Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Chemically diverse polymer microarrays and high throughput surface characterisation: a method for discovery of materials for stem cell culture†
A. D.
Celiz
a,
J. G. W.
Smith
b,
A. K.
Patel
b,
R.
Langer
c,
D. G.
Anderson
c,
D. A.
Barrett
d,
L. E.
Young
b,
M. C.
Davies
a,
C.
Denning
b and
M. R.
Alexander
*a
aLaboratory of Biophysics and Surface Analysis, School of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, UK. E-mail: morgan.alexander@nottingham.ac.uk
bWolfson Centre for Stem Cells, Tissue Engineering and Modelling Centre for Biomolecular Sciences, University of Nottingham, Nottingham, NG7 2RD, UK
cDepartment of Chemical Engineering, Harvard-MIT Division of Health Sciences and Technology, David H. Koch Institute for Integrative Cancer Research, Massachusetts Institute of Technology, 500 Main Street, Cambridge, MA 02139, USA
dSchool of Pharmacy, University of Nottingham, Nottingham, NG7 2RD, UK
First published on 12th May 2014
Abstract
Materials discovery provides the opportunity to identify novel materials that are tailored to complex biological environments by using combinatorial mixing of monomers to form large libraries of polymers as micro arrays. The materials discovery approach is predicated on the use of the largest chemical diversity possible, yet previous studies into human pluripotent stem cell (hPSC) response to polymer microarrays have been limited to 20 or so different monomer identities in each study. Here we show that it is possible to print and assess cell adhesion of 141 different monomers in a microarray format. This provides access to the largest chemical space to date, allowing us to meet the regenerative medicine challenge to provide scalable synthetic culture ware. This study identifies new materials suitable for hPSC expansion that could not have been predicted from previous knowledge of cell-material interactions.
Introduction
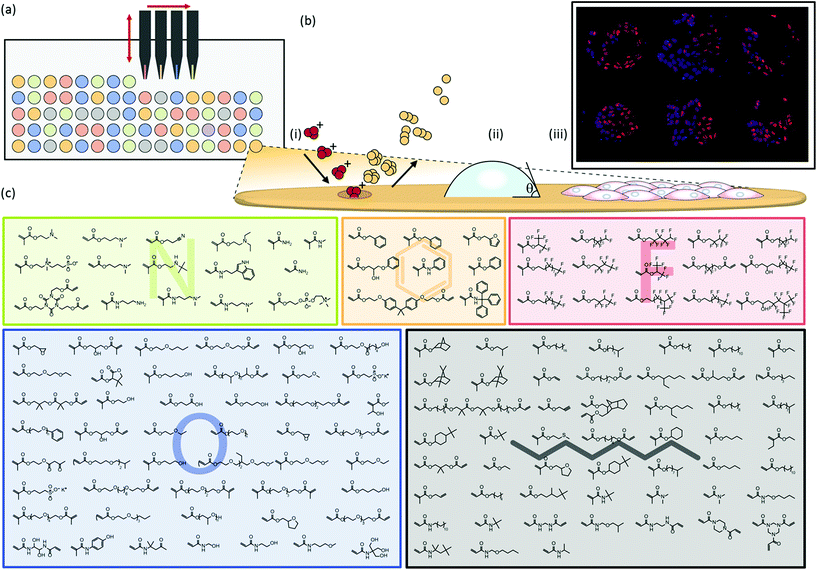
The materials community has been challenged by biologists and clinicians to provide materials which are suitable for both in vitro and in vivo applications, ranging from coronary drug-eluting stents to defined growth substrates for stem cell culture. To discover new synthetic materials, high throughput methodologies have been employed to screen a large chemical combinatorial space. Polymer microarrays are a powerful approach on which thousands of materials can be investigated on a single slide for specific biological interactions.4–6 Such microarrays have been successfully used to discover new materials for human pluripotent stem cell (hPSC) culture using in situ polymerisation methods to enable the rapid evolution of large combinatorial material libraries from one slide generation to the next to screen for different biological interactions.1–3 Array manufacture can be performed by ink-jet or contact printing monomer solutions onto a low fouling poly(2-hydroxyethyl methacrylate) (pHEMA) coated substrate and polymerised in situ via UV irradiation.7 This approach has been used to produce an array of 496 unique copolymers by mixing 22 (meth)acrylate monomers to screen for materials that support the clonal growth of hPSCs from very low initial cell seeding densities.2The microarray format is amenable to high throughput surface characterisation (HT-SC) which can be carried out in parallel to biological assays.8,9 Analysis of the actual surface chemistry rather than presumption of its composition using the monomer identity is essential to ensure the controlling moieties are identified since both surface segregation of certain monomers and contamination can occur in spots.8 Identification of structure–property relationships between material surface chemistry and biological response can be achieved using multivariate analysis (MVA).10,11 In this study a 141 (meth)acrylate and (meth)acrylamide monomer library was explored as the most chemically diverse polymer microarray to date utilising >90% of photo curable monomers that are readily commercially available. The attachment of HUES7 human embryonic stem cells in mitotically inactivated mouse embryonic fibroblast-conditioned medium (MEF-CM) is screened to illustrate the power of MVA with this high throughput screening tool. This greatly expands the chemical space for the discovery of new materials for hPSC culture from our previous library of 22 monomers, 10 of which are included in the current screen alongside 131 (meth)acrylate and (meth)acrylamide monomers, the majority of which have not been previously explored as growth substrates for hPSC culture.
