two well-known MSC markers, STRO-1 and CD105 (Fig. 1B, ESI Fig. S7 and S8†).29–32 Staining of hES-MPs with DAPI, CellTracker Green, STRO-1 and CD105 on the ‘focused array’ after 7 days incubation identified several polymers that could maintain growth, showed good cell viability, and conserved/enhanced the expression of the two markers. To select substrates that would favour the multipotent stem cells over less potent progenitors, criteria for selecting candidates for larger scale studies took into consideration marker intensities as well as cell count and viability. The best polymers in terms of number of cells mm−2, cell viability, STRO-1 intensity and CD105 intensity were ranked, with the 30 highest ranking polymers selected for more detailed studies (ESI, Table S1†).
Lead identification
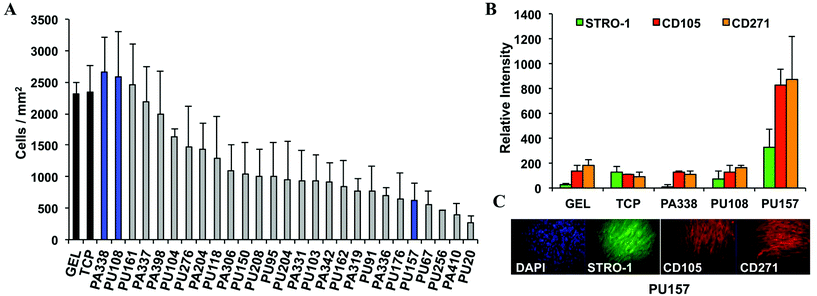
The top 30 polymer candidates were coated onto glass coverslips and incubated with hES-MPs for 7 days using gelatin and tissue culture plastic as controls, followed by staining with DAPI, CD105, STRO-1, and an additional marker CD271. CD271 is considered to be a good marker for multipotent characterisation33 and was included to provide additional criteria for lead polymer selection. Overexpression of CD271 inhibits the differentiation of MSCs into osteogenic, chondrogenic, and myogenic lineages, clearly an important role in maintaining the MSC state.34After 7 days of culture, 5 polymers showed better or similar levels of cell growth compared to gelatin or tissue culture plastic (Fig. 2A). The two top polymers, PA338 and PU108, closely resembled gelatin in terms of marker expression and were chosen as ‘lead’ substrates (Fig. 2 and 3). On larger surfaces, PU157 showed relatively modest cell growth but considerably enhanced STRO-1, CD105, and CD271 intensities (Fig. 2B), and was therefore taken forward as the third ‘lead’ polymer for long-term culture and characterisation with hES-MPs and ADMCSs.
Long-term culture and characterisation of hES-MPs
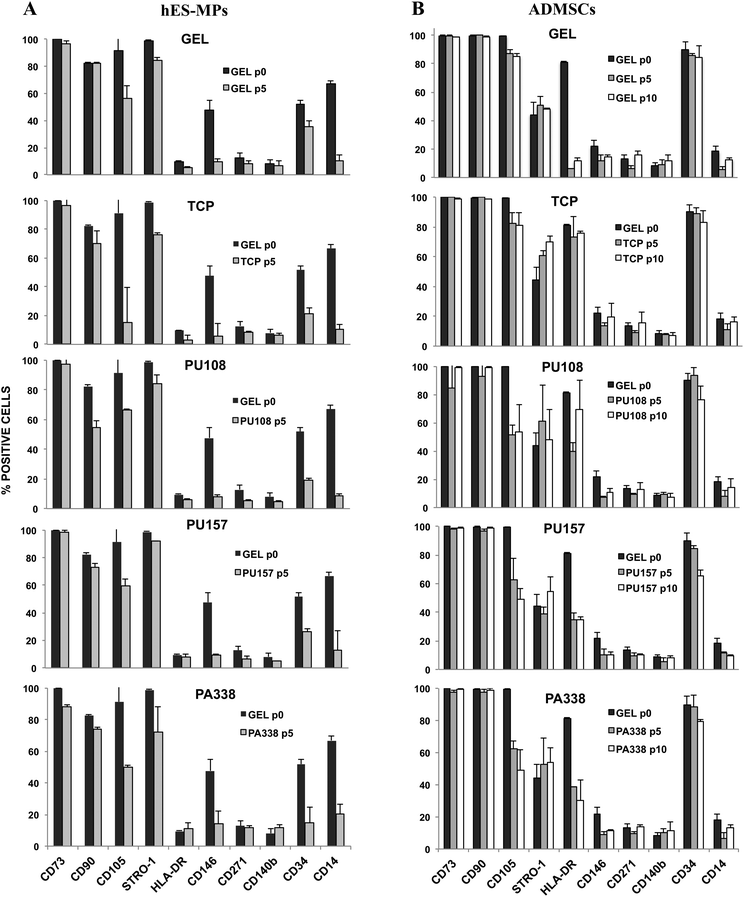
