Open Access Article
Open Access ArticleIn vitro simulated semi-dynamic gastrointestinal digestion: evaluation of the effects of processing on whey proteins digestibility and allergenicity†
Alessandra
Gasparini‡
a,
Sara
Benedé
 a,
Tullia
Tedeschi
a,
Tullia
Tedeschi
 b,
Stefano
Sforza
b,
Stefano
Sforza
 b,
Isidra
Recio
b,
Isidra
Recio
 a and
Beatriz
Miralles
a and
Beatriz
Miralles
 *a
*a
aInstituto de Investigación en Ciencias de la Alimentación, CIAL (CSIC-UAM, CEI UAM+CSIC), Nicolás Cabrera, 9, 28049 Madrid, Spain. E-mail: beatriz.miralles@csic.es
bFood and Drug Department, University of Parma, Parco Area delle Scienze, 27/A, 43124, Parma, Italy
First published on 17th January 2022
Abstract
The effect of thermal processing on digestibility of milk proteins should be better understood as this can greatly affect their immunoreactivity. The aim of this study was to evaluate the effects of thermal processing and lactosylation on digestibility and allergenicity, by comparing non heat-treated with industrially processed whey proteins. A semi-dynamic model was used to mimic the kinetics of digestion, and ELISA inhibition tests against human specific serum IgE were performed on the mass-spectrometry characterized products. A quicker gastric digestion of the industrially treated sample produced a lower immunogenic response in comparison with the raw sample, where intact conformational epitopes remained. In later digests, greater IgE reactivity was shown in the heat treated product, probably due to the release of linear epitopes, while at intestinal level the immunogenic response was negligible. Moreover, transepithelial transport of a reported β-lactoglobulin-derived allergen, KIDALNENVLVL, produced during digestion was assayed. It was found that the epitope-belonging peptide could be transported through the cell monolayer, both in the native and mono-lactosylated forms, with a favored passage of the native peptide.
1. Introduction
Whey ingredients are used to increase food nutritional value, e.g. in sports supplements and infant formulas, or as technological agents for thermal stability, emulsification, gelation and foam formation. During the production of whey protein concentrates (WPCs) and whey protein isolates (WPIs), there is a range of pre-treatment processes (clarification, pasteurization, pre-heating), in addition to the ultrafiltration and diafiltration processes that are likely to cause denaturation of minor heat-sensitive proteins.1 Drying of the retentate by atomization is used to dehydrate and control the moisture content of the concentrates but it favours the initiation of lipid oxidation and non-enzymatic browning reactions (Maillard reactions) during storage.2 Maillard reaction in milk products is initiated by the interaction between lactose and protein amino groups. Recent studies showed that practically all lysine residues of milk proteins can be lactosylated.3,4Lactosylation of milk proteins has been related to a decrease of the protein nutritional value.5 In addition, lactosylated proteins are less susceptible to enzymatic hydrolysis and thus, this may affect gastrointestinal digestion of proteins. For example, it has been shown that lysine blockage induced a significant change in the peptide pattern after simulated infant gastrointestinal digestion.6 Heat treatment reduced significantly available lysine of skim milk powder measured in growing rats.7–9 However, the effect of thermal processing on protein digestibility is still controversial since protein denaturation may enhance digestibility by the exposure new cleavage sites to the action of the digestive enzymes, but protein aggregation and glycation impairs protein digestibility.10 Although in vivo studies offer valuable information about digestion and protein digestibility, these are costly and generate ethical issues and are often replaced by in vitro models. Recently, a semi-dynamic model was developed,11,12 which is a compromise between the dynamic and static models. The gastric phase is performed in a thermostatically controlled vessel from which it is possible to remove samples from the digestion mimicking the gastric emptying and to control the addition of the enzyme. Moreover, gradual addition of the gastric solution is conducted. This model was based on in vivo digestion parameters obtained for the digestion of dairy products.13
Commonly, the resistance of the protein to the gastrointestinal digestion was considered fundamental to allergenicity, as the potential allergic response was attributed to intact proteins or large protein fragments.14,15 More recently, it was reported that also smaller peptides may still induce an allergic response.16,17 In this scenario, Maillard reaction has been found to be sometimes beneficial by damaging β-lactoglobulin epitopes and reducing their allergenic potential,18,19 but on the contrary, by reducing protein digestibility, the generation of neo-epitopes has also been reported.20 Recently, lactosylated β-lactoglobulin was found to decrease both, IgG binding and degranulation of mast cells, although this study was performed with the whole protein molecule.21 Moreover, peptide transport through the intestinal wall may be also affected since it is influenced by hydrophobicity, charge and molecular size.22 Indeed, small peptides (i.e. composed by two or three amino acids residues) are more easily absorbed as compared to long-chain ones.23 Post-translational modifications can affect brush border membrane peptidases activity, and transepithelial transport. For instance, it was also reported that the glycation of caseinomacropeptide limited the activity of brush border endopeptidases.24 However, the effect of lactosylation on the enzyme activities and peptide intestinal absorption needs a deeper investigation. Different studies on the intestinal absorption of free and peptide-bound Maillard Reaction Products (MRPs) have been performed,25–27 but only small peptides (i.e. dipeptides) were investigated.
The aim of this study was to investigate the effects of thermal processing and lactosylation on whey protein digestibility and allergenicity. A rennet whey fraction isolated from raw milk was compared to a commercial whey protein isolate. To evaluate the actual digest arriving to the small intestine, an in vitro semi-dynamic digestion model was employed. ELISA inhibition tests against human specific serum IgE were conducted on the digests over time. Transepithelial transport studies of a lactosylated peptide reported as a β-lactoglobulin-derived epitope on Caco-2 cells were performed.
