Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quantification by SIFT-MS of volatile compounds emitted by Aspergillus fumigatus cultures and in co-culture with Pseudomonas aeruginosa, Staphylococcus aureus and Streptococcus pneumoniae
Thomas W. E.
Chippendale
a,
Francis J.
Gilchrist
ab,
Patrik
Španěl
ac,
Alice
Alcock
d,
Warren
Lenney
ab and
David
Smith
*a
aInstitute for Science and Technology in Medicine, School of Medicine, Keele University, Thornburrow Drive, Hartshill, ST4 7QB, Stoke-on-Trent, UK. E-mail: d.smith@keele.ac.uk
bAcademic Department of Child Health, University Hospital of North Staffordshire, Newcastle Road, ST4 6QG, Stoke-on-Trent, UK
cJ. Heyrovský Institute of Physical Chemistry, Academy of Sciences of the Czech Republic, Dolejškova 3, 182 23, Prague 8, Czech Republic
dDepartment of Microbiology, University Hospital of North Staffordshire, Newcastle Road, ST4 6QG, Stoke-on-Trent, UK
First published on 31st July 2014
Abstract
Following our recent in vitro study of the volatile compounds emitted into the gas phase by the respiratory pathogens Pseudomonas aeruginosa (PA), and most recently Staphylococcus aureus (SA), Streptococcus pneumoniae (SP) and Haemophilus influenzae (HI), we have extended this work to the investigation of the volatile compounds emitted by in vitro cultures of the common respiratory fungus Aspergillus fumigatus (AF). The measurements were achieved using selected ion flow tube mass spectrometry (SIFT-MS) by which real time analyses of trace volatile compounds can be achieved without disturbing the cultures. It is seen that copious amounts of ammonia and the organosulphur compounds methanethiol, dimethyl sulphide and dimethyl disulphide are produced by AF cultures. These may be sufficient to allow for non-invasive detection of the AF in the airways of infected patients by breath analysis. AF also efficiently absorbs and metabolises the aldehydes acetaldehyde, butanal and pentanal from the supportive medium (brain-heart infusion broth). Preliminary studies of the volatile compounds emitted by co-cultures of AF with PA, SA and SP revealed that the biomarker HCN (for PA) is not compromised by the presence of AF, and the organosulphur compounds (for AF) are not compromised by the presence of SA or SP.
Introduction
Lower respiratory tract infection (LRTI) with bacteria or fungus is a major cause of morbidity and mortality in children across the world. Early diagnosis is vital to allow appropriate management and prevent serious complications. Unfortunately, it is often difficult to gain a microbiological diagnosis in children with a LRTI as they are rarely able to expectorate sputum. Diagnosis therefore relies on cough swabs, which are unreliable, or bronchoalveolar lavage (BAL) samples taken during bronchoscopy, which is an invasive procedure and requires a general anaesthetic. Due to these issues, a non-invasive and child-friendly method of detecting lower airway pathogens is very appealing. Breath analysis for volatile biomarkers, specific to certain lung and airway pathogens, is a promising approach to non-invasive diagnosis. There is a now a body of experimental data on the generation of volatile compounds by specific pathogens cultured in vitro that is providing growing optimism.1,2 Exhaled breath analysis is a painless procedure that is acceptable to children and adults alike.3,4 It potentially offers the prospect of early, accurate diagnosis that will promote optimal clinical care of respiratory infections.The above in vitro work has mostly been pursued by studying the volatile compounds emitted by a number of pathogens using gas chromatography mass spectrometry (GC-MS) often combined with trace compound extraction and pre-concentration techniques, most commonly solid phase microextraction (SPME). Thus, a wide variety of volatile organic compounds (VOCs) have been seen to be generated, the patterns of which are commonly analysed using statistical methods such as multivariate analyses, including principal component analysis (PCA) by which distinctions between different in vitro bacterial cultures have been identified.5,6 Our work in this area has involved a detailed investigation of the bacterium Pseudomonas aeruginosa (PA) that colonises the lungs and airways of cystic fibrosis (CF) sufferers, and over a decade of research involving in vitro and in vivo (breath analysis) studies, we have established that gaseous hydrogen cyanide is a true biomarker of PA infection.7–10 Also, the volatile metabolite methylthiocyanate has also been detected in PA cultures and is thus an additional potential breath indicator of PA infection in CF patients.11 The details of these wide-ranging studies have recently been published.12 This work has been greatly facilitated by our development of the selected ion flow tube mass spectrometry analytical technique (SIFT-MS) by which real time, on-line analyses of single breath exhalations can be performed, obviating sample collection, and also analysis of other humid air media, including mammalian13,14 and bacterial15–17 culture headspace. Analyses using SIFT-MS are accurately quantitative and rapid such that time variations in trace compounds/metabolites can be followed as in human pharmacokinetics,18,19 and analyses of culture headspace that can be followed with time avoiding direct sampling or otherwise disturbing the cultures.16,17
In a very recent paper we have reported the results of a study, using SIFT-MS, of the volatile compounds emitted by in vitro cultures of three common respiratory bacteria, Staphylococcus aureus (SA), Streptococcus pneumoniae (SP) and Haemophilus influenzae (HI).15 The unique aspect of this work is that accurate quantification was achieved for several common organic compounds, mainly alcohols, ketones and aldehydes, which were found to be greatly elevated in the headspace of the SA and SP cultures. Although some of these compounds are present in the exhaled breath of healthy individuals, the emission rates of these compounds by SA and SP are so great that when present in the airways the compounds are likely to be well above their endogenously generated levels. Hence, real time breath analysis by SIFT-MS may well allow detection of these bacteria in airways. Studies are now planned to investigate this possible diagnostic application.
As an extension to this work we have carried out a similar study of the volatile emissions from in vitro cultures of the common fungus Aspergillus fumigatus (AF). AF is a potent cause of airways infection and associated diseases known as aspergilloses.20 These particularly affect immunocompromised individuals and are a major cause of morbidity and mortality.21 Inhalation of the airborne fungal spores by patients with cystic fibrosis (CF) or asthma can result in primary airway infection (Aspergillus bronchitis) or a disease related to an allergic response to the spores (allergic bronchopulmonary aspergillosis – ABPA).20,22 AB affects between 6–25% of CF patients and 1–2% of asthma patients.20 The diagnosis is based on the culture of AF from sputum or BAL samples, chest X-ray changes and the detection of elevated antibody levels in the serum.20 The ability to confirm the detection of AF in the airways by breath analysis is very exciting, especially in children and adults who cannot expectorate sputum.
