Open Access Article
Open Access ArticleRapid and simultaneous detection of ricin, staphylococcal enterotoxin B and saxitoxin by chemiluminescence-based microarray immunoassay†
A.
Szkola
a,
E. M.
Linares
a,
S.
Worbs
b,
B. G.
Dorner
b,
R.
Dietrich
c,
E.
Märtlbauer
c,
R.
Niessner
a and
M.
Seidel
*a
aChair for Analytical Chemistry & Institute of Hydrochemistry, TU München, Marchioninistrasse 17, 81377 Munich, Germany. E-mail: michael.seidel@ch.tum.de; Fax: +49 (89) 2180-78255; Tel: +49 (89) 2180-78238
bCentre for Biological Threats and Special Pathogens, Biological Toxins, Robert Koch-Institut, Nordufer 20, 13353 Berlin, Germany
cChair of Hygiene and Technology of Milk, LMU München, Schönleutnerstraße 8/219, 85764 Oberschleißheim, Germany
First published on 5th September 2014
Abstract
Simultaneous detection of small and large molecules on microarray immunoassays is a challenge that limits some applications in multiplex analysis. This is the case for biosecurity, where fast, cheap and reliable simultaneous detection of proteotoxins and small toxins is needed. Two highly relevant proteotoxins, ricin (60 kDa) and bacterial toxin staphylococcal enterotoxin B (SEB, 30 kDa) and the small phycotoxin saxitoxin (STX, 0.3 kDa) are potential biological warfare agents and require an analytical tool for simultaneous detection. Proteotoxins are successfully detected by sandwich immunoassays, whereas competitive immunoassays are more suitable for small toxins (<1 kDa). Based on this need, this work provides a novel and efficient solution based on anti-idiotypic antibodies for small molecules to combine both assay principles on one microarray. The biotoxin measurements are performed on a flow-through chemiluminescence microarray platform MCR3 in 18 minutes. The chemiluminescence signal was amplified by using a poly-horseradish peroxidase complex (polyHRP), resulting in low detection limits: 2.9 ± 3.1 μg L−1 for ricin, 0.1 ± 0.1 μg L−1 for SEB and 2.3 ± 1.7 μg L−1 for STX. The developed multiplex system for the three biotoxins is completely novel, relevant in the context of biosecurity and establishes the basis for research on anti-idiotypic antibodies for microarray immunoassays.
Introduction
Biotoxins are substances produced by microorganisms, fungi, plants or animals, which cause harmful effects on other organisms. They include a variety of substances ranging from 0.14 kDa to 150 kDa in molecular weight (Mw).1 Their toxicity depends on the dose, the application route and the specific mechanism of action within the organism. The existence of highly toxic biotoxins makes them powerful candidates for being used as biological warfare agents.2 Among the highly toxic lectins, ricin (60 kDa) is considered a potential biological weapon due to its low lethal dose (LD50: 3 μg kg−1); accidental and intentional intoxications using ricin have been reported.3 Ricin is produced in seeds of castor oil plant4 and consists of a heterodimeric glycoprotein.5 It is an attractive biotoxin for potable water supply and food processing due to the easy extraction from the plant and the resistance to chlorination and disinfection methods. Together with ricin, saxitoxin (0.3 kDa, STX) is listed in the Chemical Weapon Convention6 and in the War Weapon Control Act, safety category B.7 Saxitoxin is a neurotoxin produced by cyanobacteria and dinoflagellates and consists of a highly polar alkaloid.8,9 Due to its high toxicity (LD50: 10 μg kg−1) and stability, saxitoxin has also high potential as a biowarfare agent, only limited by difficult synthesis.10,11 Another powerful toxin and possible agent biowarfare is the staphylococcal enterotoxin B (30 kDa, SEB) produced by a gram-positive and facultative anaerobic bacterium, Staphylococcus aureus. SEB is a protein with no potential for high mortality, but its high emetic potency (LD50: 0.02 μg kg−1) and fast action (2 to 8 h) raised an interest as incapacitating agent.12,13 SEB has high stability even resisting a few minutes at temperatures above 100 °C and is frequently involved in food poisoning outbreaks.14The use of biotoxins as biological weapon displays a permanent risk for humans.15 The described biotoxins are relatively easy to spread, causing moderate to high mortality. Due to their characteristics, these toxins are probable candidates for a warfare use and therefore need to be verified in case of terrorist attack suspicion. This potential risk leads to a demand on the simultaneous diagnosis of the biotoxins by a fast, cheap and reliable assay. Different techniques have been described to detect those toxins,3,16 including chromatography,17 spectrophotometry,18,19 mass spectrometry,20,21 electrochemistry,22 as well as assays addressing functional activity.23 While a wide range of methods is used, many assays rely on immunological detection of target molecules due to the high specificity and sensitivity.31 Microarray immunoassays (MIAs) in particular have gained attention, as they benefit of the capability to test a wide variety of analytes in a single assay, reducing time of analysis and costs.32 MIA for biotoxins is a promising tool for the identification and detection of an eventual contamination and different approaches have been described in the literature.33–35 An overview of recent detection limits for biotoxins in microarrays is given in the Table 1.