hPSCs hold promise towards application in regenerative medicine and drug development as well as providing new in vitro models of genetic disease due to their long term self-renewal and inherent ability to differentiate into all three germ layers.12 However, for these applications to be realised, scalable culture systems capable of generating the necessary numbers of cells are needed.13,14 Currently, the most widely used growth substrates for hPSC culture are mouse embryonic fibroblast (MEF) feeder layers or Matrigel™ (an undefined extracellular matrix (ECM) protein mixture harvested from Engelbreth–Holm–Swarm mouse sarcoma cells).15 To improve scalability of growth substrates and avoid the problems associated with complex biologically derived material, fully synthetic polymer-based growth substrates have been under investigation.1–3,16–20 However, commercial synthetic alternatives to Matrigel such as Synthemax, a peptide–acrylate conjugate coating containing a cell adhesion motif derived from vitronectin, have not proved cost-effective for large scale hPSC culture. A number of groups are therefore engaged in the search for scalable and cost-effective synthetic polymer substrates on which to expand stem cells.2,3,14,16–20 However, an effective and scalable polymeric substrate for hPSC expansion has yet to be found.21 Here we show the utility of employing a chemically diverse, high throughput screening tool combined with a chemometrics approach to discover new synthetic substrates for hPSC expansion.
Experimental
Polymer microarray synthesis
Polymer microarrays were synthesised using methods previously described.1,22 Briefly, polymer microarrays were formed using a XYZ3200 dispensing station (Biodot) and metal pins (946MP3B, Arrayit). The printing conditions were O2 < 2000 ppm, 25 °C, and 35% humidity. Polymerisation solution was composed of monomer (50% v/v) in dimethylformamide with photoinitiator 2,2-dimethoxy-2-phenyl acetophenone (1% w/v). Six replicates were printed on each slide. Monomers were purchased from Aldrich, Scientific Polymers and Polysciences and printed onto epoxy-coated slides (Xenopore) dip-coated with pHEMA (4% w/v, Sigma) in ethanol (95% v/v in water).Sterilisation and pre-treatment of arrays
Top and bottom array surfaces were sterilised by exposure to UV light for 15 minutes each. Arrays were washed with sterile PBS then surface pre-treated by incubation in Mouse Embryonic Fibroblast-Conditioned Media (MEF-CM) containing 20% Fetal Bovine Serum (FBS) (Invitrogen, Paisley, UK) for 1 hour at 37 °C with 5% CO2.23,24Human embryonic stem cell (hESC) culture and adhesion to arrays
The hESC line HUES7, was cultured on a Matrigel™ (BD Biosciences, Oxford, UK) coated substrate in MEF-CM, for no higher than passage 35, prior to seeding on polymer microarrays.25 Passaging of cells was achieved by incubation with 0.05% trypsin (Invitrogen, Paisley, UK) for 1 min at 37 °C, with tapping of the flasks to dissociate cells. 1 × 106 HUES7 cells were seeded in MEF-CM on each array and incubated at 37 °C with 5% CO2 for 24 hours to allow cell adhesion.Immunostaining
Adherent cells were fixed in 4% paraformaldehyde (Sigma–Aldrich, Poole, UK) and permeabilised with 0.1% Triton-X 100 (Sigma–Aldrich, Poole, UK). Non-specific binding was blocked with 8% goat serum (Sigma–Aldrich, Poole, UK) for 1 hour at room temperature. Samples were incubated with diluted mouse primary OCT4 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200; Santa Cruz Biotech, Heidelberg, Germany) overnight at room temperature. Cy3-conjugated goat anti-mouse IgG + IgM secondary antibody (1
200; Santa Cruz Biotech, Heidelberg, Germany) overnight at room temperature. Cy3-conjugated goat anti-mouse IgG + IgM secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250; Jackson Immuno Research, Inc., West Grove, PA) was applied for 1 hour at room temperature. Samples were incubated with 4′,6-diamidino-2-phenylindole (DAPI) (1
250; Jackson Immuno Research, Inc., West Grove, PA) was applied for 1 hour at room temperature. Samples were incubated with 4′,6-diamidino-2-phenylindole (DAPI) (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000; Sigma–Aldrich, Poole, UK) for 10 minutes at room temperature and then mounted in Vectorshield mounting medium (Vector Labs, Peterborough, UK). Arrays were imaged using an automated fluorescence microscope (IMSTAR) and stem cell attachment determined using CellProfiler cell image analysis software to identify the number of positively stained nuclei (http://www.cellprofiler.org/) from four array replicates.