There has been some previous exploratory work on AF culture emissions. In a study by Bazemore and coworkers using GC-MS,23 AF was cultured on media with elastin, a connective tissue protein found in the lung, and the fungus was also used to infect human lung cell cultures. It was found that sesquiterpenes, including both α-farnesene and β-farnesene isomers, were emitted by the AF cells in both cases. Similarly, SPME-GC-MS was used to analyse volatile compounds emitted by AF and several other microbes cultured on Sabouraud dextrose agar. AF was found to produce only β-farnesene and it was speculated that this might be a suitable biomarker for AF detection. In a SIFT-MS study, the headspace of AF cultures on malt extract agar were analysed after 48 and 72 h culture.24 Methanol, acetone, isoprene and dimethyl sulphide were produced and in some cases the application of the antifungal agents benomyl and tebuconazole prevented the release of these volatile compounds. An electronic nose hybrid sensor array has also been used to distinguish between the volatiles produced by AF with and without antifungal agents.24
Using SPME-GC-MS, the volatile compound 2-pentylfuran has been detected in the headspace of cultures of several different Aspergillus species and Streptococcus pneumoniae. It was also detected in the exhaled breath of four CF patients with AF lung colonization.25 The same group extended their work by a recent, more extensive study in which 2-pentylfuran was again detected in the headspace above AF cultures and the breath exhalations of 10 patients with neutropenia and 32 patients with lung disease. Fourteen healthy controls were also analysed. The detection of 2-pentlyfuran using the group's SPME-GC-MS methods detected AF-lung infection in the subjects with sensitivity and specificity of 77% and 78% respectively.26 Note, however, that 2-pentylfuran was not detected in AF culture headspace using SPME-GC-MS in the studies by Bazemore et al.23 and Lin et al.27
Thus, the present study is a continuation of our own SIFT-MS studies and similar work outlined above. The ultimate aim is the development of non-invasive methods for the detection of AF in the airways of patients with CF, asthma and diseases causing immunodeficiency. The compounds present in the headspace above cultures of clinical AF isolates have been identified and quantified using SIFT-MS. Additionally, some AF cultures were also infected/co-cultured with PA, SA and SP to investigate if the differing volatile species identified by our recent SIFT-MS research7–11,15 could be identified within such co-cultures or whether they had been modified.
Experimental
Preparation of fungal and bacterial cultures
The microbes included in this study were clinical isolates of AF fungus and the three bacteria species, viz. PA, SA and SP. The microbes were initially cultured overnight on agar for positive identification. Saline suspensions of the AF spores were produced. The turbidity of the suspensions was adjusted to approximately 0.5 optical density units, visually assessed by comparison with McFarland standards. 0.5 mL of these saline suspensions was used to inoculate 10 mL volumes of brain heart infusion (BHI) broth (Fisher Scientific) contained inside 150 mL glass bottles. In total, 5 clinical isolates were used to seed 15 AF cultures while 15 bottles containing BHI broth alone, without microbes, were prepared for comparison/controls. The bottles were immediately sealed with rubber septa and incubated at 37 °C without agitation. This method of culture preparation is adapted from that described in a recent paper.15 Some experiments were carried out on binary co-cultures of AF with PA, SA or SP, the volatile biomarkers from which have been determined by previous SIFT-MS studies under similar conditions.7–11,15 Each of the fungal/bacterial species was a clinical isolate and was first cultured overnight on agar plates and identified. These co-cultures were produced by preparing saline solutions for each (0.5 optical density units), and transferring 0.25 mL to the glass bottles containing 10 mL of BHI broth. The pH values for some of the cultures were measured following incubation; prior to incubation the pH of the BHI broth alone was measured as 7.5.Culture headspace analysis by SIFT-MS
The measurements of the concentrations of volatile biomarkers in the headspace of the microbial cultures were carried out using a Profile 3 selected ion flow tube mass spectrometer (SIFT-MS; Instrument Science, UK). The principle of the SIFT-MS technique has been fully described in previous articles.28,29 In short, a mixture of reagent ions is created in a microwave cavity discharge ion source and from this mixture a current of reagent ions of a given mass-to-charge ratio (m/z) is obtained using a quadrupole mass filter. Thus, the reagent ions (separately, H3O+, NO+ or O2+˙ ions) are injected into a fast-flowing inert carrier gas, usually pure helium, through a Venturi-type inlet. Hence, a swarm of thermalized ions is created that is convected along a flow tube. A sample of humid air (e.g. the fungal cell culture headspace) is introduced into the ion swarm via a heated sampling line coupled directly to a sample inlet port. Importantly, sample collection into bags or pre-concentration onto traps is avoided. The reagent ions then react with the trace volatile analyte compounds present in the sample during a well-defined reaction time. Bimolecular (binary) and termolecular (ternary) reactions occur that lead to conversion of a small fraction of the reagent ions to the analyte ions characteristic of the neutral analytes. The remaining (large fraction) of the reagent ions together with the analyte ions are sampled from the flowing swarm via a pinhole orifice (∼0.3 mm diameter) located at the downstream end of the flow tube and are directed into a differentially-pumped quadrupole mass spectrometer. After m/z analysis, the ions are detected and counted by an electron multiplier/pulse counting system, the characteristic analyte ions identify the neutral analyte compounds and the on-board computer immediately calculates their concentrations in the sample with the aid of a kinetics library compiled from numerous studies of ion–molecule reactions.30In the present experiments, sampling of the culture headspace was achieved by penetrating the septa that sealed the glass bottles (containing the cell cultures) with a hypodermic needle connected directly to the heated sampling line of the SIFT-MS instrument. The liquid culture volume was typically 10 mL and the headspace volume was typically 140 mL. The sample flow rate, around 30 mL min−1 in these experiments, is established by a short capillary in the sampling line. To maintain a steady constant sample flow rate, it was necessary to maintain the pressures inside the bottles at 1 bar by balancing the flow loss of headspace gas/vapour using a flexible bag containing dry, sterile cylinder air. During the sampling period, typically 100 seconds in the present experiments, some dilution of the headspace occurs due to the balancing dry air introduction resulting in a small reduction in the volatile compound concentrations. These are quickly re-established following the completion of the brief sampling when the bottle/culture remains sealed during subsequent incubation periods.
Analysis of the headspace of the cultures was performed with the SIFT-MS instrument operated in the Full Scan (FS) mode28 by which a complete mass spectrum of both reagent and analyte ions is obtained by scanning the analytical quadrupole mass spectrometer over a selected m/z range for a chosen time whilst the sample to be analysed is introduced into the helium carrier gas. The resulting mass spectrum (see Fig. 1 later) is interpreted by relating the characteristic analyte ions to the neutral trace volatile compounds present in the sample using the acquired knowledge of the ion chemistry. The concentrations of the individual trace gases can be calculated using the in-built kinetics library, which is constructed from the known ion chemistry. The FS mode of data acquisition provides sufficiently accurate analyses when the spectral line intensities are high, as is the case for the fungal/bacterial headspace samples involved in the present study. Each sample (AF cultures/co-cultures and BHI medium alone) was scanned using each of the reagent ions (H3O+, NO+ or O2+˙) for 100 seconds across an m/z range of 10 to 180.