| Biotoxins | Application | Limit of detection (μg L−1) | Type of detection | Reference |
|---|---|---|---|---|
| Ricin | Biosecurity | 0.1 | Antibody/fluorescence | 24 |
| SEB | 0.01 | |||
| Ricin | Food safety | 0.5 | Antibody/fluorescence | 25 |
| SEB | 0.5 | |||
| Ricin | Protein screening | 15 | Aptamer/fluorescence | 26 |
| Ricin | Biosecurity | 80 | Carbohydrate/chemiluminescence | 27 |
| SEB | Proof of principle for microarray development | 3.10–6 | Antibody/electrochemistry | 28 |
| STX | Food safety | 0.4 | Antibody/chemiluminescence | 29 |
| STX | Food safety | 0.82 | Antibody/surface plasmon resonance | 30 |
Although some microarrays have been successfully developed, a few challenges still need to be faced according to the broad diversity of samples and particularities of each biotoxin. In order to overcome these problems, different approaches have been described combining different assay principles on the microarray. Hartmann et al.36 described a novel assay format that combines competitive and direct immunoassay principles into one system to overcome dilution sample problems of proteins. Molecules present in high concentrations as well as those occurring at low concentrations could be quantified within the same assay. A greater challenge is the wide range of molecular weight. Small molecules with less than 1000 Da in molecular weight are not considered amenable to sandwich immunoassays due to their difficulty of simultaneous recognition by two antibodies.37 In this case, other arrangements can be used, including competitive assays. Parro et al.38 and Fernández-Calvo et al.39 described the development of protein microarray technologies for automatic in situ detection and identification combining sandwich and competitive immunoassays. The assay was developed to analyze liquid and solid samples from extraterrestrial origin, ranging from small molecules and proteins to whole cells and spores. Although the direct immobilization of analytes on the microarray for the competitive assay was successfully performed, this is not always the case. There are small molecules whose structure does not have enough functional groups for immobilization or are not available in the required amounts.40 In this case, the immobilization may affect the antibody recognition, the regenerability of the microarray or does not provide concentrated spots, as already described for STX.29 Additionally, the direct immobilization of molecules on the microarray may require previous coupling to other larger molecules (e.g. albumin) or different chemical functionalities on the microarray surface, which increases the work and cost of production.
Based on this challenge, this work describes the development of a microarray for ricin, SEB and STX detection, combining sandwich and indirect competitive immunoassays in one platform. The combination of both methods is possible through the use of anti-idiotypic antibodies for small molecules. These antibodies are immunoglobulins, whose paratope mimics the structure of an antigen and recognize the epitopes of the antibody produced for the antigen.41 It represents a powerful alternative for direct immobilization of small molecules, in this case STX, on the microarray. The proteotoxins, ricin and SEB, are detected using sandwich based immunoassay, where labeled antibodies bind to the antigen and the antibody–antigen pairs are captured by the immobilized antibodies. For STX detection, a competition between its labeled antibodies and the immobilized anti-idiotypic antibodies take place on the chip. Thus, it is produced a microarray containing anti-idiotypic antibodies for STX and conventional antibodies for ricin and SEB. This strategy avoids the need of analyte coupling to large molecules or different surface chemistry for immobilizing antibodies and analyte on the same microarray surface. The microarray is placed in an automated system, the Munich Chip Reader (MCR3), which allows precise and fast on-site analysis. For the first time, MIA is capable to detect proteotoxins and small toxins, simultaneously. The microarray can be used as a tool for monitoring biotoxins in samples as a preventive protection of the population against natural or deliberate contaminations. Moreover, this technology shows the potential of anti-idiotypic antibodies for the simultaneous detection of small and large molecules on the same microarray.