2. Materials and methods
2.1 Samples
Raw milk was purchased from a local milk producer to prepare a cheese whey sample (CW). Whey protein isolate (WPI) was purchased on a local store. Serum samples from milk allergic patients were obtained from our human blood serum bank. Human samples were collected with written consent from the next of kin, caretakers, or guardians on the behalf of the minors/children involved in the performed studies. All experiments were conducted according to the principles expressed in the Declaration of Helsinki and the Bioethics Committee from the Consejo Superior de Investigaciones Científicas (CSIC), Spain. The researchers involved in the process handled and used such material sensitively and responsibly. Individual features of human sera are shown in ESI Table S1.†2.2 Whey production from cheese (CW sample)
Raw milk was defatted centrifuging at 4500g at 5 °C for 20 min and then removing fat manually. Then it was filtered on wool glass. Nearly 2 L of sample was warmed at 33 °C. Then 50.62 mg of CaCl2 was added and, after 2 min under stirring, 500 μL of rennet was added. It was stirred for 3 min and then left at 33 °C for 1 h. Cheese was cut in small pieces and filtered on a strainer covered with the proper cloth. Whey was then centrifuged at 4500g at 5 °C for 10 min and filtered on glass wool. Aliquots were stored at −20 °C.2.3 MALDI-TOF characterization
A Bruker Autoflex Speed spectrometer (Bruker Daltonics GmbH, Bremen, Germany) was used. Samples were dissolved in MilliQ water at 1% of protein concentration and then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100. One μL was loaded on a Bruker MTP 384 Polished Steel target and 1 μL of sinapinic acid in 1% formic acid/50% ACN (Bruker Daltonics, Billerica, MA, USA) was used as matrix. Ions were detected in positive linear mode at a mass range of m/z 5–25 kDa. For calibration, 0.5 μL of Protein Calibration Standard I (Bruker Daltonics) was loaded on the target with 0.5 μL, of sinapinic acid solution.
100. One μL was loaded on a Bruker MTP 384 Polished Steel target and 1 μL of sinapinic acid in 1% formic acid/50% ACN (Bruker Daltonics, Billerica, MA, USA) was used as matrix. Ions were detected in positive linear mode at a mass range of m/z 5–25 kDa. For calibration, 0.5 μL of Protein Calibration Standard I (Bruker Daltonics) was loaded on the target with 0.5 μL, of sinapinic acid solution.
2.4 Semi-dynamic in vitro gastric digestion
The in vitro semi-dynamic digestion was performed according to the protocol of Mulet-Cabero et al.12 In order to use equal amount of protein, the WPI sample was diluted in Milli-Q water at the 0.95% of protein concentration, as determined from Kjeldahl analysis for the CW sample. The digestion was performed in a double wall glass vessel controlled at 37 °C and under stirring. Sixty mL of sample were mixed with the Simulated Salivary Fluid (SSF) without the addition of amylase. After 2 min, the gradual addition of the Simulated Gastric Fluid (SGF) and of pepsin (4000 U mL−1 of digest, Sigma Aldrich, St Luis, MO, USA) was started. In order to mimic the gastric emptying, five aliquots were collected from the digestion as reported in Table 1. The pH of these aliquots was measured and increased to pH 7 with 2 M NaOH in order to inactivate pepsin activity. Samples were stored at −20 °C. From gastric emptying points G1, G3 and G5 (Table 1), 10 mL were used for the static intestinal digestion following the INFOGEST protocol.12,13 The Simulated Intestinal Fluid (SIF) was added to the samples with pancreatin (200 U mL−1 TAME units of digest, Sigma Aldrich) and bile salts (2.5 mM in the final digest, Sigma Aldrich).| Digestion time (min) | Volume (mL) | |
|---|---|---|
| Gastric emptied point 1 (G1) | 18.08 | 25 |
| Gastric emptied point 2 (G2) | 36.15 | 25 |
| Gastric emptied point 3 (G3) | 54.23 | 25 |
| Gastric emptied point 4 (G4) | 72.30 | 25 |
| Gastric emptied point 5 (G5) | 90.38 | 25 |
Digestion was performed for 2 hours at 37 °C in a shaking incubator at 120 rpm. Then, enzymatic activity was stopped by heating at 85 °C and centrifuging at 5000g for 30 min. The supernatant was collected and freeze-dried. Digestion was performed in duplicate.
2.5 HPLC-MS/MS analysis
HPLC-MS/MS analyses and data processing was performed according to Santos-Hernández et al.28 Prior to the analyses, the reduction step was performed with 146 mM DTT. Gradient elution was performed with and Agilent 110 HPLC separation system (Agilent, CA, USA). Detection was performed with a Bruker Daltonics Esquire 3000 Ion Trap mass spectrometer (Bruker, MA; USA) in a full scan acquisition mode from 100 to 2300 m/z, and setting target mass at 450, 750 and 1200. Biotools version 3.2 was used for interpreting the matched MS/MS spectra. For peptide profile visualization, the web application Peptigram was used.292.6 SDS-PAGE analysis
The electrophoretic separation was performed according to the protocol of Sanchón et al.30 Samples and the marker (Precision Plus Protein™ Unstained protein standards, Biorad, Hercules, CA, USA) were loaded on 12% bis–tris Criterion™ XT gel (Biorad). Gels where then stained with Coomassie Blu (Instant ble, Expedeon, Swavesey, UK). Images were acquired with a Molecular Imager VersaDocTM MP 5000 system (Bio-Rad) and using the software QuantityOne® 1-D (Bio-Rad).2.7 UPLC-UV analysis
UPLC-UV analyses were performed on an ACQUITY UPLC separation system (Waters Technologies, Cerdanyola del Vallés, Spain). The analytical column used was an Aeris WIDEPORE XB-C18 LC column (Phenomenex, 200 Å, 3.6 μm, 2.1 mm × 150 mm, CA, USA). Eluent A was 90% water + 10% acetonitrile + 0.1% formic acid and eluent B was 90% acetonitrile + 10% water + 0.1% formic acid. Flow rate was set at 0.20 mL min−1, injection volume 3 μL. Detection was performed at 214 nm. Samples were previously diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 in water deionized, then 1
4 in water deionized, then 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with eluent A + Urea 6 M and centrifuged 3 min at 11
1 with eluent A + Urea 6 M and centrifuged 3 min at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. For protein quantification two external calibration curves were used: for α-lactalbumin (87.12% of purity, Sigma Aldrich) 0.1, 0.2, 0.4, 0.8, 1 mg mL−1 dilutions were used; for β-lactoglobulin (86.35% of purity, Sigma Aldrich) 0.25, 0.5, 1, 2, 4 mg mL−1 dilutions were used.