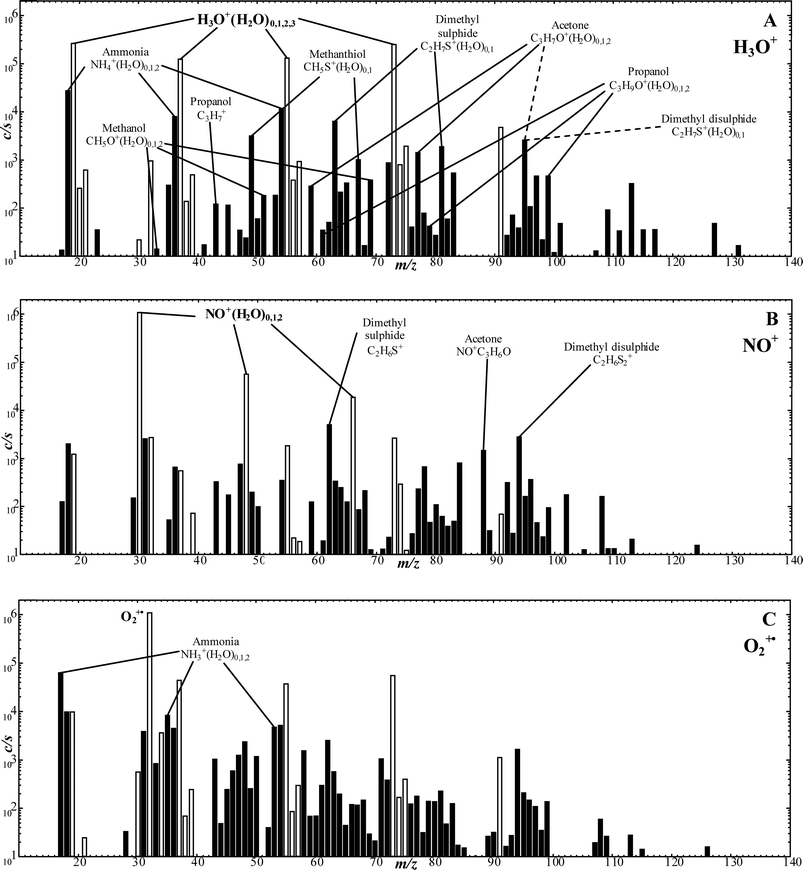 | ||
| Fig. 1 SIFT-MS full scan mass spectra (ion mass-to-charge ratio, m/z, plotted against the ion counts-per-second, c/s) showing the compounds present in the headspace of A. fumigatus cultures when analysed employing H3O+ (A), NO+ (B) and O2+˙ (C) as the reagent ion. The cultures were incubated at 37 °C for 72 h prior to the headspace analysis. The analyte ions of the trace compounds produced by AF, as referred to in Table 1 and Fig. 2, are indicated on the mass spectra. Note that small fractions of the hydronium ion and its hydrates ((H3O+)0,1,2,3) at m/z 19, 37, 55, 73 are also present in the NO+ and O2+˙ spectra (B and C respectively), which are shown with open bars. | ||
Principal component analysis
Principal component analysis (PCA) represents the most basic and well understood approach to multivariate analysis of complex data. PCA methods have previously been used for the analysis of bacterial culture headspace data obtained using secondary electrospray ionisation mass spectrometry (SESI-MS) when it was seen that the species were clearly separated based on the volatile compounds they emitted.6,31,32 Data interpretation using PCA has previously been used in similar PTR-MS analyses.33 The use of multivariate methods for data analysis in SIFT-MS is still in its infancy but has recently been described by others34 and in our recent paper.15The aim of PCA is to reduce the dimensions of the data, which means reducing the number of variables involved (thereby allowing graphical representation of combinations of concentrations of several compounds in a two dimensional plot, for example). PCA describes the relationship between variables and observations/cases and also identifies data outliers. It can be used for visualisation of multidimensional data (in our case profiles of concentrations of several VOCs) by rotating the multidimensional coordinate system and projecting the data points into fewer dimensions. PCA is very useful for two-dimensional (and sometimes, somewhat misleadingly and questionably, three-dimensional) graphical visualization of multidimensional data, which can be used to reveal different groups of data points. The input data for PCA is in the form of a matrix containing concentrations of compounds (rows correspond to individual observations and columns to different compounds). The eigenvalues calculated as a result of the PCA provide a measure of the variation that is described by each principal component. The largest eigenvalue is related to the first principal component, PC1. For each principal component a corresponding eigenvector is calculated as the projection of the individual compounds to the resulting principal component coordinates and these eigenvectors can be included in the plots of the transformed data points as arrows indicating the projected directions corresponding to the original individual compounds.
In the present study, for PCA analyses the sets of the full scan SIFT-MS mass spectral data for different cultures (i.e. ion peak intensity tables obtained from mass spectra such as those shown in Fig. 1) were first used to calculate concentrations of volatile compounds from the counts rates of the reagent and characteristic analyte ions.30T-tests were then used to identify those volatile compounds with headspace concentrations that were significantly altered in bacterial cultures relative to medium alone at the three culture incubation time points (24, 48 and 72 h). Compounds for which the differences were more significant as indicated by p < 0.05 were included in the PCA analysis (using the pcaMSwin software by Dryahina and Španěl35). The results for these PCA analyses of the current data are presented and discussed later.