Materials and methods
Chemicals and materials
Di-potassium hydrogen phosphate, di-sodium hydrogen phosphate, sodium hydroxide, absolute ethanol 99.8%, N,N-dimethylformamide (DMF), di(N-succinimidyl) carbonate (DSC), 4-dimethylaminopyridine (DMAP), anti-rabbit HRP, ethylenediamine, 3-glycidyloxypropyltrimethoxysilane (GOPTS), 3,3′,5,5′-tetramethylbenzidine potassium hydrogen phosphate (TMB), pluronic F127, D(+)-trehalose dihydrate, casein, sodium chloride were obtained from Sigma-Aldrich (Taufkirchen, Germany). Sulfo-NHS-LC-biotin and Hellmanex were obtained from Fisher Scientific (Schwerte, Germany) and Hellma GmbH (Mannheim, Germany), respectively. Jeffamine ED-2003 polyetheramine was obtained from Huntsman (Salt Lake City, USA). Westar supernova ELISA luminol and hydrogen peroxide were purchased from Cyanagen (Bologna, Italy). The ARcare 90106 adhesive film was obtained from Adhesive Research Ireland (Limerick, Ireland). The poly(methyl methacrylate) support for the chip was produced in our laboratory. The glass slides (76 mm × 26 mm × 1 mm) were purchased from Carl Roth (Karlsruhe, Germany). Microplates with 96- and 384-wells were obtained from Greiner GmbH (Frickenhausen, Germany).Mouse monoclonal antibody (mAb) production against ricin (R109, R18, R21) and SEB (S1001, S419) was described elsewhere.31 Clone S3849 against SEB was produced similarly. Highly purified agglutinin and ricin were produced as described.42 Anti-idiotypic antibodies from mouse mAb 1F8, anti-STX from mouse mAb 7H11 and biotinylated anti-STX from mouse mAb 7H11 were obtained from the Chair of Hygiene and Technology of Milk (LMU Munich, Germany). Anti-idiotypic antibody production and antibody biotinylation are described in more details in the ESI (S1†). HRP-labeled streptavidin was purchased from Axxora Germany GmbH (Lörrach, Germany). Horseradish peroxidase (HRP) was purchased from Roche Diagnostics GmbH (Mannheim, Germany). Saxitoxin (STX) was obtained from Institute of Agri-Food and Land Use (Belfast, Ireland). Staphylococcus enterotoxin B (SEB) was purchased from Diavita GmbH (Heidelberg, Germany). Poly(horseradish peroxidase)–streptavidin (SA–PolyHRP40) was obtained from Senova GmbH (Jena, Germany).