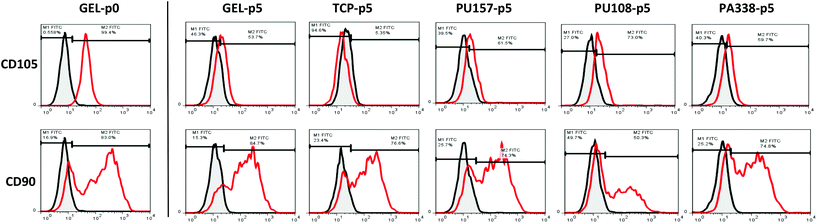
For extended culture on the lead polymers PU157, PU108 and PA338 (Fig. 3), a variety of MSC markers were used to fully probe the suitability of the substrates for long-term MSC culture. In addition to CD105, STRO-1 and CD271, seven additional markers were used. These included positive markers CD90 and CD73, and negative markers CD14 and CD34, chosen because they are used as part of the minimal criteria for defining MSCs.29 However, CD34 positive populations are present in MSCs obtained from adipose tissue35 and MSC marker expression in general has been shown to depend on the source.36 CD146 and CD140b were selected as these are considered to be in vivo MSC markers,5 whereas HLA-DR is a standard negative marker for MSCs as well as an indicator of the immune modulatory state of MSCs (HLA-DR is up regulated when MSCs are primed into their immune modulatory state).37Prior to the long-term culture on the ‘lead’ polymers, hES-MPs were characterised at ‘passage 0’, i.e., the point of removal from gelatin, for the expression of CD105, STRO-1, CD271, CD90, CD73, CD34, CD14, CD146, CD140b, and HLA-DR (Fig. 4A). Here, cells were positive for CD73 and STRO-1 (100% and 99%, respectively), positive for CD105 and CD90 (91% and 83%, respectively), and moderately positive for CD34, CD146 and CD14 (CD14 was subsequently down regulated). hES-MPs grown on the ‘lead’ polymers were assessed with these 10 markers at passage 5 to monitor marker conservation or changes in the marker expression levels.
Flow cytometry analysis at passage 5 showed that hES-MPs cultured on PU108, PU157 and PA338 expressed similar levels of the markers to cells grown on gelatin (Fig. 4A, ESI Fig. S9†). However CD105, which is considered one of the essential markers for MSCs, expression was reduced on cells grown on all ‘lead’ polymers, but levels were comparable with gelatin. Interestingly, hES-MPs are currently derived through serial passaging of hESCs on gelatin.38 Notably, CD105 was lost at passage 5 on tissue culture plastic (Fig. 4A and 5).