Results
Analysis of the headspace above cultures of A. fumigatus (AF)
In accordance with the recipe described above, 15 BHI broth samples and 15 AF cultures derived from 5 isolates were prepared in the glass bottles over an experimental period of a few weeks. Three separate, ostensibly identical cultures were prepared from each isolate and the headspace of each was analysed after 24, 48 and 72 hours of culture at 37 °C in the sealed glass bottles without agitation to allow the headspace to develop and the volatile compounds to equilibrate with their liquid concentrations. Three BHI broth samples were also held at the same temperature and the headspace of each was similarly analysed concomitant with the AF cultures. Each BHI broth and each AF culture headspace was analysed by SIFT-MS using the three available reagent ions (H3O+, NO+, O2+˙), since it is known from numerous previous studies36–39 that, for example, alcohols are best analysed using H3O+, most organosulphur compounds with NO+ and ammonia with O2+˙, as indicated later in Table 1. Thus, 9 analytical spectra were obtained for each AF isolate culture at each incubation time realising 45 spectra for the AF cultures at each incubation time. Sample spectra are shown in Fig. 1 as obtained for all three reagent ion species; note that characteristic analyte are identifiable for some compounds on both the H3O+ and NO+ reagent ion spectra.| Compound | Sample | Concentration (ppbv) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | ||||||||
| Mean | SE | Range | Mean | SE | Range | Mean | SE | Range | ||
| a Note that sophisticated analytical methods are required to quantify acetaldehyde in the presence of dimethyl sulphide by SIFT-MS.40 | ||||||||||
| Ammonia | BHI | 2071 | 136 | 1566–3373 | 2112 | 103 | 1618–2994 | 2096 | 74 | 1557–2607 |
| (O2+˙) | ||||||||||
| NH3+(H2O)0,1 | AF | 5626 | 635 | 1668–9051 | 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 329 329 |
1166 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 199–28 199–28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 017 017 |
38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 382 382 |
2338 | 23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 312–60 312–60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 655 655 |
| 17, 35 | ||||||||||
| Methanol | BHI | 74 | 7 | 35–124 | 82 | 12 | 12–201 | 72 | 8 | 24–128 |
| (H3O+) | ||||||||||
| CH5O+(H2O)0,1,2 | AF | 361 | 69 | 55–858 | 695 | 47 | 380–1002 | 721 | 55 | 450–118 |
| 33, 51, 69 | ||||||||||
| Ethanol | BHI | 117 | 6 | 86–162 | 106 | 15 | 46–274 | 88 | 7 | 44–154 |
| (H3O+) | ||||||||||
| C2H7O+(H2O)0,1,2 | AF | 309 | 126 | 86–2018 | 434 | 57 | 61–844 | 262 | 50 | 107–720 |
| 47, 65, 83 | ||||||||||
| Propanol | BHI | 33 | 3 | 11–56 | 27 | 4 | 6–52 | 32 | 3 | 16–56 |
| (H3O+) | ||||||||||
| C3H7+; C3H9O+(H2O)0,1,2 | AF | 27 | 6 | 0–92 | 246 | 33 | 50–419 | 277 | 36 | 0–451 |
| 43, 61, 79, 97 | ||||||||||
| Acetone | BHI | 500 | 41 | 283–835 | 482 | 40 | 322–800 | 477 | 41 | 303–852 |
| (NO+) | ||||||||||
| NO+(C3H6O) | AF | 723 | 26 | 579–959 | 778 | 46 | 517–1126 | 1109 | 121 | 509–2001 |
| 88 | ||||||||||
| Methanthiol | BHI | 11 | 2 | 0–31 | 7 | 2 | 0–21 | 7 | 2 | 0–20 |
| (H3O+) | ||||||||||
| CH4OS+(H2O)0,1,2 | AF | 1787 | 390 | 7–4367 | 2744 | 211 | 1633–4360 | 1668 | 192 | 938–3332 |
| 49, 67, 85 | ||||||||||
| Dimethyl sulphide | BHI | 5 | 1 | 0–10 | 4 | 1 | 0–11 | 3 | 1 | 0–8 |
| (NO+) | ||||||||||
| C2H6S+ | AF | 184 | 63 | 3–841 | 2776 | 311 | 298–5436 | 1571 | 210 | 745–3061 |
| 62 | ||||||||||
| Dimethyl disulphide | BHI | 37 | 4 | 8–56 | 24 | 3 | 0–45 | 25 | 4 | 2–51 |
| (NO+) | ||||||||||
| C2H6S2+ | AF | 290 | 45 | 21–599 | 983 | 94 | 519–1931 | 963 | 94 | 367–1596 |
| 94 | ||||||||||
| Acetaldehydea | BHI | 865 | 42 | 603–1206 | 944 | 51 | 642–1258 | 929 | 54 | 563–1414 |
| (H3O+) | ||||||||||
| C2H5O+(H2O)0,1,2 | AF | 11 | 4 | 0–43 | 59 | 26 | 0–316 | 32 | 21 | 0–298 |
| 45, 63, 81 | ||||||||||
| Butanal | BHI | 134 | 9 | 86–199 | 137 | 9 | 97–238 | 136 | 6 | 100–174 |
| (NO+) | ||||||||||
| C4H7O+; NO+C4H8O | AF | 2 | 1 | 0–10 | 6 | 2 | 0–26 | 4 | 1 | 0–14 |
| 71, 102 | ||||||||||
| Pentanal | BHI | 532 | 37 | 317–793 | 536 | 35 | 364–725 | 497 | 27 | 308–704 |
| (NO+) | ||||||||||
| C5H9O+; NO+C5H10O | AF | 3 | 1 | 0–12 | 14 | 6 | 0–77 | 5 | 1 | 0–15 |
| 85, 116 | ||||||||||
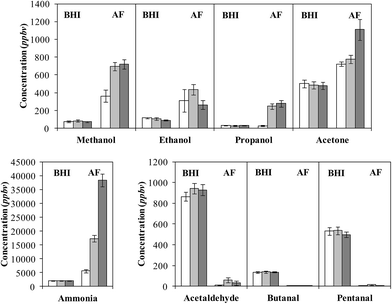
No attempt was made to explore differences between the headspace data for the 5 separate isolates, but cursory inspections revealed no major differences. Obvious is the consistent appearance in the spectra of characteristic analyte ions of 10 compounds in the headspace, the concentrations of which varied appreciably between the 5 isolates. The detailed data, including the mean concentration values for all 5 isolates, the standard error of the mean and the concentration ranges for each compound at the three incubation times, in parts-per-billion by volume (ppbv) are given in Table 1. To facilitate interpretation of these data, vertical bar charts of the AF culture headspace concentrations at the three incubation time vis-à-vis the BHI medium headspace concentrations alone are shown in Fig. 2, which immediately reveals clear groupings and trends that will now be summarised.