Glass slide and microarray preparation
Before antibody immobilization, glass slides were treated and functionalized according to procedures described elsewhere.43,44 The microarray preparation was performed as described in Wolter et al.43 More details are described in the ESI (S1†). The spots were produced using the Spotter BioOdyssey Calligrapher MiniArrayer from Bio-Rad Laboratories GmbH (Munich, Germany). The solid pin SNS 9 was purchased from ArrayIt (Sunnyvale, USA).Measurement of the antibody microarray with MCR3
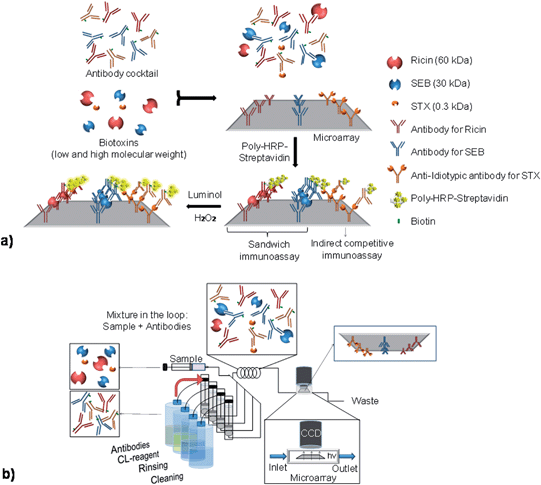
The microarray was connected to the fluidic system of the microarray analysis platform in the MCR3 from GWK Präzisionstechnik (Munich, Germany). More details are described by Kloth et al.45 and in the ESI, S1.† An aliquot of 0.3 mL of standard solution or sample was placed in the incubation loop of the MCR3. Then, 0.7 mL of standard or sample was mixed with 0.7 mL of detection antibody solution. The antibodies were used in the following concentrations: anti-ricin R18 1.6 mg L−1, anti-SEB 419 0.5 mg L−1, anti-STX mAb 7H11 0.1 mg L−1. The concentrations were previously optimized for each antibody. This mixture was pumped into the 50 μL unit with a flow rate of 10 μL s−1 and an interaction time of 10 s in the flow cell. The chip was then washed with 2 mL of PBST (10 mM KH2PO4, 70 mM K2HPO4, 145 mM NaCl with 0.05% of Tween 20, v/v) at 500 μL s−1. Subsequently, 1 mL conjugate solution (SA–PolyHRP40 1 mg L−1) was added in two portions into the flow cell: 200 μL at 100 μL s−1 and 800 μL at 5 μL s−1. After a further washing step with PBST (2 mL at 500 μL s−1), 250 μL of luminol and hydrogen peroxide were simultaneously added into the flow cell with a flow rate of 10 μL s−1. The CL signal was recorded for 60 s with a CCD camera. After the image acquisition, the microarray was rinsed with PBST buffer (2 mL at 500 μL s−1 and 3 mL at 250 μL s−1). The total running time of the assay was 18 min. The background image is previously recorded before the standard solutions or samples are added to the microarray. The optimizing experiments were performed only once and the error bars are the average of five spots. The calibration curves were measured three times with different microarray chips; hence the error bars are the standard deviation of 3 values obtained by the average of five spots per chip. The limit of detection (LOD) was calculated by the blank average plus three times the standard deviation of the blank. The three toxins are listed by the U.S. Department of Health and Human Services (HHS) as “HHS select agents and toxins”, which affect humans. During the experiments, samples were handled in a extractor hood and waste was collected separate from usual waste (solid waste was deactivated with 5% NaOH and liquid waste was deactivated with 5% NaOH and then autoclaved for 1 h at 121 °C).Results and discussion
Assay principle
To perform a multiplex immunoassay with different molecular weight biotoxins, an antibody microarray is designed and produced by the immobilization of anti-ricin and anti-SEB capture antibodies on the glass slide with anti-idiotypic antibodies for STX, as observed in Fig. 1a.Prior to the detection, the biotinylated antibodies and the biotoxins are incubated together to promote the interaction between the respective antigen–antibody pairs. This step is especially important for the competitive assay for STX detection.
On the microarray, the pairs of antibody–biotoxin bind to the respective capture antibodies for ricin and SEB, producing a sandwich. In contrast, the detection of STX occurs through a competition between the free STX molecules and the immobilized anti-idiotypic antibodies to the biotinylated anti-STX antibodies. The detection signal is provided by enzyme catalyzed chemiluminescence reaction with luminol and hydrogen peroxide, using peroxidase–streptavidin conjugates. The signal for ricin and SEB are directly proportional to the biotoxin concentrations. For STX, the chemiluminescence intensity is inversely proportional to the concentration of the antigen. The pre-incubation of the biotinylated antibodies with the biotoxins is performed by the injection of the solutions into the loop of the MCR3 system (Fig. 1b). The solution is then automatically driven to the microarray surface, where the reactions take place and the signal is registered. The spots on the microarray are identified by their location on the recorded image. The antibodies are spotted on a defined location and order, which allows the fast recognition of the correspondent system.