The marker CD90, which is bimodal in hES-MPs,38,39 showed a substantial increase in the negative fraction with culture on PA108 when compared to other substrates (Fig. 5). However, hES-MPs cultured on PU157 and PA338 maintained CD90 at levels similar to tissue culture plastic and gelatin (73%, 74% 70% and 82% of positive cells for PU157, PA338, tissue culture plastic and gelatin, respectively). Decreasing levels of CD90 have been associated with the loss of the immune modulatory properties of MSCs.40 The comparatively low expression of this marker in hES-MPs in general, compared to adult derived MSCs, may explain the fact why hES-MPs do not possess the property of immune modulation associated with adult derived MSCs.39 The ability of PU157 and PA338 to maintain marker expression (including CD105 and CD90) similar to gelatin indicates that they may act as chemically defined alternatives to gelatin for serial passaging of hESCs to produce hES-MPs.
Long-term culture and characterisation of ADMSCs
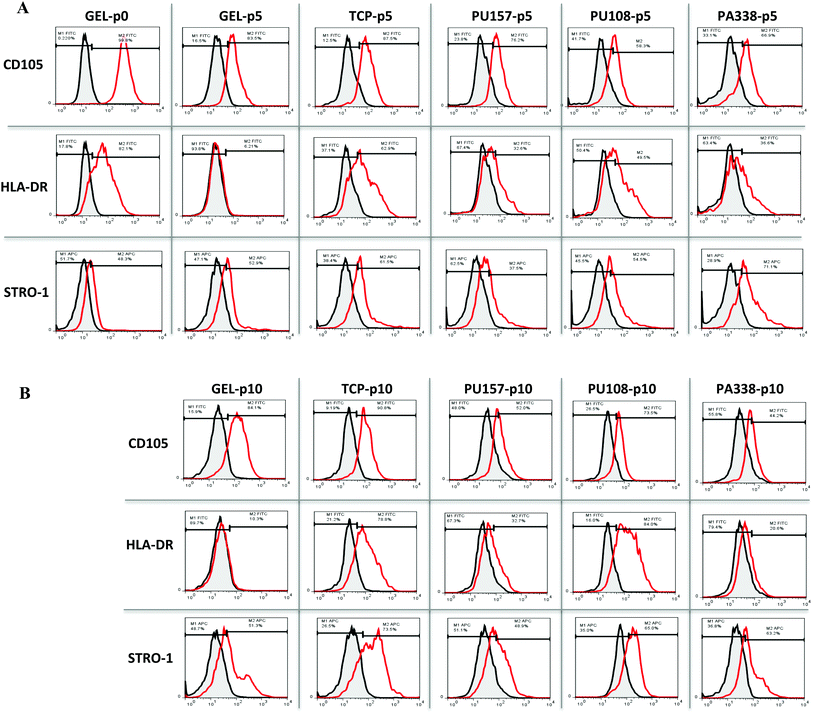
ADMSCs grown on PU157, PU108 and PA338, closely resembled the gelatin control with most of the MSC markers at passage 5 (Fig. 4B, ESI Fig. S10†). At passage 5, ADMSCs grown on tissue culture plastic, gelatin, PU157, PU108 and PA338 were positive for CD105, although a loss in marker intensity was observed with all substrates when compared to ‘passage 0’ (Fig. 6A).A further reduction in the CD105 marker intensity was observed by passage 10 with PU157, PU108 and PA338 (49%, 54% and 49% of positive cells, respectively), although with cells grown on gelatin and tissue culture plastic the levels were maintained (85% and 81%, respectively) (Fig. 4B, 6B and ESI S11†).
Surprisingly, the ADMSCs clearly expressed HLA-DR at ‘passage 0’, however, on gelatin this marker was subsequently down regulated by passage 5. The cells grown on tissue culture plastic and PU108 continued to be positive for this marker up to passage 10 (76% and 69%, respectively). However, PU157 and PA338 showed reduced expression of HLA-DR by passage 5 (35% and 38% positive, respectively), with low levels maintained by passage 10 (Fig. 4B, 6A and B). The expression of HLA-DR at ‘passage 0’ may indicate an inflammatory environment from where the ADMSCs were sourced (IFN-γ upregulates this receptor in MSCs). Prior to their application in therapy, MSCs are treated with IFN-γ to prime them into the immune modulatory state, corresponding to upregulation of the HLA-DR receptor.41 Whether upregulation of the receptor through substrate interaction is an indicator that the substrate could prime the cells into the immune modulatory state is an interesting question.