1000; Sigma–Aldrich, Poole, UK) for 10 minutes at room temperature and then mounted in Vectorshield mounting medium (Vector Labs, Peterborough, UK). Arrays were imaged using an automated fluorescence microscope (IMSTAR) and stem cell attachment determined using CellProfiler cell image analysis software to identify the number of positively stained nuclei (http://www.cellprofiler.org/) from four array replicates.
Time-of-flight secondary-ion mass spectrometry
Measurements were conducted using a ToF-SIMS 4 (IONTOF GmbH) instrument operated using a 25 kV Bi3+ primary ion source exhibiting a pulsed target current of ∼1 pA. Samples were scanned at a pixel density of 100 pixels per mm, with 8 shots per pixel over a given area. An ion dose of 2.45 × 1011 ions per cm2 was applied to each sample area ensuring static conditions were maintained throughout. Both positive and negative secondary ion spectra were collected (mass resolution of >7000), over an acquisition period of fifteen scans (the data from which were added together). Owing to the non-conductive nature of the samples, charge compensation, in the form of a low energy (20 eV) electron flood gun, was applied.Water contact angle measurements
WCAs were measured using the sessile drop method on a fully automated Krüss DSA 100 instrument. A water drop with a volume of ∼400 pL was used. Ultrapure water was used for the WCA measurements (18.2 MΩ cm resistivity at 25 °C). The side profiles of the drops were recorded for image analysis. WCA calculations were performed using a circle segment function intersecting with a straight baseline representing the surface from three replicates.Multivariate analysis
The ToF-SIMS spectral data were analysed using principal component analysis (PCA), and the correlation between ToF-SIMS spectra and WCA or hPSC cell adhesion was analysed using PLS regression analysis as previously described.26 580 positive and 602 negative ions were selected from a group of polymers from the array containing all 141 monomers to form the peak lists. All peak intensities in a ToF-SIMS spectrum were normalized to the total secondary ion counts to remove the influence of primary ion beam fluctuation. The positive and negative ion intensity data were arranged into one concatenated data matrix. Both multivariate analysis methods were carried out using the Eigenvector PLS Toolbox 5.2. The SIMPLS algorithm was used for the PLS analysis. A “leave one out” cross validation method was used in the PLS regression analysis. The number of latent variables used to construct the PLS model was determined as the first minimum in the root mean square error of cross validation (RMSECV) curve (Fig. S1†). All datasets were mean-centred and square root mean scaled before analysis as determined using PCA analysis (Fig. S2 and S3†).Results and discussion
Polymer microarrays were synthesised by robotically spotting monomers via contact printing onto a pHEMA substrate and subsequent in situ polymerisation using UV irradiation to form polymer spots with diameters of up to 400 μm (Fig. 1a). Contact printed monomers of certain chemistries can be susceptible to spreading prior to UV curing. Despite the wide chemical diversity of the present array, no chemical spreading was observed using time-of-flight secondary-ion mass spectrometry (ToF-SIMS) imaging (Fig. S4†). This was achieved by contact printing monomer solutions at a higher dilution than previous studies.7To maximise the chemical diversity that can be explored on a single microarray we printed an array using monomers comprising a variety of nitrogen, fluorine, oxygen, aromatic and aliphatic side chain functionalities (Fig. 1c). Subsequent mixing of hits from this array will allow these commercially available off-the-shelf compounds to explore nearly a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 member material library employing only pairwise mixing. Monomers were selected based upon their ability to be polymerised in situ initiated by UV irradiation, low volatility and wide diversity in side-chain and main-chain chemistry to achieve the largest chemical combinatorial space yet published using a polymer microarray.