Ammonia, a very basic compound, is present at easily measurable but stable levels in the BHI broth media, but continuously increases in the AF headspace during the incubation period reaching a very high concentration around 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ppbv at 72 hours of incubation. For ammonia, the partition between the gas phase (headspace) concentration and the liquid phase concentration is critically dependent on the pH of the liquid. After 72 h incubation at 37 °C the pH of the AF culture was somewhat lower (6.9, slightly acidic) than that of the BHI broth alone (7.5, alkaline). Given the high concentration of ammonia in the headspace at pH 6.9, and knowing that the Henry's law coefficient for ammonia is approximately 34 mol kg−1 bar−1 at 37 °C (determined using figures given in ref. 41), the liquid phase concentration of ammonia, NH3, must be at the millimolar level after 72 h of culture together with a very high concentration of ammonium ions, NH4+. AF is known to produce ammonia/ammonium via a number of different pathways42 and indeed plays an important role as a nitrogen recycler in the environment.43 Ammonia is not produced at such high levels by the common respiratory bacterium species SA and SP;15 these bacteria are also involved in the present study in co-cultures with AF, as is seen later. Thus, it is conceivable that AF infection of the airways could lead to elevated levels of nose-exhaled ammonia and, ipso facto, of mouth-exhaled ammonia.44
000 ppbv at 72 hours of incubation. For ammonia, the partition between the gas phase (headspace) concentration and the liquid phase concentration is critically dependent on the pH of the liquid. After 72 h incubation at 37 °C the pH of the AF culture was somewhat lower (6.9, slightly acidic) than that of the BHI broth alone (7.5, alkaline). Given the high concentration of ammonia in the headspace at pH 6.9, and knowing that the Henry's law coefficient for ammonia is approximately 34 mol kg−1 bar−1 at 37 °C (determined using figures given in ref. 41), the liquid phase concentration of ammonia, NH3, must be at the millimolar level after 72 h of culture together with a very high concentration of ammonium ions, NH4+. AF is known to produce ammonia/ammonium via a number of different pathways42 and indeed plays an important role as a nitrogen recycler in the environment.43 Ammonia is not produced at such high levels by the common respiratory bacterium species SA and SP;15 these bacteria are also involved in the present study in co-cultures with AF, as is seen later. Thus, it is conceivable that AF infection of the airways could lead to elevated levels of nose-exhaled ammonia and, ipso facto, of mouth-exhaled ammonia.44
The alcohols methanol and ethanol are often produced efficiently by bacteria, but they are at low levels in the BHI broth medium headspace and increase to only modest levels in the AF cultures headspace, as can be seen in Fig. 2. Production apparently ceased after 48 h incubation, which may be indicative of a change in metabolism. This contrasts with the SA and SP bacteria that produce both alcohols, especially ethanol at very high levels,15 which differentiates these bacteria from the AF fungus. As with SA and SP, propanol was produced to a much lesser extent,15 and it was only observed after 48 h of incubation. Acetone was also present in the BHI medium headspace and only increased by about 50% in the AF culture headspace, although this ketone actually decreased from medium levels in the headspace of SA and SP cultures.
Two other groups of compounds are detected in the AF headspace; three organosulphur compounds and three aldehydes, as are listed in Table 1 and represented in the bar charts in Fig. 2. The clear distinction between these two groups is that whilst the organosulphur compounds are essentially absent in the medium headspace and clearly increase during culture in the AF headspace, the reverse is true for the aldehydes in that the headspace levels of the aldehydes in the medium headspace is rapidly reduced to close to nothing in the AF culture headspace.
The organosulphur compounds methanethiol, dimethyl sulphide (DMS) and dimethyl disulphide (DMDS) reach their maximum headspace concentration, or close to, after 48 h of culture; indeed, it is apparent that the methanethiol and dimethyl sulphide were apparently depleted post 48 h of culture. It was noticeable during the course of the study that the characteristic green AF spores were only visible following 48 h of culture, and so the production of these organosulphur compounds may be indicative of a change in metabolism due to sporulation. Production of organosulphur compounds was not seen in our similar SIFT-MS studies of the SA and SP cultures15 which, in this way, differentiates the fungal AF from the bacterial SA and SP.
The aldehydes acetaldehyde, butanal and pentanal are apparently consumed by the AF cells. Aldehyde dehydrogenase (ALDH) enzymes, which convert aldehydes to carboxylic acids, are expressed by AF cells,42 and these are the likely catalysts for this aldehyde metabolism. AF is also capable of converting aldehydes to alcohols by fermentation,42 and given the small but obvious increase in ethanol concentration, it is also possible that fermentation was occurring during the incubation period. The destruction/metabolism of these aldehydes often occurs in mammalian cultures,45–48 but bacteria very often generate these aldehydes as is the case for the SA and SP bacterial cultures that efficiently produce acetaldehyde in particular.15 This might well be another distinguishing property of these SA and SP bacterial cultures and the AF fungal culture.
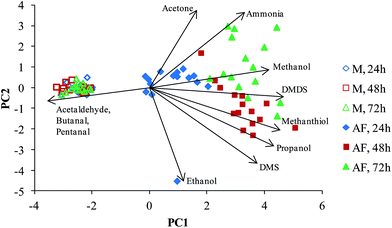
The PCA analysis of the concentrations data of the selected compounds (using the criterion of the significance of the difference p < 0.05) were carried out as described in the Experimental section resulting in the plot shown in Fig. 3. A clear separation between the culture and media headspace samples is observed. The arrows (eigenvectors) indicate that the media headspace is largely characterised by relatively high aldehydes concentrations, which is due to the aldehyde consumption by the AF cells, as discussed earlier. Interestingly, the PCA plot separates the AF samples by their incubation period. Propanol, methanethiol and DMS in particular are shown to be produced during the first 48 hours, and ammonia, methanol and DMDS the major compounds produced after 72 hours, but these trends are clearly evident on inspection of the data given in Table 1 and Fig. 2. Note the apparent outlier for the AF culture after 24 h incubation, which is a result of the ethanol concentration in one culture being 2018 ppbv as opposed to the mean level of 309 ppbv in all the 24 h cultures. The ethanol was apparently depleted from this culture during the second 24 h of culture.