000g. For protein quantification two external calibration curves were used: for α-lactalbumin (87.12% of purity, Sigma Aldrich) 0.1, 0.2, 0.4, 0.8, 1 mg mL−1 dilutions were used; for β-lactoglobulin (86.35% of purity, Sigma Aldrich) 0.25, 0.5, 1, 2, 4 mg mL−1 dilutions were used.
2.8 ELISA inhibition test
Human IgE binding of undigested WPI and CW, on samples G1, G3 and G5 from the gastric digestion and samples I1, I3, and I5 from the intestinal digestion, was assessed by inhibition ELISA as previously reported,31 using a pool of sera from allergic patients (ESI Fig. S1†). A reference sample of standard whey proteins was used as a control.2.9 Peptide synthesis
Synthesis of peptide KIDALNENKVLVL and its lactosylated forms were performed according to Gasparini et al.32 Peptide was synthesized using the Syro I automated synthesizer (Biotage, Uppsala, Sweden). The resin Fmoc-Leu-Wang LL was used and the acetylation step prior to peptide cleavage from the resin was not performed. The obtained peptide was then dissolved with lactose (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 ratio) in DMF under nitrogen atmosphere. The solution was incubated at 70 °C for 40 h and then solvent was removed under vacuum. Peptides mixture was then freeze-dried and analysed with HPLC-MS analysis as described in section 2.5. Proteomics Toolkit (Institute for Systems Biology, Seattle, WA, USA) was used for the identification of the MS ions (ESI Table S2†). The mixture was dissolved in water and ultrafiltration was performed with a 1 kDa cut off (Microsep advanced centrifugal device, Pall Corporation, Port Washington, NY, USA) to remove the unreacted lactose.
50 ratio) in DMF under nitrogen atmosphere. The solution was incubated at 70 °C for 40 h and then solvent was removed under vacuum. Peptides mixture was then freeze-dried and analysed with HPLC-MS analysis as described in section 2.5. Proteomics Toolkit (Institute for Systems Biology, Seattle, WA, USA) was used for the identification of the MS ions (ESI Table S2†). The mixture was dissolved in water and ultrafiltration was performed with a 1 kDa cut off (Microsep advanced centrifugal device, Pall Corporation, Port Washington, NY, USA) to remove the unreacted lactose.
2.10 Transepithelial transport studies
Caco-2 cells cultivation was performed as described in literature.33 For transport studies the Hubatsch et al., protocol was followed.34 Cells were seeded at a density of 2.6 × 105 cells per cm2 in 12-well filter support inserts (Costar® Transwell® Permeable Supports 12 mm Diameter insert 12 well 0.4 μm polycarbonated Membrane, Corning, NY, USA). The integrity of cell monolayers was tested measuring the transepithelial electrical resistance (TEER) with Evom2 Epithelial Voltammeter (World Precision Instruments, FL, USA). Only Caco-2 monolayer with TEER values more than 500 Ω cm−2 were used. Experiments were performed 21–29 days after seeding and medium was changed 24 h before the assay. Samples were dissolved in HBSS at 1.7 mM final concentration and filtered on 0.2 μm filters.35 Collected samples were freeze-dried. The assay was performed in triplicate. For the HPLC-MS/MS analysis (section 2.5), samples were reconstituted in eluent A at different concentrations: the apical side collected before the incubation was reconstituted in 100 μL and then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 with eluent A; the apical side after 60 min was reconstituted in 300 μL and then diluted 1
12 with eluent A; the apical side after 60 min was reconstituted in 300 μL and then diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 with eluent A; the basolateral samples were reconstituted in 100 μL of eluent A. Lucifer Yellow assay was performed to test the integrity of the Caco-2 monolayers as described by Fernández-Tomé et al.33 Fluorescence was measured using a Fluostar Optima microplate reader (BMG Labtech, Ortenberg, Germany) setting 480 nm of excitation and 520 nm of emission. The analysis was performed in triplicate.
12 with eluent A; the basolateral samples were reconstituted in 100 μL of eluent A. Lucifer Yellow assay was performed to test the integrity of the Caco-2 monolayers as described by Fernández-Tomé et al.33 Fluorescence was measured using a Fluostar Optima microplate reader (BMG Labtech, Ortenberg, Germany) setting 480 nm of excitation and 520 nm of emission. The analysis was performed in triplicate.
3. Results and discussion
3.1. Protein characterization of substrates
In order to evaluate the effects of processing on whey proteins, untreated whey from raw milk cheese (CW) was compared with a commercial whey protein isolate (WPI). The products were analysed with MALDI-TOF mass spectrometry (Fig. 1). As opposed to CW, in the WPI sample, the peak relative to α-lactalbumin (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 178 Da) was accompanied by signals corresponding to lactosylated forms of the protein at m/z 14
178 Da) was accompanied by signals corresponding to lactosylated forms of the protein at m/z 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 502 and 14
502 and 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 826 (lactosylation 324 Da).
826 (lactosylation 324 Da).
 | ||
| Fig. 1 MALDI-TOF spectrum of CW (blue) and of WPI (red) samples (α-LA = α-lactalbumin; β-LG = β-lactoglobulin). Intensity is on the y axis; mass-to-charge ratio is on the x axis. | ||
In the spectra, variants A and B from β-lactoglobulin were distinguished (at m/z 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 358 and 18
358 and 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 271, respectively) in addition to the corresponding mono- and di-lactosylated forms of each variant. In the CW spectrum also peaks corresponding to monolactosylation could be detected but at a lower extent. This was not surprising as similar signals have been previously observed in unheated samples using mass spectrometry.36 Overall, the spectra were consistent with an increased lactosylation degree in processed samples.