000 member material library employing only pairwise mixing. Monomers were selected based upon their ability to be polymerised in situ initiated by UV irradiation, low volatility and wide diversity in side-chain and main-chain chemistry to achieve the largest chemical combinatorial space yet published using a polymer microarray.
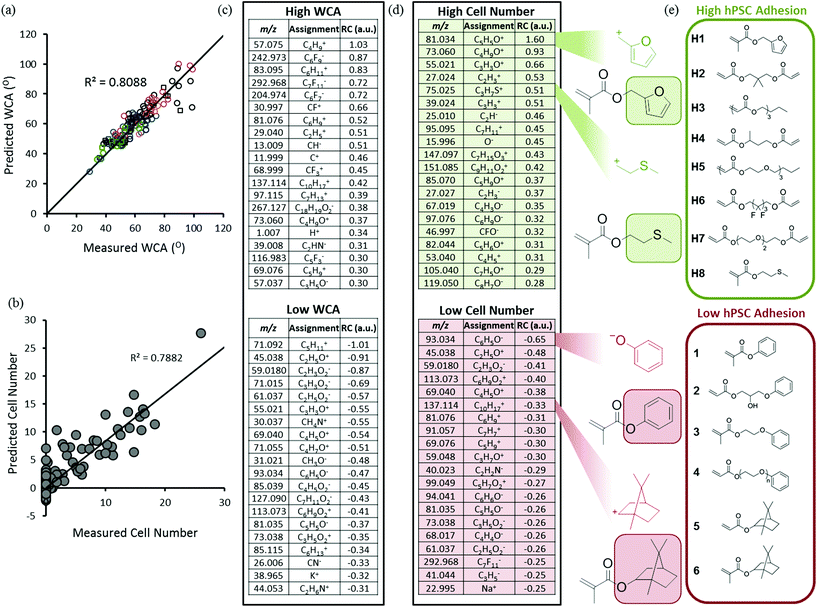
High throughput surface characterisation (HT-SC) was performed on the array to provide a chemical analysis of the outermost surface of each polymer (Fig. 1b). Automated acquisition ToF-SIMS was used to characterise the surface chemistry and pico litre sessile drop water contact angle (WCA) measurements were performed to probe the surface wettability.27 WCAs measured on the materials varied from 29 to 99°, reflecting the chemical diversity of the array (Fig. S5†). Previously, it has been shown that partial least-squares (PLS) regression analysis, a chemometric method that enables us to model correlations between univariate (cell response) and multivariate (ToF-SIMS spectra) datasets, can be employed to predict surface wettability of an array comprising 24 monomers to form 576 copolymers based upon their ToF-SIMS spectra.10 Correlation with WCA was used as a validation of the approach prior to subsequent application to the question of whether PLS could find chemistries in ToF-SIMS data that controlled cells response to the polymers. The same PLS regression analysis approach was applied to the present array to determine if a correlation can be identified between the surface chemistry and wettability of this far more diverse library. A PLS regression model was found to successfully predict the wettability of the materials within the polymer microarray from the concatenated positive and negative spectra acquired from the spots indicating a relationship between surface chemistry and wettability which includes all the spots on the array. This is shown by the linear relationship between predicted and experimental WCA values, with an R2 of 0.81 (Fig. 2a). To challenge this correlation, the data were split into a training and a test set, which produced an R2 for the test set of 0.61 (Fig. S1†), confirming on comparison of the R2 in Fig. 2 that this correlation was not a product of over fitting. Key explanatory variables (secondary ions) in the multivariate dataset can be identified which correlate either positively or negatively with the prediction (WCA). The 15 largest positive and negative regression coefficients (RCs) have been identified as those ions which are most significant in describing the WCA for the whole library of materials (Fig. 2c). Positive RCs associated with high WCA were found to be assigned (according to their m/z values) largely to ions indicative of hydrophobic surface species such as hydrocarbons and fluorocarbons. Similarly, negative RCs associated with low WCA were found to be secondary ions associated with hydrophilic surface species such as oxygenated hydrocarbons and amine fragments. The physicochemical sense of this finding indicates the utility of multivariate PLS regression analysis to identify correlations between the ToF-SIMS spectra of such a large number of components in a single library of materials and a univariate dataset (in this case WCA).