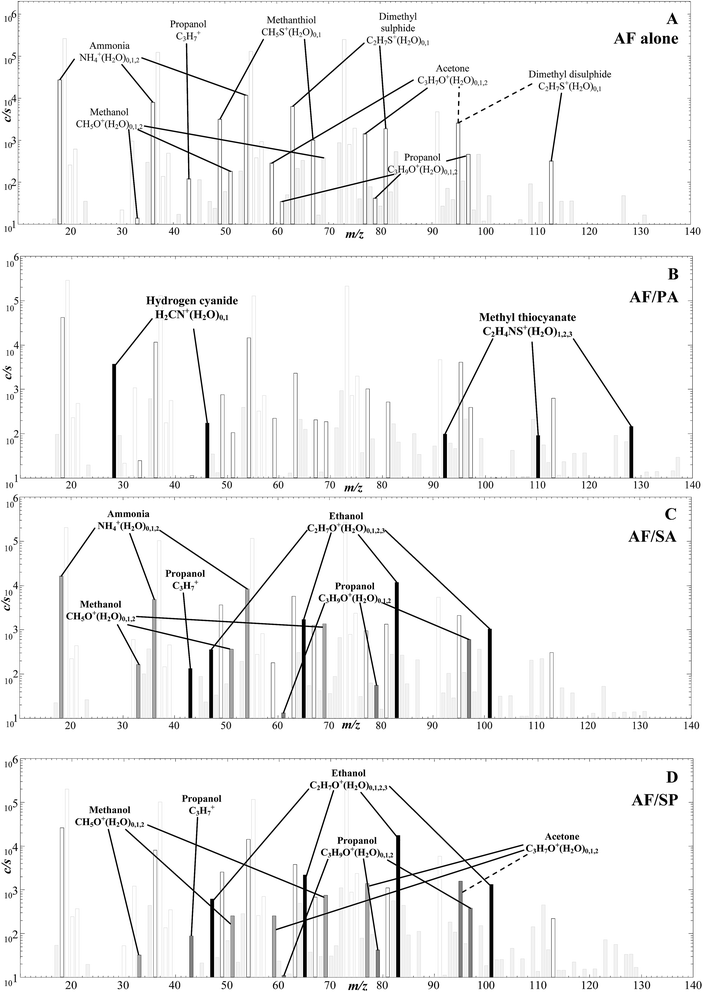
Headspace analysis of AF co-cultures with three species of respiratory bacteria
In this limited exploratory study, just 3 co-cultures of AF with each of the respiratory bacteria Pseudomonas aeruginosa (PA), Staphylococcus aureus (SA) and Streptococcus pneumoniae (SP) were produced to investigate if any obvious interactions between the bacterial and fungal cells were manifest and if discreet biomarkers of the separate species were still identifiable. Thus, the headspaces of the co-cultures were analysed using only H3O+ reagent ions and sample spectra are shown in Fig. 4. The characteristic analyte ions for the AF culture alone in Fig. 4A are as given in the H3O+ reagent ion spectrum shown in Fig. 1A, outstanding being the analyte ions of the organosulphur compounds indicated.The spectrum for the AF/PA co-culture in Fig. 4B clearly shows additional and discreet analyte ions for the presence of biomarkers for PA, notably hydrogen cyanide and methyl thiocyanate as identified in previous SIFT-MS studies,7–9,11 which are easily distinguished from those produced by AF. The absolute concentrations of these several metabolites as measured in the co-culture headspace are given in Table 2, but it is not profitable at this stage to attempt to compare absolute concentration of the metabolites released from the AF and PA simply because no effort was made to standardise the number of cells of each pathogen. This may also explain the apparent lack of propanol production in the AF/PA co-cultures when the alcohol was emitted by AF when cultured alone.
| Compound | Sample | Concentration (ppbv) | Compound | Sample | Concentration (ppbv) | ||||
|---|---|---|---|---|---|---|---|---|---|
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | ||||
| Ammonia | AF | 5600 |
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
38![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Acetone | AF | 720 | 780 | 1100 |
| (O2+˙) | AF/PA | 8800 | 26![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
42![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
NO+ | AF/PA | 440 | 460 | 620 |
| NH3+(H2O)0,1 | AF/SA | 620 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
NO+(C3H6O) | AF/SA | 510 | 1000 | 680 |
| 17, 35 | AF/SP | 1800 | 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
33![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
88 | AF/SP | 700 | 550 | 1200 |
| Methanol | AF | 360 | 700 | 720 | Methanthiol | AF | 1800 | 2700 | 1700 |
| (H3O+) | AF/PA | 200 | 430 | 450 | (H3O+) | AF/PA | 550 | 1500 | 900 |
| CH5O+(H2O)0,1,2 | AF/SA | 530 | 1720 | 2300 | CH4OS+(H2O)0,1,2 | AF/SA | 200 | 2200 | 4100 |
| 33, 51, 69 | AF/SP | 200 | 2000 | 2900 | 49, 67, 85 | AF/SP | 5 | 5700 | 3800 |
| Ethanol | AF | 310 | 430 | 260 | Dimethyl sulphide | AF | 180 | 2800 | 1600 |
| (H3O+) | AF/PA | 160 | 500 | 190 | (NO+) | AF/PA | 200 | 1100 | 1500 |
| C2H7O+(H2O)0,1,2 | AF/SA | 490 | 2800 | 3900 | C2H6S+ | AF/SA | 25 | 560 | 2800 |
| 47, 65, 83 | AF/SP | 720 | 4600 |
14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
62 | AF/SP | 10 | 5400 | 3300 |
| Propanol | AF | 30 | 250 | 280 | Dimethyl disulphide | AF | 290 | 980 | 960 |
| (H3O+) | AF/PA | 45 | 80 | 35 | (NO+) | AF/PA | 390 | 1900 | 3500 |
| C3H7+; C3H9O+(H2O)0,1,2 | AF/SA | 220 | 100 | 460 | C2H6S2+ | AF/SA | 890 | 870 | 1700 |
| 43, 61, 79, 97 | AF/SP | 90 | 460 | 420 | 94 | AF/SP | 70 | 1700 | 1740 |
| Acetaldehyde | AF | 10 | 60 | 30 | Hydrogen cyanide | AF | 0 | 5 | 5 |
| (H3O+) | AF/PA | 5 | 40 | 20 | (H3O+) | AF/PA | 6300 | 4400 | 4500 |
| C2H5O+(H2O)0,1,2 | AF/SA | 1300 | 25 | 120 | CH2N+(H2O)0,1 | AF/SA | 0 | 0 | 5 |
| 45, 63, 81 | AF/SP | 20 | 25 | 0 | 28, 46 | AF/SP | 15 | 10 | 10 |
| Butanal | AF | 0 | 5 | 5 | Methyl thiocyanate | AF | 10 | 5 | 10 |
| (NO+) | AF/PA | 20 | 10 | 5 | (H3O+) | AF/PA | 20 | 80 | 150 |
| C4H7O+; NO+C4H8O | AF/SA | 20 | 5 | 5 | C2H4NS+(H2O)1,2,3 | AF/SA | 25 | 5 | 10 |
| 71, 102 | AF/SP | 0 | 5 | 5 | 92, 110, 128 | AF/SP | 15 | 0 | 5 |
| Pentanal | AF | 5 | 15 | 5 | |||||
| (NO+) | AF/PA | 5 | 10 | 10 | |||||
| C5H9O+; NO+C5H10O | AF/SA | 210 | 15 | 25 | |||||
| 85, 116 | AF/SP | 20 | 5 | 15 | |||||
The spectra for the AF/SA and AF/SP co-cultures are shown in Fig. 4C and D. Again, the analyte ions of ammonia and organosulphur compounds are obvious and, additionally, there are large signals of analyte ions due to ethanol and smaller signals due to propanol analyte ions. Our previous detailed study15 showed that when both SA and SP are cultured alone they emit ethanol and propanol together with the aldehydes acetaldehyde, butanal and pentanal, but none of these aldehydes are detected in these AF/SA and AF/SP co-cultures. Thus, it appears that the removal of aldehydes from the co-cultures by AF fungus metabolism is very efficient. This apparent modification of the aldehydes emitted by the SA and SP bacteria when co-existing with AF fungus needs to be considered when assessing the prospect of utilizing these aldehydes in exhaled breath for detection and diagnosis of SA and SP infection.