At the start (‘passage 0’), ADMSCs showed a 44% positive population for STRO-1. By passage 5 there was a distinct positive shift in STRO-1 intensity with tissue culture plastic, which became even more pronounced by passage 10 (Fig. 6A and B).42 High expression of STRO-1 has been associated with an increase in adipogenic and osteogenic markers PPAR-G and RUNX2, respectively, and upregulation of either of these markers could reduce the ability of the cells to differentiate down the other lineage.43
Differentiation of ADMSCs
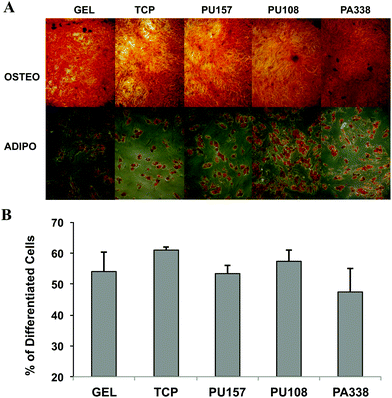
At passage 5, ADMSCs grown on PU157, PU108, PA338, gelatin, and tissue culture plastic were differentiated down the adipogenic and osteogenic lineages to evaluate if they retained their differentiation potential. Histological staining of the cells with Alizarin Red and Oil red O showed that the cells had differentiated down the osteogenic and adipogenic lineages, respectively, proving that the cells had clearly maintained their multi-lineage potential (Fig. 7A). Also, the ADMSCs grown on various substrates showed no major difference in the proportion of cells capable of differentiating down the adipogenic lineage (Fig. 7B). Image analysis of the Alizarin Red S stained cells showed that PA338, gelatin and tissue culture plastic had comparable levels of staining, whereas PU157 and PU108 showed somewhat lower levels of staining (ESI Fig. S12†).26 A similar ability of ADMSCs to differentiate at passage 5, regardless of which substrate they were grown on, suggests that the differences seen in marker expression at this passage has no significant effect on their differentiation capacity.Polymer analysis
To investigate if the chemical composition of the polymer surface can be correlated to cell binding/growth and stem cell maintenance, the monomers present in the top candidates from the scale up studies were analysed. The top ten performing polymers from the PA-library showed some monomers to be highly preferred over others (ESI Fig. S13 and S14†). Methyl methacrylate was present in 8 out of the 10 top polymers, and 9 polymers contained a monomer with a tertiary amine, with aliphatic amines (6 out of 9) rendering the surface positively charged at physiological pH. In addition, the presence of an aryl ring seemed beneficial (PA338 is a copolymer of methyl methacrylate and N-methylaniline functionalised glycidyl methacrylate.Analysis of the top 10 polyurethanes from the scale-up experiments showed 4,4′-methylenebis(phenylisocyanate) and poly(butyleneglycol) were present in 8 and 5 polymers, respectively, both of which were present in the lead PU157 along with 3-dimethylamino-1,2-propanediol (ESI Fig. S15 and S16†). Certain monomers, e.g. poly(ethylene glycol) (PEG), seemed disadvantageous in terms of binding and promotion of growth.
Adhesion of a cell on the surface is a complex process depending on a combination of cell–protein and polymer–protein interactions, and the physico-chemical properties of the surface. In stem cell culture, the ability of the substrate to maintain the cells in their stem cell state adds another level of complexity to the material–cell interactions. Therefore, although some common features could be found on the composition of non-binding versus binding polymers, drawing direct ‘structure–function relationships’ for polymer surfaces and stem cell maintenance remains a challenge, further highlighting the advantages of a high-throughput approach for the discovery of new substrates.