To investigate the ability of these 141 homopolymers to support hPSC attachment, polymer microarrays were first preconditioned with MEF-CM containing 20% fetal bovine serum (FBS) and then cultured for 24 hours in the same medium. Non-adherent cells were aspirated with the culture medium at 24 hours and the remaining adherent cells were fixed and stained with DAPI to identify nuclei and OCT4 as a pluripotency marker. Of the 141 materials screened in the array, 47 polymers supported hPSC attachment. ‘Hit’ monomers which supported the greatest hPSC attachment are shown in Fig. 2e. Only cells which displayed both DAPI and OCT4 markers were used to determine ‘hit’ materials. The proportion of OCT4 positive cells on these polymers showing the greatest attachment was between 65–92%. The 25 best performing materials ranked by cell attachment are plotted in Fig. S6† and monomer structures are identified in Fig. S7.†
Previous studies have observed that polymers with WCAs between 65° and 80° are optimal for hPSC interaction.2 However, across a chemically diverse library, WCA has been demonstrated to be a poor predictor of cell-material interactions in comparison to the more specific measurement of the surface chemistry provided by ToF SIMS.11 Consistent with these previous observations, a relationship between WCA and cell attachment could not be observed for the 141 materials in this array (Fig. S8†). Therefore, PLS regression analysis was used to search for correlations between the polymer surface chemistry captured by ToF-SIMS and the cell attachment to the arrayed materials. This analysis successfully predicted cell attachment per polymer spot for the library of materials as shown by the linear relationship identified between predicted and experimental cell adhesion, with an R2 of 0.79 (Fig. 2b). The top 15 positive RCs displayed in Fig. 2d are most significant in describing cell adhesion for the polymer library. The secondary ion with top positive RC, C5H5O+, is strongly associated with fragmentation of the side-chain functionality of the monomer that promoted the highest cell adhesion; furfuryl methacrylate (hit monomer H1 in Fig. 2e). Another positive regression vector (RV) loading, C3H7S+, was attributed to methylthioethyl methacrylate (monomer H8 in Fig. 2e).
The secondary ion fragment with largest negative RC, C6H5O−, is strongly associated with the aromatic side-chain functionality of monomers that promoted no cell adhesion including phenyl methacrylate (Fig. 2e). The next four most negative RVs, C2H5O+, C2H3O−, C6H9O2− and C4H5O+ are all associated with ethylene glycol moieties which are known to resist protein adsorption from the culture medium and prevent cell adhesion in general. The ion C10H17+ can be assigned to the norbornyl moiety, which has highest intensity arising from the two monomers norbornyl acrylate and norbornyl methacrylate (monomers 5 and 6 respectively in Fig. 2e), which both failed to support cell adhesion.
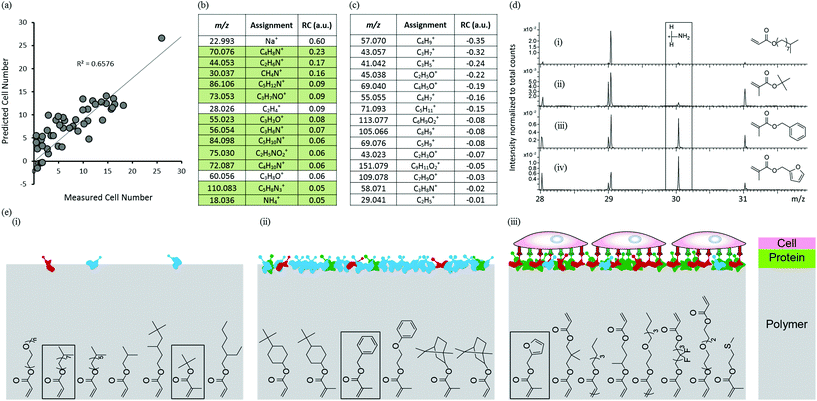
To investigate the mechanism governing hPSC attachment to the arrayed materials, arrays were analysed by ToF-SIMS after conditioning with culture medium for 1 hour to mirror the protocol used prior to the hPSC adhesion assay. A rinsing procedure intended to remove excess or loosely adhered proteins was applied prior to analysis using ToF-SIMS. The medium used, MEF-CM, is a very complex mixture of ECM proteins, growth factors and cellular metabolites. Secondary ions assigned to amino acids are detected which arise from surface bound proteins through fragmentation in the primary ion bombardment process. PLS regression analysis was used to identify correlations between the chemistry of the conditioned surface and cell attachment for the polymer library. Samples that did not support cell adhesion were omitted from the model leaving 46 materials. Only positive ion spectra were used to construct the model as they contain peaks that are useful to identify amino acids.28,29 PLS regression analysis successfully predicted cell attachment for the 46 materials as shown by the linear relationship between predicted and experimental cell adhesion to the micro array, with an R2 of 0.66 (Fig. 3a). The 15 most positive RCs displayed in Fig. 3b are most significant in describing cell adhesion for the medium-conditioned polymer library; all but 3 of these ion fragments originate from proteins adsorbed from the culture medium (green background in Fig. 3b). This indicates the importance of protein adsorption in achieving cellular attachment to these materials. However, identification of individual proteins from the complex conditioned media is not possible using ToF-SIMS. The ion fragment with largest positive RC, Na+, originates from the medium which was