271, respectively) in addition to the corresponding mono- and di-lactosylated forms of each variant. In the CW spectrum also peaks corresponding to monolactosylation could be detected but at a lower extent. This was not surprising as similar signals have been previously observed in unheated samples using mass spectrometry.36 Overall, the spectra were consistent with an increased lactosylation degree in processed samples.
3.2. Semi-dynamic gastric digestion
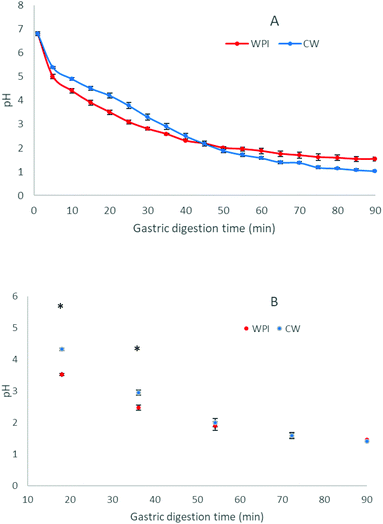
Fig. 2 shows the pH curves during the gastric phase for CW and WPI, measured in the digestion vessel (Fig. 2A) and in the withdrawn emptying aliquot (Fig. 2B). Gastric digestions of both substrates showed a pH value ca. 6.8 after the initial addition of the gastric juice and food, and reached a final pH of 1.05 ± 0.02 (CW) and 1.47 ± 0.09 (WPI). The profile showed a rapid pH decrease in the first 5 min, followed by a slower, continuous pH reduction, with no visible coagulation. A similar pH trend was previously reported in other semi-dynamic gastric digestions performed on milk proteins, but the previously studied products contained caseins and showed coagulation.11,37 However, gastric emptying reduced the buffering capacity differently for both products in the first period. The pH at the emptied digest in G1 and G2 was higher for CW while from G3, an overlay of the determined values was observed.3.3. Protein degradation during gastric emptying
The aliquots from gastric emptying were analysed with SDS-PAGE in order to follow whey proteins degradation during digestion. SDS-PAGE bands at around 18 kDa corresponding to the most abundant protein, β-lactoglobulin, were clearly detectable at all-time points although a decrease in the intensity of the signals with the progress of the digestion could be observed (Fig. 3). This is due to both, the increasing dilution with SGF and to a partial digestion of the protein as further confirmed. The band corresponding to α-lactalbumin, at around 14 kDa, was only detected in the first emptying point (G1) of both products. Being absent in G2, it can be assumed that this protein is completely digested during the first 36 min of gastric digestion. Indeed, from this time the pH is below 4, and it has been reported that, at this pH, α-lactalbumin undergoes changes in conformation, thus being more susceptible to pepsin hydrolysis.37–39 Additional higher molecular bands between 150 and 50 kDa corresponded to lactoferrin, the polymeric immunoglobulin receptor, bovine serum albumin and other minor whey proteins according to the identification by Bär et al.40 These proteins were more abundant in CW and could be detected in CW-G1, in contrast to WPI-G1. Comparing substrates, the band relative to α-lactalbumin has lower intensity in WPI than in CW, concomitant with the appearance of a new band in WPI-G1 with a MW under 10 kDa. Because no intact proteins were expected at the intestinal phase, these aliquots have been analysed for peptide release by HPLC-MS/MS.3.4. Peptide release during semi-dynamic gastrointestinal digestion
Fig. 4 shows the peptide profile in gastric digests G1, G3 and G5 and their corresponding intestinal digests, i.e., I1, I3, I5, of the two whey proteins, α-lactalbumin and β-lactoglobulin for CW and WPI samples. The complete list of identified peptides is given in ESI.†In CW, only the region 41–52 from α-lactalbumin was identified in G1, whereas peptides covering 47% the protein sequence were identified in WPI, showing a delayed digestion in the non-treated CW. In contrast, both samples showed similar coverage at G3. Most of the identified peptides belonged to α-lactalbumin regions 1–52 and 76–123. The amount of peptides and covered region were lower in G5 suggesting the progress of digestion and the subsequent decrease of the peptide size. It has to be noted that peptides <5 amino acids were not detected under our analysis conditions. Interestingly, α-lactalbumin domains 1–19, 15–34, 45–64 and 90–123 are reported in literature to be IgE-binding epitopes,41,42 and some identified peptides released during the gastric phase, such as, 1–10, 26–31, 41–50, 118–123 belonged to these previously reported IgE-binding epitopes.43
Regarding β-lactoglobulin-derived peptides, these were absent in G1 due to the low degree of hydrolysis at this time point. In CW, a low number of peptides appeared in G3 but they increased in G5. However, for WPI, peptides at G3 are comparable to those identified in G5. For both products, peptide identification confirmed a partial digestion of β-lactoglobulin in the gastric phase, but a higher number of peptides were observed in WPI. Peptides covering regions 1–54, 74–108, 123–131 (only in G5), 150–162 were identified. From them, peptides within the amino acid residues 1–16, 31–60, 76–95, 121–140 and 136–150 are known to be IgE-binding epitopes.42,44,45 Overall, lower number of peptides could be identified at the intestinal phase, with a remarkable change in the profile, some of them corresponding to newly released regions not being detected in the gastric phase. In the case of α-lactalbumin, peptides derived mainly from regions 61–68 and 81–89, that had been previously found resistant to gastrointestinal in vivo digestion.30 Interestingly, peptides from region 80–90 were previously found in human intestinal digests with remarkable abundance.46 α-Lactalbumin fragment (81–88), LDDDLTDD, and various derived fragments were identified in both products. Regarding β-lactoglobulin, in the intestinal phase collected from CW, it is possible to observe peptides deriving from region 106–119, while they are not present in the gastric emptied points. At positions 106 and 119 there are two cysteine residues that can bind forming a disulphide bond in the folded protein affecting the digestibility in the gastric phase. Interestingly, peptides deriving from this region were observed in the gastric fractions of the WPI sample, probably due to a higher denaturation of the protein in this sample.