Surface characterisation
Of the lead polymers, PA338 and PU157 had a water contact angle of 81° and 70°, respectively (representative control polymers, which showed no or poor cell binding or growth, had contact angles of 27–67°) (ESI Table S2†). However, PU108 had a relatively small water contact angle of 46°, suggesting that water contact angle or the surface energy is a poor indicator of stem cell adhesion/growth in the polymer libraries investigated.To investigate global protein binding on the polymer surfaces, polymer coated cover slips were treated with culture media (supplemented with 10% FCS) for 24 h, and adhered proteins analysed by gel electrophoresis (ESI Fig. S17†). With the exception of PU108, which showed slightly different protein binding pattern, all the studied polymers, including poor cell binder controls, showed protein binding similar to gelatin and tissue culture plastic, thus suggesting that the surface chemistry plays an important role in the cell adhesion and growth, although subtle changes in binding of low abundance proteins cannot be ruled out.
Conclusions
By using hES-MPs in a ‘three-step’ screening strategy, which consisted of two iterative rounds of polymer microarray screens followed by a large scale 30 polymer study, a number of polymers were identified that could potentially support long-term MSC culture. Based on cell growth and the intensity of markers CD105, STRO-1 and CD271, two polyurethanes (PU157 and PU108) and one polyacrylate (PA338), were identified as ‘lead’ substrates. hES-MPs and ADMSCs were cultured on PU157, PU108 and PA338 for 5 and 10 passages, respectively, and analysed for 10 MSC markers. All substrates showed similar growth rates for both cell types. After 5 passages on the polymer substrates, both cell types expressed similar marker levels as control cells grown on gelatin, whereas on tissue culture plastic the marker CD105 was lost (hES-MPs). hES-MPs cultured on PU108 showed an increase in the negative fraction for CD90 and ADMSCs cultured on PU108 continued to express HLA-DR at passage 10. Irrespective of some variation in the marker expression between ADMSCs grown on different substrates, the cells were successfully differentiated into osteoblasts and adipocytes at passage 5. Overall, PU157 and PA338 showed the most similarity to gelatin and efficiently maintained growth and conserved phenotype over multiple passages for both hES-MPs and ADMSCs. As a potential alternative to gelatin, PU157 and PA338 offer chemically defined synthetic polymer substrates for efficient long term MSC culture.Acknowledgements
This work was supported by EU FP7 funding (project 223410, BEST Stem Cells). We thank Kay Samuel for help with the flow cytometry experiments, Drs Anne Hansen and Mei Wu for assistance with the microarray fabrication, Dr Steve Pells for input on figure preparation, and Drs Chris Armit and Marieke Hoeve for helpful discussions.Notes and references
- A. J. Friedenstein, U. F. Deriglasova, N. N. Kulagina, A. F. Panasuk, S. F. Rudakowa, E. A. Luriá and I. A. Ruadkowet, Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method, Exp. Hematol., 1974, 2, 83–92 CAS.
- M. W. Lee, J. Choi, M. S. Yang, Y. J. Moon, J. S. Park, H. C. Kim and J. K. Young, Mesenchymal stem cells from cryopreserved human umbilical cord blood, Biochem. Biophys. Res. Commun., 2004, 16, 273–278 CrossRef PubMed.
- H. Wang, S. Hung, S. Peng, C. Huang, H. Wei, Y. Guo, F. Yu-Show, C. Mei-Lai and C. Chin-Chang, Mesenchymal stem cells in the Wharton's jelly of the human umbilical cord, Stem Cells, 2004, 22, 1330–1337 CrossRef PubMed.
- P. A. Zuk, M. Zhu, H. Mizuno, J. I. Huang, W. J. Futrell, A. J. Katz, P. Benhaim, H. P. Lorenz and M. H. Hedrick, Multilineage cells from human adipose tissue: implications for cell-based therapies, Tissue Eng., 2001, 7, 211–226 CrossRef CAS PubMed.