Assay optimization
In order to develop a multi-analyte detection system for biotoxins, it is necessary to optimize the parameters which influence the assay sensitivity. Antibody selection and concentration, interaction time, continuous/stopped flow, sequential/parallel addition, addition rate and reaction volume are the main parameters to be optimized. The optimization was exemplarily performed for ricin detection. R. communis agglutinin is a 120 kDa lectin with about 90% sequence identity to ricin and was chosen for the initial tests due to its lower toxicity.3 Different antibodies were available for ricin detection that cross-react with R. communis agglutinin, including monoclonal (mAb R109 and mAb R21) and a biotinylated monoclonal detection antibody (mAb R18).To enhance the assay sensitivity, the conventional label based on enzyme horseradish peroxidase–streptavidin (SA–HRP) conjugates was replaced by poly(horseradish peroxidase)–streptavidin (SA–PolyHRP40) conjugates. SA–PolyHRP40 is a supramolecular complex composed of five identical covalent HRP homopolymer blocks covalently coupled to streptavidin molecules. For the SA–PolyHRP40, there is an average of 200 monomer HRP molecules per complex unit.46 The comparison of both labels was performed using a sandwich ELISA immunoassay with TMB substrate, monoclonal capture antibody R109 and monoclonal detection antibody R18. The calibration curves for SA–HRP and SA–PolyHRP40 (available at the ESI, S2†) were obtained in the range of 0 to 1000 μg L−1 of agglutinin. The SA–PolyHRP40 curve showed higher sensitivity with a working range saturating at 100 μg L−1, where the SA–HRP curve only started its working range with 10 times lower absorbance. PolyHRP40 conjugates quantitatively delivered a large number of signal-generating enzyme molecules per one bound analyte molecule, resulting in a considerable signal enhancement. As a result, the assay using SA–PolyHRP40 produces higher sensitivity and therefore it was chosen for the next optimizations. For all further measurements ricin has been used instead of agglutinin, which is recognized by the same set of antibodies with high affinity.47
To combine sandwich and competitive immunoassay principles in the flow system, it is important to meet the requirements of the individual assays. The competitive immunoassay requires the pre-incubation of the antibody and the antigen prior to the detection with parallel addition on the microarray. For the sandwich assay, the pre-incubation is not necessary, but it can also be favorable for the detection. In order to investigate the effects of the sequential or parallel addition on the assay performance, four conditions were compared for ricin: (I) sequential addition of reactants (ricin and biotinylated detection antibody) at 1 μL s−1 with 10 s of interaction time (duration: 1 h 15 min), (II) parallel addition in continuous flow at 1 μL s−1 (duration: 26 min), (III) parallel addition with pre-incubation step of the detection antibody and the ricin for 1 min in the MCR3 loop, injecting 50 μL of sample at 1 μL s−1 with 20 s of incubation time (duration: 34 min) and (IV) parallel addition with pre-incubation step, injecting 5 μL of sample at 1 μL s−1 with 10 s of incubation time (duration: 1 h 15 min). The assay was tested with two different capture antibodies, R109 and R21, and the biotinylated mAb R18 as the detection antibody. The results (see ESI, S3†) indicated that the parallel addition produces faster results with higher chemiluminescence signal for all conditions in comparison to the sequential addition. The evaluation of the three conditions using parallel addition (II–IV) indicated better performance with the use of stopped-flow principle (III and IV) as an approximation to a stationary system. In this case, a defined volume is pumped into the chip and remains there for a certain time. The method (IV) results in higher CL signal (8421 a.u. for R109 and 7827 a.u. for R21) than the method (III) with 2988 a.u. for R109 and 3756 a.u. for R21, indicating that a lower unit volume in a shorter time interaction leads to higher signals. Although the incubation time for the method IV is half of the method III, the additional interaction time for the program IV was 41 minutes. This means that the ricin molecules had more time to come in contact with the immobilized antibodies and interact with them, justifying the higher CL-signal intensity. It is also observed that the monoclonal R109 antibody produces higher CL signal in comparison to the R21 antibody. The results showed that the use of parallel addition with stopped flow principle is favorable to enhance the assay sensitivity for the sandwich assay and it indicates the promising combination with the competitive assay in the multiplex system.