3.5. IgE binding of digested products
In the present study, the possible structural modifications induced by the processing did not affect the IgE binding capacity of undigested CW and WPI samples evaluated by inhibition ELISA (Fig. 5A). However, gastrointestinal digestion decreased the IgE binding capacity of both samples, but also differences among substrates were found. The WPI-G1 showed lower IgE binding capacity than CW-G1 (Fig. 5A), which could be related with the higher digestibility of α-lactalbumin observed in WPI, as indicated by the lower amount of intact α-lactalbumin identified by the UPLC-UV analysis (Fig. 5B) and the higher number of α-lactalbumin-derived peptides from WPI (Fig. 4). In fact, it has been described that the allergenicity of α-lactalbumin is mainly related to its conformational epitopes.42,47 Contrary to G1, WPI-G3 and WPI-G5 exhibited higher IgE reactivity than the corresponding CW counterparts (Fig. 5A). This cannot be ascribed to a higher degree of hydrolysis of CW, as indicated by the remaining intact protein determined by UPLC. Intact α-lactalbumin was not detected at this stage of digestion (Fig. 5B) but β-lactoglobulin was present to a greater extent in CW than in WPI in G3 samples (Fig. 5C). Higher degree of hydrolysis of β-lactoglobulin for the heated sample could have exposed linear IgE-binding epitopes. In fact, it has been shown that the allergenicity of β-lactoglobulin is mainly related to the presence of linear epitopes.42,47 In G5 the amount of β-lactoglobulin was similar in both digested meals (Fig. 5C), confirming the assumption that the higher IgE binding capacity observed for WPI could be related to the released peptides. Indeed, the β-lactoglobulin peptide f (83–95), KIDALNENKVLVL was exclusively found in the WPI gastric digests, while in CW digests, only fragments from the referred peptide were found. This peptide is comprised in the β-lactoglobulin epitope fragment 76–95, TKIPAVFKIDALNENKVLVL.43 Low IgE binding and no significant differences between substrates were observed in the intestinal samples except for digest I3 where the IgE binding capacity was higher for CW – than for WPI-I3 (Fig. 5A). Although a qualitative similar pattern of released peptides was observed for CW- and WPI-I3 samples, the IgE response could be attributed to quantitative differences of peptide epitopes.Differences in the digestibility of the main whey proteins, α-lactalbumin and β-lactoglobulin, between the two substrates could explain the IgE binding at the different time points. α-Lactalbumin was detected by UPLC only in the first gastric emptying point, showing a four times lower amount in the WPI sample. Indeed, the IgE binding of WPI resulted far below CW. For the gastric points G3 and G5, the peptides released, rather than the amount of intact protein, seemed to impact the immunological response, with prominent differences in the samples with medium-size (14-amino acid residues) fragments from reported epitopes. The higher digestibility of WPI could be related to the processing involved that may affect protein structures, denaturing and making them more accessible for the digestive enzymes. Differences in β-lactoglobulin digestibility between heated and unheated skimmed milk powder were reported in a dynamic gastric digestion study, resulting in higher resistance to digestion of the unheated sample.48 In another example, a digestion study performed with the semi-dynamic model on raw milk and UHT milk, reported a lower degree of hydrolysis of β-lactoglobulin in raw milk during the gastric phase.37
3.6. Transepithelial transport study of a lactosylated β-lactoglobulin peptide
Peptide derived from the previously mentioned β-lactoglobulin epitope, KIDALNENKVLVL, shows two lysine residues very likely lactosylated due to the thermal treatment (ESI Fig. S2†).4,32 This peptide was identified in the WPI gastric digests, with lactosylation at one or both lysine residues in addition to the unlactosylated molecule. Thus, the possible transport of this peptide under different forms through the intestinal barrier was tested. The peptide was synthesized obtaining a mixture, as confirmed by HPLC-MS/MS: mono-lactosylated peptide (36%), di-lactosylated peptide (10%), and unglycated peptide (53%).In the apical chamber from the Caco-2 culture, the three forms were identified with the following proportions: 42% of mono-lactosylated, 8% of di-lactosylated, and 50% of unmodified peptide. After 60 min of incubation the distribution in the apical side was: 57% lactosylated, 8% di-lactosylated, 35% unmodified peptide. This difference could be related to an easier transepithelial transport of the native peptide with respect to the glycated forms, in accordance to what has been described for other dietary Maillard reaction products26 or to greater susceptibility of the non-lactosylated peptide to the activity of the Caco-2 cells peptidases. Interestingly, in the basolateral side, both the unmodified (54%) and the mono-lactosylated (46%) peptides were found, while the di-lactosylated form could not be detected. Again, the proportion between the two forms suggests the native peptide to be more easily transported. However, the passage of the peptide linked to the carbohydrate moiety is not prevented. Table 2 shows the sequences of the parent peptide and fragments identified in the apical or basolateral sides in the Caco-2 cell culture. Although at least ten different forms were observed in the apical side, only five sequences were unequivocally detected in the basolateral chamber. The translocated forms correspond to the native peptide, its mono-lactosylated form, and three sequences with an asparagine at the N-terminal end. The C-terminus contains leucine or valine, where the single residue disappearance suggests the exopeptidase action by the brush border peptidases. The favoured transport of these three peptides with common N-terminal residue is consistent with an earlier observation of the importance of this amino acid for the transport of oligopeptides across the Caco-2 cell monolayer.49 The shorter forms were always identified as non-lactosylated products pointing again to a promoted transport of unglycated forms.