- M. Crisan, S. Yap, L. Casteilla, C. W. Chen, M. Corselli, T. S. Park, G. Andriolo, B. Sun, B. Zheng, L. Zhang, C. Norotte, P. N. Teng, J. Traas, R. Schugar, B. M. Deasy, S. Badylak, H. J. Buhring, J. P. Giacobino, L. Lazzari, J. Huard and B. Péault, A perivascular origin for mesenchymal stem cells in multiple human organs, Cell Stem Cell, 2008, 3, 301–313 CrossRef CAS PubMed.
- M. F. Pittenger, A. M. Mackay, S. C. Beck, R. K. Jaiswal, R. Douglas, J. D. Mosca, M. A. Moorman, D. W. Simonetti, S. Craig and D. R. Marshak, Multilineage potential of adult human mesenchymal stem cells, Science, 1999, 284, 143–147 CrossRef CAS.
- S. Wakitani, T. Saito and A. I. Caplan, Myogenic cells derived from rat bone marrow mesenchymal stem cells exposed to 5-azacytidine, Muscle Nerve, 1995, 18, 1417–1426 CrossRef CAS PubMed.
- P. Tropel, N. Platet, J. C. Platel, D. Noël, M. Albrieux, A. L. Benabid and F. Berger, Functional neuronal differentiation of bone marrow-derived mesenchymal stem cells, Stem Cells, 2006, 24, 2868–2876 CrossRef CAS PubMed.
- L. Wang, L. Ott, K. Seshareddy, M. L. Weiss and M. S. Detamore, Musculoskeletal tissue engineering with human umbilical cord mesenchymal stromal cells, Regen. Med., 2011, 6, 95–109 CrossRef CAS PubMed.
- H. M. Lazarus, O. N. Koc, S. M. Devine, P. Curtin, R. T. Maziarz, H. K. Holland, E. J. Shpall, P. McCarthy, K. Atkinson, B. W. Cooper, S. L. Gerson, M. J. Laughlin, F. R. Loberiza Jr., A. B. Moseley and A. Bacigalupo, Cotransplantation of HLA-identical sibling culture-expanded mesenchymal stem cells and hematopoietic stem cells in hematologic malignancy patients, Biol. Blood Marrow Transplant., 2005, 11, 389–398 CrossRef PubMed.
- P. Fiorina, M. Jurewicz, A. Augello, A. Vergani, S. Dada, S. La Rosa, M. Selig, J. Godwin, K. Law, C. Placidi, R. N. Smith, C. Capella, S. Rodig, C. N. Adra, M. Atkinson, M. H. Sayegh and R. Abdi, Immunomodulatory function of bone marrow-derived mesenchymal stem cells in experimental autoimmune type 1 diabetes, J. Immunol., 2009, 183, 993–1004 CrossRef CAS PubMed.
- J. Cohen, Mesenchymal stem cell transplantation in multiple sclerosis, J. Neurol. Sci., 2013, 333, 43–49 CrossRef CAS PubMed.
- A. Augello, R. Tasso, S. M. Negrini, R. Cancedda and G. Pennesi, Cell therapy using allogeneic bone marrow mesenchymal stem cells prevents tissue damage in collagen-induced arthritis, Arthritis Rheum., 2007, 56, 1175–1186 CrossRef CAS PubMed.
- J. R. Mauney, D. L. Kaplan and V. Volloch, Matrix-mediated retention of osteogenic differentiation potential by human adult bone marrow-derived mesenchymal stem cells during ex vivo expansion, Biomaterials, 2004, 25, 3233–3243 CrossRef CAS PubMed.
- J. R. Mauney, V. Volloch and D. L. Kaplan, Matrix-mediated retention of adipogenic differentiation potential by human adult bone marrow-derived mesenchymal stem cells during ex vivo expansion, Biomaterials, 2005, 26, 6167–6175 CrossRef CAS PubMed.
- M. Pevsner-Fischer, S. Levin and D. Zipori, The origins of mesenchymal stromal cell heterogeneity, Stem Cell Rev., 2011, 7, 560–568 CrossRef CAS PubMed.
- H. Mizomoto, The synthesis and screening of polymer libraries using a high throughput approach, PhD Thesis, University of Southampton, 2004 Search PubMed.