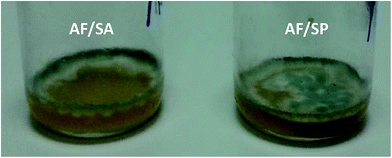
The photographs of the AF/SA and AF/SP co-cultures in Fig. 5 display the unusual growth of the AF/SA co-cultures around the sides of the container, which is evidence of a competitive interaction between the two microbes, as compared to the normal growth of the AF/SP co-cultures. Only three AF/SA co-cultures were produced for this study, but this unusual growth occurred in each of these three co-cultures, whereas in each of the AF alone cultures and the AF/PA and AF/SP co-cultures the AF covered the surface of the liquid. This “sporulation” – the dark spores (as are shown in Fig. 5) – became visible prior to the 48 h culture headspace analysis experiments. Should this occur in the lungs and airways of infected individuals it would, presumably, affect volatile compounds emitted into the exhaled breath of infected patients. The growth patterns of these co-cultures require further investigation.
Discussion and concluding remarks
Following our successful identification of hydrogen cyanide as an in vitro and in vivo biomarker of PA infection in patients with CF,11,12,49 the research described in this paper is intended to be a prelude to similar studies of potential biomarkers for AF, SA and SP. The present in vitro studies of AF cultures reveal easily detectable emissions of ammonia and some organosulphur compounds in the headspace. These observations provide incentive for in vivo studies of the exhaled breath of patients, especially children known to be infected with this fungal pathogen. However, it must be recognised that some organosulphur compounds are generated in the oral cavity by bacterial action50 and so, to bypass this source of mouth-exhaled breath contamination, it would be important to study nose-exhaled breath to detect AF in the lower airways.44 If the low level production of methanol, ethanol and acetone by in vitro AF cultures is mirrored in vivo, it is unlikely to contribute much to the systemically generated breath levels of these compounds.51,52 The relatively small increase in acetone seen above the AF cultures is such that in the AF-infected lungs any increase in acetone would not be sufficient to cover the endogenous variations in this compound in exhaled breath even in healthy individuals.53It is also important to report that the co-existence of AF and PA does not disguise the presence of HCN as a biomarker of PA in vitro and so it is unlikely to distort HCN levels in nose-exhaled breath in vivo. Thus, HCN can still be used as a definitive biomarker of PA in the lungs of cystic fibrosis patients even when they are also infected with AF.
Concerning the further information that can be gleaned for the co-culture studies, both SA and SP produce only two compounds (ethanol and propanol) that are visible and not produced by AF alone. However, the very efficient generation of ethanol and acetaldehyde by SA and SP hold promise for the detection of these bacteria in the lungs and airways even in the presence of AF fungus. So the detection of co-existing pathogens by breath analysis is not out of the question, although much more research is needed, and especially detailed breath analysis investigations involving infected patients are required.
Finally, a cautionary note is required. Whilst these studies indicate the potential of VOC analysis in diagnosing the presence of particular bacterium in the lungs and airways by breath analysis, it must be recognised that there are obvious differences between the physiological states of microbes grown in vitro and those growing in vivo. For example, in vitro cultures are closed batch planktonic culture systems, whereby the growing environment is changing due to the activities of the species themselves, whereas in vivo it is more likely that microbes are attached as biofilms and continuously fed by lung-tissue fluid. Moreover, there is a marked difference in nutrient composition of BHI broth and human tissue-fluid. One difference could be in the amino acid content (cysteine, methionine and glutathione), which could alter the production rate of volatile sulphur compounds. These possible confounding variables can only be assessed by further in vitro experiments and by detailed breath analysis studies.
Acknowledgements
We are grateful to the North Staffordshire Medical Institute and the University Hospital of North Staffordshire Charitable Trust Funds for financial support of this project.References
- Breath Analysis For Clinical Diagnosis And Therapeutic Monitoring, ed. A. Amann and D. Smith, World Scientific Publishing Co. Pte. Ltd., Singapore, 2005 Search PubMed.
- Volatile Biomarkers, ed. A. Amann and D. Smith, Elsevier, Boston, USA, 2013 Search PubMed.
- P. Španěl, K. Dryahina and D. Smith, J. Breath Res., 2007, 1, 026001 CrossRef PubMed.
- D. Smith, P. Španěl, B. Enderby, W. Lenney, C. Turner and S. J. Davies, J. Breath Res., 2010, 4, 017101 CrossRef PubMed.
- M. E. Dolch, C. Hornuss, C. Klocke, S. Praun, J. Villinger, W. Denzer, G. Schelling and S. Schubert, Eur. J. Clin. Microbiol. Infect. Dis., 2012, 31, 3007–3013 CrossRef CAS PubMed.
- J. J. Zhu, H. D. Bean, Y. M. Kuo and J. E. Hill, J. Clin. Microbiol., 2010, 48, 4426–4431 CrossRef CAS PubMed.
- W. Carroll, W. Lenney, T. S. Wang, P. Španěl, A. Alcock and D. Smith, Pediatr. Pulmonol., 2005, 39, 452–456 CrossRef PubMed.
- F. J. Gilchrist, A. Alcock, J. Belcher, M. Brady, A. Jones, D. Smith, P. Španěl, K. Webb and W. Lenney, Eur. Respir. J., 2011, 38, 409–414 CrossRef CAS PubMed.
- V. Shestivska, P. Španěl, K. Dryahina, K. Sovová, D. Smith, M. Musilek and A. Nemec, J. Appl. Microbiol., 2012, 113, 701–713 CrossRef CAS PubMed.
- F. J. Gilchrist, H. Sims, A. Alcock, J. Belcher, A. M. Jones, D. Smith, P. Španěl, A. K. Webb and W. Lenney, Anal. Methods, 2012, 4, 3661–3665 RSC.
- V. Shestivska, A. Nemec, P. Dřevínek, K. Sovová, K. Dryahina and P. Španěl, Rapid Commun. Mass Spectrom., 2011, 25, 2459–2467 CrossRef CAS PubMed.
- D. Smith, P. Španěl, F. J. Gilchrist and W. Lenney, J. Breath Res., 2013, 7, 044001 CrossRef PubMed.
- J. Sulé-Suso, A. Pysanenko, P. Španěl and D. Smith, Analyst, 2009, 134, 2419–2425 RSC.
- D. Smith, T. Wang, J. Sulé-Suso, P. Španěl and A. J. El Haj, Rapid Commun. Mass Spectrom., 2003, 17, 845–850 CrossRef CAS PubMed.