| Analyzed chamber | Theoretical massa | Observed mass | Sequence |
|---|---|---|---|
| a Monoisotopic. | |||
| Apical | 1467.87 | 1468.3 | KIDALNENKVLVL |
| 1793.87 | 1792.3 | KIDALNENKVLVL + Lact | |
| 2115.87 | 2113.0 | KIDALNENKVLVL + 2 Lact | |
| 1354.78 | 1355.1 | KIDALNENKVLV | |
| 813.50 | 813.8 | ENKVLVL | |
| 927.54 | 927.9 | NENKVLVL | |
| 1040.62 | 1040.9 | LNENKVLVL | |
| 1111.66 | 1112.0 | ALNENKVLVL | |
| 814.46 | 814.8 | NENKVLV | |
| 715.39 | 715.6 | NENKVL | |
| Basolateral | 1467.87 | 1467.2 | KIDALNENKVLVL |
| 1793.87 | 1793.4 | KIDALNENKVLVL + Lact | |
| 927.54 | 927.8 | NENKVLVL | |
| 814.46 | 814.7 | NENKVLV | |
| 715.39 | 715.7 | NENKVL | |
4. Conclusions
The semi-dynamic in vitro gastrointestinal digestion model was applied to study the effects of processing on whey protein digestion rate and allergenicity. Comparing the results obtained for the untreated CW sample and WPI, a more rapid digestion of the WPI sample was observed, resulting in more extensive protein digestibility in the form of released peptides. At 20 min of gastric digestion, intact α-lactalbumin was present at a higher amount in CW, showing higher IgE-binding, while in later digests, WPI showed greater reactivity, probably due to the appearance of linear epitopes. This supports the effects of processing on digestion and, as a consequence, on protein allergenicity, underlining the importance of kinetic digestion studies to evaluate this aspect. Interestingly it was found that peptide β-lactoglobulin KIDALNENKVLVL, a potential IgE-binding peptide, can be transported across the cell monolayer, both in the native and lactosylated forms, and the activity of the cell peptidases was observed by the generation of new shorter peptides. A deeper investigation on the role of lactosylated residues in the different absorption of the peptides would help to ascertain the route involved in the transport of the unmodified and modified peptides.Author contributions
A. G. Investigation, writing-original draft. S. B. Investigation, writing-review editing; T. T. supervision, writing-review editing. S. S. Supervision, writing-review editing, I. R. conceptualization, writing-review&editing. B. M. Conceptualization, writing-review&editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has received financial support from grant PID2019-107663RB-I00 and AGL2017-88864-R funded by MCIN/AEI/10.13039/501100011033. S. B. is supported by a Juan de la Cierva Incorporación contract (Ref. IJCI-2017-31345) funded by MCIN/AEI/10.13039/501100011033.References
- O. L. Ramos, R. N. Pereira, R. M. Rodrigues, J. A. Teixeira, A. A. Vicente and F. X. Malcata, Whey and whey powders: production and uses. Encyclopedia of food and health, Elsevier Ltd, 1st edn, 2015 Search PubMed.
- C. Morr and E. A. Foegeding, Composition and functionality of commercial whey and milk protein concentrated and isolates: A status report, Food Technol., 1990, 44, 100–112 CAS.
- J. Meltretter, J. Wüst and M. Pischetsrieder, Modified peptides as indicators for thermal and nonthermal reactions in processed milk, J. Agric. Food Chem., 2014, 62, 10903–10915 CrossRef CAS PubMed.
- S. Milkovska-Stamenova and R. Hoffmann, Identification and quantification of bovine protein lactosylation sites in different milk products, J. Proteomics, 2016, 134, 112–126 CrossRef CAS PubMed.
- M. A. J. S. Van Boekel, Effect of heating on Maillard reactions in milk, Food Chem., 1998, 62, 403–414 CrossRef CAS.
- H. E. Zenker, H. J. Wichers, M. M. M. Tomassen, S. Boeren, N. W. De Jong and K. A. Hettinga, Peptide release after simulated infant in vitro digestion of dry heated cow's milk protein and transport of potentially immunoreactive peptides across the caco-2 cell monolayer, Nutrients, 2020, 12, 1–24 CrossRef PubMed.
- A. Rérat, R. Calmes, P. Vaissade and P. A. Finot, Nutritional and metabolic consequences of the early Maillard reaction of heat treated milk in the pig. Significance for man, Eur. J. Nutr., 2002, 41, 1–11 CrossRef PubMed.
- S. M. Rutherfurd and P. J. Moughan, A new method for determining digesitble reactive lysines in foods, J. Agric. Food Chem., 1996, 44, 2202–2209 CrossRef.
- S. M. Rutherfurd and P. J. Moughan, Application of a new method for determining digestible reactive lysine to variably heated protein sources, J. Agric. Food Chem., 1997, 45, 1582–1586 CrossRef CAS.
- G. A. A. Van Lieshout, T. T. Lambers, M. C. E. Bragt and K. A. Hettinga, How processing may affect milk protein digestion and overall physiological outcomes: A systematic review, Crit. Rev. Food Sci. Nutr., 2020, 60, 2422–2445 CrossRef CAS PubMed.
- A. I. Mulet-Cabero, N. M. Rigby, A. Brodkorb and A. R. Mackie, Dairy food structures influence the rates of nutrient digestion through different in vitro gastric behaviour, Food Hydrocolloids, 2017, 67, 63–73 CrossRef CAS.
- A. I. Mulet-Cabero, L. Egger, R. Portmann, O. Ménard, S. Marze, M. Minekus, S. Le Feunteun, A. Sarkar, M. M. Grundy, F. Carrière, M. Golding, D. Dupont, I. Recio, A. Brodkorb and A. Mackie, A standardised semi-dynamic in vitro digestion method suitable for food- an international consensus, Food Funct., 2020, 11, 1702–1720 RSC.