not removed despite rinsing-SIMS is very sensitive to low levels of sodium. The next two highest positive RVs, C4H8N+ and C2H6N+, are both related to amino acid fragments of adsorbed proteins but cannot be assigned to a specific amino acid.29 The next highest positive RV, CH4N+ (CH2NH2+), is most prevalent in the amino acid glycine, but is common to all amino acids. The 15 most negative RCs displayed in Fig. 3c are most significant in describing low cell adhesion for the medium conditioned polymer library; none of these ion fragments are related to amino acids but all are characteristic of the surface chemistry of the materials that supported low cell adhesion. The ion fragment with highest negative RC, C4H9+ (+CH2CH(CH3)2/+C(CH3)3), is characteristic of hydrophobic monomers containing side-chains of isopropyl or t-butyl groups e.g. Isooctyl acrylate. This monomer supported low hPSC attachment. The next highest negative RV, C3H7+ (CH(CH3)2), can be attributed to hydrophobic monomers containing side-chains of isopropyl groups. These findings have enabled the identification of monomers and chemical moieties that exhibit low protein adsorption and low cell attachment.
To further probe the relationship between protein adsorption from the medium and hPSC adhesion to the arrayed materials, ToF-SIMS spectra were compared using the CH4N+ ion fragment (m/z = 30.03) which is commonly found in all amino acids which makes this a good general indicator for protein adsorption. The ToF-SIMS spectra of four polymers, isodecyl acrylate, t-butyl methacrylate, benzyl methacrylate and furfuryl methacrylate are shown in Fig. 3d(i–iv) respectively. The m/z = 30 region of the ToF-SIMS spectra for isodecyl acrylate and t-butyl methacrylate (Fig. 3d(i–ii)) showed very little intensity for this ion fragment, indicating that these materials resist protein adsorption and are therefore unable to support hPSC adhesion (also shown inset in Fig. 3e(i)). This supports the negative RVs for cell attachment found in Fig. 3c, the two most negative RCs are moieties characteristic of isodecyl and t-butyl groups. The ToF-SIMS spectra of benzyl methacrylate and furfuryl methacrylate (structures are shown inset in Fig. 3e(ii) and (iii) respectively) showed a significant intensity for the CH4N+ ion fragment which indicates that protein adsorption had occurred on these materials. However, despite the chemical similarity of these two monomers, cellular attachment to these two materials differed significantly. Benzyl methacrylate did not support any hPSC adhesion and furfuryl methacrylate was the best performing material within the library and supported high hPSC adhesion. This suggests that adsorption of proteins from a complex biological milieu such as MEF-CM is not sufficient alone to support hPSC adhesion. The adsorption of specific proteins in the correct conformation that can engage with cell surface integrins are essential to facilitate cell adhesion to a variety of surfaces.30,31 This is schematically illustrated in Fig. 3e. These findings suggest that subtle changes in surface chemistry of these materials have a profound effect upon protein adsorption and consequently cellular attachment. Furthermore, diverse protein adsorption to the chemically diverse array of materials was observed and no relationship between WCA and protein was observed (Fig. S9†). Blocking experiments have previously been used to determine specific integrins essential for hPSC attachment on a variety of surfaces.2,32,33 Human-serum coated surfaces have been shown to engage the vitronectin-binding integrins, αvβ3 and αvβ5 to facilitate hPSC adhesion.2 We suggest the same integrins allow cell adhesion to our preconditioned surfaces as vitronectin is highly abundant in serum.34
PLS regression analysis has identified chemical moieties that controlled material wettability and hPSC attachment. However, the polymer moieties revealed by PLS regression analysis to control wettability and hPSC adhesion were very different suggesting that wettability alone is not sufficient to predict hPSC adhesion. PLS regression analysis successfully used ToF-SIMS data from the polymer surface to predict the wettability. The hPSC adhesion was predicted in the same way using analysis of the polymer surface revealing very different controlling surface moieties. Molecular descriptors have proved useful to model cell adhesion to polymer surfaces.35 However, these descriptors cannot describe a protein conditioned polymer surface adequately to model cell adhesion. But PLS regression analysis of cell response using the ToF-SIMS data of the protein conditioned surfaces was successful in revealing insights into the role of protein adsorption in HUES7 cell attachment to this library of materials.