- T. W. E. Chippendale, F. J. Gilchrist, P. Španěl, A. Alcock, W. Lenney and D. Smith, Anal. Methods, 2014, 6, 2460–2472 RSC.
- K. Sovová, J. Čepl, A. Markoš and P. Španěl, Analyst, 2013, 138, 4795–4801 RSC.
- T. W. E. Chippendale, P. Španěl and D. Smith, Rapid Commun. Mass Spectrom., 2011, 25, 2163–2172 CrossRef CAS PubMed.
- D. Smith, T. Wang, P. Španěl and R. Bloor, Physiol. Meas., 2006, 27, 437 CrossRef PubMed.
- D. Smith, A. Pysanenko and P. Španěl, Rapid Commun. Mass Spectrom., 2010, 24, 1066–1074 CrossRef CAS PubMed.
- P. D. Barnes and K. A. Marr, Infect. Dis. Clin. North Am., 2006, 20, 545–561 CrossRef PubMed.
- T. M. Hohl and M. Feldmesser, Eukaryotic Cell, 2007, 6, 1953–1963 CrossRef CAS PubMed.
- D. A. Stevens, R. B. Moss, V. P. Kurup, A. P. Knutsen, P. Greenberger, M. A. Judson, D. W. Denning, R. Crameri, A. S. Brody, M. Light, M. Skov, W. Maish and G. Mastella, Clin. Infect. Dis., 2003, 37, S225–S264 CrossRef PubMed.
- R. A. Bazemore, J. Feng, L. Cseke and G. K. Podila, J. Breath Res., 2012, 6, 016002 CrossRef CAS PubMed.
- N. P. Pont, C. A. Kendall and N. Magan, Mycopathologia, 2012, 173, 93–101 CrossRef PubMed.
- M. Syhre, J. M. Scotter and S. T. Chambers, Med. Mycol., 2008, 46, 209–215 CrossRef CAS PubMed.
- S. T. Chambers, M. Syhre, D. R. Murdoch, F. McCartin and M. J. Epton, Med. Mycol., 2009, 47, 468–476 CrossRef CAS PubMed.
- J. C. Lin, M. Li, W. M. Xu, B. Peng, Z. L. Guo, W. Shui, Y. L. Xin and C. R. Zhang, J. Immunol. Tech. Infect. Dis., 2013, 2, 1000112 Search PubMed.
- D. Smith and P. Španěl, Mass Spectrom. Rev., 2005, 24, 661–700 CrossRef CAS PubMed.
- D. Smith, A. Pysanenko and P. Španěl, Int. J. Mass Spectrom., 2009, 281, 15–23 CAS.
- P. Španěl, K. Dryahina and D. Smith, Int. J. Mass Spectrom., 2006, 249–250, 230–239 Search PubMed.
- J. J. Zhu, H. D. Bean, M. J. Wargo, L. W. Leclair and J. E. Hill, J. Breath Res., 2013, 7, 016003 CrossRef PubMed.
- J. J. Zhu, H. D. Bean, J. Jimenez-Diaz and J. E. Hill, J. Appl. Physiol., 2013, 114, 1544–1549 CrossRef CAS PubMed.
- M. Bunge, N. Araghipour, T. Mikoviny, J. Dunkl, R. Schnitzhofer, A. Hansel, F. Schinner, A. Wisthaler, R. Margesin and T. D. Mark, Appl. Environ. Microbiol., 2008, 74, 2179–2186 CrossRef CAS PubMed.
- C. Turner, C. Batty, E. Escalona, J. O. Hunter and C. Proudman, Curr. Anal. Chem., 2013, 9, 614–621 CrossRef CAS.
- K. Dryahina and P. Španěl, Windows application for direct analysis of SIFT-MS data using Principal component analysis (PCA), Source code in Delphi, accessed November 2013.
- P. Španěl and D. Smith, Int. J. Mass Spectrom. Ion Processes, 1997, 167–168, 375–388 CrossRef.
- P. Španěl, Y. Ji and D. Smith, Int. J. Mass Spectrom. Ion Processes, 1997, 165–166, 25–37 CrossRef.
- P. Španěl and D. Smith, Int. J. Mass Spectrom., 1998, 176, 167–176 CrossRef.
- P. Španěl and D. Smith, Int. J. Mass Spectrom., 1998, 176, 203–211 CrossRef.
- D. Smith, T. W. E. Chippendale and P. Španěl, Curr. Anal. Chem., 2013, 9, 550–557 CrossRef CAS.
- R. Sander, in NIST Chemistry WebBook, NIST Standard Reference Database Number 69, National Institute of Standards and Technology, Gaithersburg MD, p. 20899 Search PubMed.
- AsperCyc, The Central Aspergillus Data REpository (CADRE), accessed 31 March 2014, http://www.aspercyc.org.uk.
- J. C. Rhodes, Med. Mycol., 2006, 44, S77–S81 CrossRef CAS PubMed.
- D. Smith, T. Wang, A. Pysanenko and P. Španěl, Rapid Commun. Mass Spectrom., 2008, 22, 783–789 CrossRef CAS PubMed.
- A. Sponring, W. Filipiak, T. Mikoviny, C. Ager, J. Schubert, W. Miekisch, A. Amann and J. Troppmair, Anticancer Res., 2009, 29, 419–426 CAS.
- C. Brunner, W. Szymczak, V. Hollriegl, S. Mortl, H. Oelmez, A. Bergner, R. M. Huber, C. Hoeschen and U. Oeh, Anal. Bioanal. Chem., 2010, 397, 2315–2324 CrossRef CAS PubMed.
- W. Filipiak, A. Sponring, A. Filipiak, C. Ager, J. Schubert, W. Miekisch, A. Amann and J. Troppmair, Cancer Epidemiol., Biomarkers Prev., 2010, 19, 182–195 CrossRef CAS PubMed.
- T. W. E. Chippendale, B. Hu, A. J. El Haj and D. Smith, Analyst, 2012, 137, 4677–4685 RSC.
- F. J. Gilchrist, R. J. Bright-Thomas, A. M. Jones, D. Smith, P. Španěl, A. K. Webb and W. Lenney, J. Breath Res., 2013, 7, 026010 CrossRef CAS PubMed.
- A. Pysanenko, P. Španěl and D. Smith, J. Breath Res., 2008, 2, 046004 CrossRef CAS PubMed.
- C. Turner, P. Španěl and D. Smith, Physiol. Meas., 2006, 27, 321–337 CrossRef PubMed.
- C. Turner, P. Španěl and D. Smith, Rapid Commun. Mass Spectrom., 2006, 20, 61–68 CrossRef CAS PubMed.
- P. Španěl, K. Dryahina, A. Rejšková, T. W. E. Chippendale and D. Smith, Physiol. Meas., 2011, 32, N23–N31 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2014 |