- A. Brodkorb, L. Egger, M. Alminger, P. Alvito, R. Assunção, S. Ballance, T. Bohn, C. Bourlieu-Lacanal, R. Boutrou, F. Carrière, A. Clemente, M. Corredig, D. Dupont, C. Dufour, C. Edwards, M. Golding, S. Karakaya, B. Kirkhus, S. Le Feunteun, U. Lesmes, A. Macierzanka, A. R. Mackie, C. Martins, S. Marze, D. J. McClements, O. Ménard, M. Minekus, R. Portmann, C. N. Santos, I. Souchon, R. P. Singh, G. E. Vegarud, M. S. J. Wickham, W. Weitschies and I. Recio, INFOGEST static in vitro simulation of gastrointestinal food digestion, Nat. Protoc., 2019, 14, 991–1014 CrossRef CAS PubMed.
- J. D. Astwood, J. N. Leach and R. L. Fuchs, Stability of food allergens to digestion in vitro, Nat. Biotechnol., 1996, 14, 1269–1273 CrossRef CAS PubMed.
- R. D. J. Huby, R. J. Dearman and I. Kimber, Why are some proteins allergens?, Toxicol. Sci., 2000, 55, 235–246 CrossRef CAS PubMed.
- K. L. Bøgh and C. B. Madsen, Food Allergens: Is there a correlation between stability to digestion and allergenicity?, Crit. Rev. Food Sci. Nutr., 2016, 56, 1545–1567 CrossRef PubMed.
- T. Eiwegger, N. Rigby, L. Mondoulet, H. Bernard, M. T. Krauth, A. Boehm, E. Dehlink, P. Valent, J. M. Wal, E. N. Mills and Z. Szépfalusi, Gastro-duodenal digestion products of the major peanut allergen Ara h 1 retain an allergenic potential, Clin. Exp. Allergy, 2006, 36, 1281–1288 CrossRef CAS PubMed.
- M. Perusko, M. van Roest, D. Stanic-Vucinic, P. J. Simons, R. H. H. Pieters, T. Cirkovic Velickovic and J. J. Smit, Glycation of the major milk allergen β-Lactoglobulin changes its allergenicity by alterations in cellular uptake and degradation, Mol. Nutr. Food Res., 2018, 62, 1–12 CrossRef PubMed.
- A. Taheri-Kafrani, J. C. Gaudin, H. Rabesona, C. Nioi, D. Agarwal, M. Drouet, J. M. Chobert, A. K. Bordbar and T. Haertle, Effects of heating and glycation of β-lactoglobulin on its recognition by ige of sera from cow milk allergy patients, J. Agric. Food Chem., 2009, 57, 4974–4982 CrossRef CAS PubMed.
- J. Zhong, H. Yu, Y. Tu, L. Zhou, W. Liu, S. Luo, C. Liu and S. Prakash, Comparison of antigenicity and conformational changes to β-lactoglobulin following kestose glycation reaction with and without dynamic high-pressure microfluidization treatment, Food Chem., 2019, 278, 491–496 CrossRef CAS PubMed.
- G. P. Bosman, S. Oliveira, P. J. Simons, J. Sastre Torano, G. W. Somsen, L. M. J. Knippels, R. Haselberg, R. J. Pieters, J. Garssen and K. Knipping, Limited lactosylation of beta-lactoglobulin from cow's milk exerts strong influence on antigenicity and degranulation of mast cells, Nutrients, 2021, 13, 1–13 CrossRef PubMed.
- B. Wang and B. Li, Charge and hydrophobicity of casein peptides influence transepithelial transport and bioavailability, Food Chem., 2018, 245, 646–652 CrossRef CAS PubMed.
- C. Giromini, F. Cheli, R. Rebucci and A. Baldi, Invited review: Dairy proteins and bioactive peptides: Modeling digestion and the intestinal barrier, J. Dairy Sci., 2019, 102, 929–942 CrossRef CAS PubMed.
- R. Boutrou, J. Jardin, A. Blais, D. Tomé and J. Léonil, Glycosylations of κ-casein-derived caseinomacropeptide reduce its accessibility to endo- but not exointestinal brush border membrane peptidases, J. Agric. Food Chem., 2008, 56, 8166–8173 CrossRef CAS PubMed.
- S. Grunwald, R. Krause, M. Bruch, T. Henle and M. Brandsch, Transepithelial flux of early and advanced glycation compounds across Caco-2 cell monolayers and their interaction with intestinal amino acid and peptide transport systems, Br. J. Nutr., 2006, 95, 1221–1228 CrossRef CAS PubMed.
- M. Hellwig, S. Geissler, R. Matthes, A. Peto, C. Silow, M. Brandsch and T. Henle, Transport of free and peptide-bound glycated amino acids: Synthesis, transepithelial flux at Caco-2 cell monolayers, and interaction with apical membrane transport proteins, ChemBioChem, 2011, 12, 1270–1279 CrossRef CAS PubMed.
- M. Hellwig, S. Geissler, A. Peto, I. Knütter, M. Brandsch and T. Henle, Transport of free and peptide-bound pyrraline at intestinal and renal epithelial cells, J. Agric. Food Chem., 2009, 57, 6474–6480 CrossRef CAS PubMed.
- M. Santos-Hernández, F. Alfieri, V. Gallo, B. Miralles, P. Masi, A. Romano, P. Ferranti and I. Recio, Compared digestibility of plant protein isolates by using the INFOGEST digestion protocol, Food Res. Int., 2020, 137, 109708 CrossRef PubMed.
- J. Manguy, P. Jehl, E. T. Dillon, N. E. Davey, D. C. Shields and T. A. Holton, Peptigram: A web-based application for peptidomics data visualization, J. Proteome Res., 2017, 16, 712–719 CrossRef CAS PubMed.
- J. Sanchón, S. Fernández-Tomé, B. Miralles, B. Hernández-Ledesma, D. Tomé, C. Gaudichon and I. Recio, Protein degradation and peptide release from milk proteins in human jejunum. Comparison with in vitro gastrointestinal simulation, Food Chem., 2018, 239, 486–494 CrossRef PubMed.