Conclusions
A material array comprising 141 unique homopolymers has been synthesised to operate as a chemically diverse high throughput screening tool. This tool was employed to screen for human pluripotent stem cell adhesion in a high throughput manner. Several novel homopolymers have been discovered that supported cell adhesion in MEF-CM. In parallel to biological assays, HT-SC has been used to characterise the surface chemistry of these materials. These new materials are excellent candidates for synthetic substrate expansion of human pluripotent stem cells. Protein resistance for certain polymers was noted under the preconditioning conditions utilised here. Analysis of the medium conditioned surfaces and its relationship to hPSC number revealed the importance of protein adsorption for cellular attachment to the materials and identified new chemical moieties that resist protein adsorption.Acknowledgements
C.D. is supported by the EPSRC, British Heart Foundation, Heart Research UK and National Centre for the Replacement, Refinement and Reduction of Animals in Research (NC3Rs). M.R.A. gratefully acknowledges the EPSRC (grant number EP/H045384/1), the Wellcome Trust for funding and The Royal Society for the provision of his Wolfson Research Merit Award.Notes and references
- D. G. Anderson, S. Levenburg and R. Langer, Nat. Biotechnol., 2004, 22, 863 CrossRef CAS PubMed.
- Y. Mei, K. Saha, S. R. Bogatyrev, J. Yang, A. L. Hook, Z. I. Kalcioglu, S. W. Cho, M. Mitalipova, N. Pyzocha, F. Rojas, K. J. Van Vliet, M. C. Davies, M. R. Alexander, R. Langer, R. Jaenisch and D. G. Anderson, Nat. Mater., 2010, 9, 768 CrossRef CAS PubMed.
- R. Zhang, A. Liberski, R. Sanchez-Martin and M. Bradley, Biomaterials, 2009, 30, 6193 CrossRef CAS PubMed.
- A. L. Hook, D. G. Anderson, R. Langer, P. Williams, M. C. Davies and M. R. Alexander, Biomaterials, 2012, 31, 187 CrossRef PubMed.
- A. L. Hook, C. Chien-Yi, J. Yang, J. Luckett, A. Cockayne, S. Atkinson, Y. Mei, R. Bayston, D. J. Irvine, R. Langer, D. G. Anderson, P. Williams, M. C. Davies and M. R. Alexander, Nat. Biotechnol., 2012, 30, 868 CrossRef CAS PubMed.
- A. L. Hook, C. Chang, J. Yang, S. Atkinson, R. Langer, D. G. Anderson, M. C. Davies, P. Williams and M. R. Alexander, Adv. Mater., 2013, 25, 2542 CrossRef CAS PubMed.
- A. D. Celiz, A. L. Hook, D. J. Scurr, D. G. Anderson, R. Langer, M. C. Davies and M. R. Alexander, Surf. Interface Anal., 2013, 45, 202 CrossRef CAS.
- A. J. Urquhart, D. G. Anderson, M. Taylor, M. R. Alexander, R. Langer and M. C. Davies, Adv. Mater., 2007, 19, 2486 CrossRef CAS.
- M. C. Davies, M. R. Alexander, A. L. Hook, J. Yang, Y. Mei, M. Taylor, A. J. Urquhart, R. Langer and D. G. Anderson, J. Drug Targeting, 2010, 18, 741 CrossRef CAS PubMed.
- A. J. Urquhart, M. Taylor, D. G. Anderson, R. Langer, M. C. Davies and M. R. Alexander, Anal. Chem., 2008, 80, 135 CrossRef CAS PubMed.
- J. Yang, Y. Mei, A. L. Hook, M. Taylor, A. J. Urquhart, S. R. Bogatyrev, R. Langer, D. G. Anderson, M. C. Davies and M. R. Alexander, Biomaterials, 2010, 31, 8827 CrossRef CAS PubMed.
- D. Rajamohan, E. Matsa, S. Kalra, J. Crutchley, A. Patel, V. George and C. Denning, BioEssays, 2012, 35, 281 CrossRef PubMed.
- R. J. Thomas, D. Anderson, A. Chandra, N. M. Smith, L. E. Young, D. Williams and C. Denning, Biotechnol. Bioeng., 2009, 102, 1636 CrossRef CAS PubMed.