- D. G. Anderson, D. Putnam, E. B. Lavik, T. A. Mahmood and R. Langer, Biomaterial microarrays: rapid, microscale screening of polymer-cell interaction, Biomaterials, 2005, 26, 4892–4897 CrossRef CAS PubMed.
- M. Wu, H. Bridle and M. Bradley, Targeting Cryptosporidium parvum capture, Water Res., 2012, 46, 1715–1722 CrossRef CAS PubMed.
- S. Pernagallo, M. Wu, M. P. Gallagher and M. Bradley, Colonising new frontiers-microarrays reveal biofilm modulating polymers, J. Mater. Chem., 2011, 21, 96–101 RSC.
- A. Hansen, L. McMillan, A. Morrison, J. Petrik and M. Bradley, Polymers for the rapid and effective activation and aggregation of platelets, Biomaterials, 2011, 32, 7034–7041 CrossRef CAS PubMed.
- D. G. Anderson, S. Levenberg and R. Langer, Nanoliter-scale synthesis of arrayed biomaterials and application to human embryonic stem cells, Nat. Biotechnol., 2004, 22, 863–866 CrossRef CAS PubMed.
- A. Hansen, H. K. Mjoseng, R. Zhang, M. Kalloudis, V. Koutsos, P. A. de Sousa and M. Bradley, High-Density Polymer Microarrays: Identifying Synthetic Polymers that Control Human Embryonic Stem Cell Growth, Adv. Healthcare Mater., 2013, 3, 848–853 CrossRef PubMed.
- C. R. E. Duffy, R. Zhang, S.-E. How, A. Lilienkampf, P. A. de Sousa and M. Bradley, Long term mesenchymal stem cell culture on a defined synthetic substrate with enzyme free passaging, Biomaterials, 2014, 35, 5998–6005 CrossRef CAS PubMed.
- R. Zhang, H. Mjoseng, M. Hoeve, N. Bauer, S. Pells, R. Besseling, S. Velugotla, G. Tourniaire, R. Kishen, Y. Tsenkina, C. Armit, C. R. E. Duffy, M. Helfen, F. Edenhofer, P. A. de Sousa and M. Bradley, A thermoresponsive and chemically defined hydrogel for long-term culture of human embryonic stem cells, Nat. Commun., 2013, 4, 1335–1342 CrossRef PubMed.
- A. Mehlem, C. E. Hagberg, L. Muhl, U. Eriksson and A. Falkevall, Imaging of neutral lipids by oil red O for analyzing the metabolic status in health and disease, Nat. Protocols, 2013, 8, 1149–1154 CAS.
- M. T. Yang, J. Fu, Y.-K. Wang, R. A. Desai and C. S. Chen, Assaying stem cell mechanobiology on microfabricated elastomeric substrates with geometrically modulated rigidity, Nat. Protocols, 2011, 6, 187–213 CAS.
- S. Pernagallo, J. J. Diaz-Mochon and M. Bradley, A cooperative polymer-DNA microarray approach to biomaterial investigation, Lab Chip, 2009, 3, 397–403 RSC.
- M. Dominici, K. Le Blanc, I. Mueller, I. Slaper-Cortenbach, F. Marini, D. Krause, R. Deans, A. Keating, D. Prockop and E. Horwitz, Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement, Cytotherapy, 2006, 8, 315–317 CrossRef CAS PubMed.
- J. E. Dennis, J. P. Carbillet, A. I. Caplan and P. Charbord, The STRO-1+ marrow cell population is multipotential, Cells Tissues Organs, 2002, 170, 73–82 CrossRef PubMed.
- P. J. Simmons and B. Torok-Storb, Identification of stromal cell precursors in human bone marrow by a novel monoclonal antibody, STRO-1, Blood, 1991, 78, 55–62 CAS.
- S. Gronthos, D. M. Franklin, H. A. Leddy, R. Storms and J. M. Gimble, Characterization of surface protein expression on human adipose tissue-derived stromal cells, J. Cell Physiol., 2001, 189, 54–63 CrossRef CAS PubMed.