- S. Benedé, I. López-Expósito, R. López-Fandiño and E. Molina, Identification of IgE-binding peptides in hen egg ovalbumin digested in vitro with human and simulated gastroduodenal fluids, J. Agric. Food Chem., 2014, 62, 152–158 CrossRef PubMed.
- A. Gasparini, S. Buhler, A. Faccini, S. Sforza and T. Tedeschi, Thermally-induced lactosylation of whey proteins: Identification and synthesis of lactosylated β-lactoglobulin epitope, Molecules, 2020, 25, 1294 CrossRef CAS PubMed.
- S. Fernández-Tomé, J. Sanchón, I. Recio and B. Hernández-Ledesma, Transepithelial transport of lunasin and derived peptides: Inhibitory effects on the gastrointestinal cancer cells viability, J. Food Compos. Anal., 2018, 68, 101–110 CrossRef.
- I. Hubatsch, E. G. E. Ragnarsson and P. Artursson, Determination of drug permeability and prediction of drug absorption in Caco-2 monolayers, Nat. Protoc., 2007, 2, 2111–2119 CrossRef CAS PubMed.
- D. Regazzo, D. Mollé, G. Gabai, D. Tomé, D. Dupont, J. Leonil and R. Boutrou, The (193–209) 17-residues peptide of bovine β-casein is transported through caco-2 monolayer, Mol. Nutr. Food Res., 2010, 54, 1428–1435 CrossRef CAS PubMed.
- R. A. Siciliano, M. F. Mazzeo, S. Arena, G. Renzone and A. Scaloni, Mass spectrometry for the analysis of protein lactosylation in milk products, Food Res. Int., 2013, 54, 988–1000 CrossRef CAS.
- A. I. Mulet-Cabero, A. R. Mackie, P. J. Wilde, M. A. Fenelon and A. Brodkorb, Structural mechanism and kinetics of in vitro gastric digestion are affected by process-induced changes in bovine milk, Food Hydrocolloids, 2019, 86, 172–183 CrossRef CAS.
- G. Miranda, G. Hazé, P. Scanff and J. P. Pélissier, Hydrolysis of α-lactalbumin by chymosin and pepsin. Effect of conformation and Ph, Lait, 1989, 69, 451–459 CrossRef CAS.
- G. Picariello, P. Ferranti, O. Fierro, G. Mamone, S. Caira, A. S. Di Luccia, S. Monica and F. Addeo, Peptides surviving the simulated gastrointestinal digestion of milk proteins: Biological and toxicological implications, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2010, 878, 295–308 CrossRef CAS PubMed.
- C. Bär, D. Mathis, P. Neuhaus, D. Dürr, W. Bisig, L. Egger and R. Portmann, Protein profile of dairy products: Simultaneous quantification of twenty bovine milk proteins, Int. Dairy J., 2019, 97, 167–175 CrossRef.
- H. Hochwallner, U. Schulmeister, I. Swoboda, M. Focke-Tejkl, V. Civaj, N. Balic, M. Nystrand, A. Härlin, J. Thalhamer, S. Scheiblhofer, W. Keller, T. Pavkov, D. Zafred, B. Niggemann, S. Quirce, A. Mari, G. Pauli, C. Ebner, N. G. Papadopoulos, U. Herz, E. A. van Tol, R. Valenta and S. Spitzauer, Visualization of clustered IgE epitopes on α-lactalbumin, J. Allergy Clin. Immunol., 2010, 125, 1279–1285 CrossRef CAS PubMed.
- K. M. Järvinen, P. Chatchatee, L. Bardina, K. Beyer and H. A. Sampson, IgE and IgG binding epitopes on α-Lactalbumin and β-Lactoglobulin in cow's milk allergy, Int. Arch. Allergy Immunol., 2001, 126, 111–118 CrossRef PubMed.
- H. Matsuo, T. Yokooji and T. Taogoshi, Common food allergens and their IgE-binding epitopes, Allergol. Int., 2015, 64, 332–343 CrossRef CAS PubMed.
- I. Cerecedo, J. Zamora, W. G. Shreffler, J. Lin, L. Bardina, M. C. Dieguez, J. Wang, A. Muriel, B. de la Hoz and H. A. Sampson, Mapping of the IgE and IgG4 sequential epitopes of milk allergens with a peptide microarray-based immunoassay, J. Allergy Clin. Immunol., 2008, 122, 589–594 CrossRef CAS PubMed.
- Y. J. Cong and L. F. Li, Identification of the critical amino acid residues of immunoglobulin E and immunoglobulin G epitopes in α-Lactalbumin by alanine scanning analysis, J. Dairy Sci., 2012, 95, 6307–6312 CrossRef CAS PubMed.
- R. Boutrou, C. Gaudichon, D. Dupont, J. Jardin, G. Airinei, R. Benamouzig and D. Tome, Sequential release of milk protein – derived bioactive peptides in the jejunum in healthy humans, Am. J. Clin. Nutr., 2013, 97, 1314–1323 CrossRef CAS PubMed.
- G. Bu, Y. Luo, Z. Zheng and H. Zheng, Effect of heat treatment on the antigenicity of bovine α-lactalbumin and β-lactoglobulin in whey protein isolate, Food Agric. Immunol., 2009, 20, 195–206 CrossRef CAS.
- L. Sánchez-Rivera, O. Ménard, I. Recio and D. Dupont, Peptide mapping during dynamic gastric digestion of heated and unheated skimmed milk powder, Food Res. Int., 2015, 77, 139–132 CrossRef.
- L. Ding, L. Wang, Z. Yu, S. Ma, Z. Du, T. Zhang and J. Liu, Importance of terminal amino acid residues to the transport of oligopeptides across the Caco-2 cell monolayer, J. Agric. Food Chem., 2017, 65, 7705–7712 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1fo04102a |
| ‡ Present address: Faculty of Science and Technology, Free university of Bozen-Bolzano, Piazza Università 5, 39100, Bolzano, Italy. |
| This journal is © The Royal Society of Chemistry 2022 |