- M. M. Mahlstedt, D. Anderson, J. S. Sharp, R. McGilvray, M. D. Barbadillo Munoz, L. D. Buttery, M. R. Alexander, F. R. A. J. Rose and C. Denning, Biotechnol. Bioeng., 2009, 105, 130 CrossRef PubMed.
- C. Xu, M. S. Inokuma, J. Denham, K. Golds, P. Kundu, J. D. Gold and M. K. Carpenter, Nat. Biotechnol., 2001, 19, 971 CrossRef CAS PubMed.
- D. A. Brafman, C. W. Chang, A. Fernandez, K. Willert and S. Varghese, Biomaterials, 2013, 31, 9135 CrossRef PubMed.
- L. G. Villa-Diaz, H. Nandivada, J. Ding, N. C. Nogueira-De-Souza, P. H. Krebsbach, K. S. O'Shea, J. Lahann and G. D. Smith, Nat. Biotechnol., 2010, 28, 581 CrossRef CAS PubMed.
- E. E. Irwin, R. Gupta, D. C. Dashti and K. E. Healy, Biomaterials, 2011, 32, 6912 CrossRef CAS PubMed.
- K. Saha, Y. Mei, C. M. Reisterer, N. K. Pyzocha, J. Yang, J. Muffat, M. C. Davies, M. R. Alexander, R. Langer, D. G. Anderson and R. Jaenisch, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 18714 CrossRef CAS PubMed.
- C. W. Chang, Y. Hwang, D. Brafman, T. Hagan, C. Phung and S. Varghese, Biomaterials, 2013, 34, 912 CrossRef CAS PubMed.
- A. D. Celiz, J. G. W. Smith, R. Langer, D. G. Anderson, D. A. Barrett, D. A. Winkler, M. C. Davies, L. E. Young, C. Denning and M. R. Alexander, Nat. Mater. DOI:10.1038/NMAT3972.
- A. L. Hook, C. Chien-Yi, J. Yang, D. J. Scurr, R. Langer, D. G. Anderson, S. Atkinson, P. Williams, M. C. Davies and M. R. Alexander, J. Visualized Exp., 2012 DOI:10.3791/3636.
- P. W. Burridge, D. Anderson and H. Priddle, Stem Cells, 2007, 25, 929 CrossRef CAS PubMed.
- C. Denning, C. Allegrucci and H. Riddle, Int. J. Dev. Biol., 2006, 50, 27 CrossRef CAS PubMed.
- C. A. Cowan, I. Klimanskaya and J. McMahon, N. Engl. J. Med., 2004, 350, 1353 CrossRef CAS PubMed.
- M. Taylor, A. J. Urquhart, D. G. Anderson, R. Langer, M. C. Davies and M. R. Alexander, Surf. Interface Anal., 2009, 41, 127 CrossRef CAS.
- M. Taylor, A. J. Urquhart, M. Zelzer, M. C. Davies and M. R. Alexander, Langmuir, 2007, 23, 6875 CrossRef CAS PubMed.
- M. S. Wagner and D. G. Castner, Langmuir, 2001, 17, 4649 CrossRef CAS.
- H. E. Canavan, D. J. Graham, X. Cheng, B. D. Ratner and D. G. Castner, Langmuir, 2007, 23, 50 CrossRef CAS PubMed.
- W. B. Tsai, J. M. Grunkemeier and T. A. Horbett, J. Biomed. Mater. Res., 1999, 44, 130 CrossRef CAS.
- B. G. Keselowsky, D. M. Collard and A. J. Garcia, Biomaterials, 2004, 25, 5947 CrossRef CAS PubMed.
- Y. Meng, S. Eshghi, Y. J. Li, R. Schmidt, D. V. Schaffer and K. E. Healy, FASEB J., 2010, 24, 1056 CrossRef CAS PubMed.
- S. Rodin, A. Domogatskaya, S. Ström, E. M. Hansson, K. R. Chien, J. Inzunza, O. Hovatta and K. Tryggvason, Nat. Biotechnol., 2010, 28, 611 CrossRef CAS PubMed.
- E. G. Hayman, M. D. Pierschbacher, Y. Ohgren and E. Ruoslahti, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 4003 CrossRef CAS.
- V. C. Epa, J. Yang, A. L. Hook, R. Langer, D. G. Anderson, M. C. Davies, M. R. Alexander and D. A. Winkler, J. Mater. Chem., 2012, 22, 20902 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c4bm00054d |
| This journal is © The Royal Society of Chemistry 2014 |