- H. J. Buhring, V. L. Battula, S. Treml, B. Schewe, L. Kanz and W. Vogel, Novel markers for the prospective isolation of human MSCs, Ann. N. Y. Acad. Sci., 2007, 1106, 262–271 CrossRef PubMed.
- Y. Mikami, Y. Ishii, N. Watanabe, T. Shirakawa, S. Suzuki, S. Irie, K. Isokawa and M. J. Honda, CD271/P75ntr Inhibits the differentiation of mesenchymal stem cells into osteogenic, adipogenic, chondrogenic and myogenic lineages, Stem Cells Dev., 2011, 20, 901–913 CrossRef CAS PubMed.
- M. Corselli, C. W. Chen, B. Sun, S. Yap, J. P. Rubin and B. Péault, The tunica adventitia of human arteries and veins as a source of mesenchymal stem cells, Stem Cells Dev., 2012, 21, 1299–1308 CrossRef CAS PubMed.
- A. A. Nery, I. C. Nascimento, T. Glaser, V. Bassaneze, J. E. Krieger and H. Ulrich, Human mesenchymal stem cells: from immunophenotyping by flow cytometry to clinical applications, Cytometry A, 2013, 83, 48–61 CrossRef PubMed.
- N. Singer and A. Caplan, Mesenchymal Stem Cells: Mechanisms of Inflammation, Annu. Rev. Pathol.: Mech. Dis., 2011, 6, 457–478 CrossRef CAS PubMed.
- C. Karlsson, K. Emanuelsson, F. Wesserg, K. Kajic, M. Z. M. Axell, P. S. Eriksson, A. Lindahl, J. Hyllner and R. Strehl, Human embryonic stem cell-derived mesenchymal progenitors-potential in regenerative medicine, Stem Cell Res., 2009, 3, 39–50 CrossRef PubMed.
- O. Li, A. Tormin, B. Sundberg, J. Hyllner, K. Le Blanc and S. Scheding, Human embryonic stem cell-derived mesenchymal stroma cells (hES-MSCs) engraft in vivo and support hematopoiesis without suppressing immune function: implications for off-the shelf ES-MSC therapies, PLoS One, 2013, 8, e55319 CAS.
- D. Campioni, R. Rizzo, M. Stignani, L. Melchiorri, L. Ferrari, S. Moretti, A. Russo, G. P. Bagnara, L. Bonsi, F. Alviano, G. Lanzoni, A. Cuneo, O. R. Baricordi and F. Lanza, A decreased positivity for CD90 on human mesenchymal stromal cells (MSCs) is associated with a loss of immunosuppressive activity by MSCs, Cytometry, Part B, 2009, 76, 225–230 CrossRef PubMed.
- W. K. Chan, A. S. Lau, J. C. Li, H. K. Law, Y. L. Lau and G. C. Chan, MHC expression kinetics and immunogenicity of mesenchymal stromal cells after short-term IFN-γ challenge, Exp. Hematol., 2008, 36, 1545–1555 CrossRef CAS PubMed.
- R. H. Lee, M. J. Seo, A. A. Pulin, C. A. Gregory, J. Ylostalo and D. J. Prockop, The CD34-like protein PODXL and α6-integrin (CD49f) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice, Blood, 2009, 113, 816–826 CrossRef CAS PubMed.
- S. Isenmann, A. Arthur, C. W. A. Zennettino, J. L. Turner, S. Shi, C. A. Glackin and S. Grunthos, TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment, Stem Cells, 2009, 27, 2457–2468 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Microarray fabrication; polymer synthesis and monomer structures; cell growth, cell viability and MSC marker intensity for top 30 polymers on the ‘focused array’; selection criteria for scale up studies; flow cytometry histograms for all the 10 MSC markers in long term culture; monomer analysis of binding polymers; polymer surface analysis. See DOI: 10.1039/c4bm00112e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2014 |