The influence of the sample volume, flow rate and interaction time on the CL signal was also investigated and the results are described in the ESI (S4†). The injected volume on the microarray was varied from 5 to 50 μL and the CL signal was compared. The signals for 5 μL (57681 a.u. for R109 and 54483 a.u. for R21) and 50 μL (49912 a.u. for R109 and 48150 a.u. for R21) showed a maximum decrease in the CL signal intensity of 13.5%. Nevertheless, the assay time is more than two times faster for 50 μL than for 5 μL. Therefore, the volume of 50 μL was chosen. The flow rate optimization showed that 10 μL s−1 was the best compromise between assay time and CL signal, reducing even more the analysis time to 18 min. Table 2 summarizes the final optimized parameters, indicating pre-incubation of biotoxins and antibodies for 1 min in the loop, injection of 50 μL units of the mixture by a stopped-flow principle at a flow rate of 10 μL s−1 with interaction times of 10 s.
| Parameters | Optimized values |
|---|---|
| Label | Poly(horseradish peroxidase)–streptavidin |
| Time of pre-incubation | 1 min |
| Injected volume | 50 μL |
| Flow rate | 10 μL s−1 |
| Interaction time | 10 s |
| Total duration | 18 min |
Calibration curves of the biotoxins
The optimized program enabled the measurement of calibration curves for ricin using the capture antibodies R21 and R109 on the flow analysis platform MCR3. For this purpose, both antibodies were immobilized together on different microarrays for each concentration and a concentration range of 1 to 2000 μg L−1 of ricin was measured. Fig. 2a shows that the capture antibody R109 provides higher CL signals, to an average factor of 2–3 in comparison to the R21. The affinity of R109 to ricin appears to be higher, because the midpoint (224.1 μg L−1) and the work area (from 48.8 to 1029.8 μg L−1) are shifted to lower concentrations compared to R21 with a midpoint at 740.7 μg L−1 and a work area between 151.6 and 1589.7 g L−1. Therefore, R109 antibody was used for the multiplex analysis.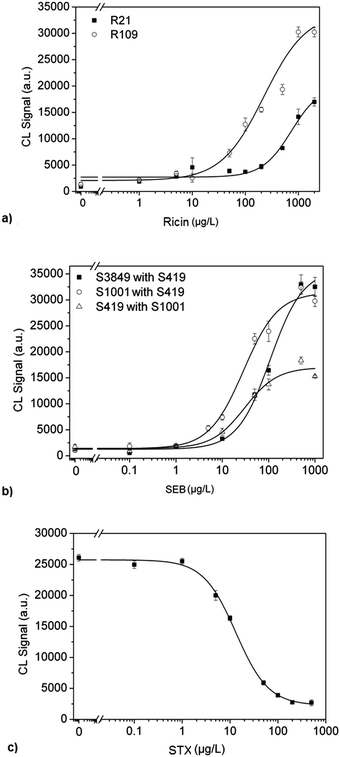 | ||
| Fig. 2 Calibration curves. Calibration curves separately obtained for ricin, SEB and STX. Different combination of antibodies were used for ricin and SEB. | ||
Staphylococcal enterotoxin B is similarly to ricin a high molecular weight toxin and is detected in a heterogeneous sandwich ELISA. Therefore, the measurement program optimized for ricin was also used for the SEB detection. The calibration curve was produced by varying the concentration from 0 to 1000 μg L−1. For the detection of SEB three monoclonal antibodies were used: S3849, S1001 and S419. These antibodies were tested as capture and detection antibodies, resulting in 6 combinations in a sandwich format. Fig. 2b shows three combinations that yielded the most sensitive calibration curves. The antibody pair S3849 and S419 provided the highest CL signals. However, the sensitivity of this antibody pair is lower because the midpoint is 108.2 μg L−1 and the working area is from 31.2 to 375.5 μg L−1 compared to the other two antibody pairs, which are shifted to higher levels. The other two pairs S1001 with S419 and S419 with S1001 showed work areas as well as the midpoint in the same order. However, the antibody pair S1001with S419 has a higher CL signal intensity by a factor of 2 for each calibration point and also a lower detection limit, 0.1 μg L−1. Thus, the antibody pair S1001 with S419 was chosen for the following multiplex measurements.
STX detection was performed with anti-idiotypic antibodies in an indirect competitive ELISA format. The antibodies were immobilized at 0.5 g L−1 on the glass surface of the chip. The saxitoxin calibration curve was obtained for the concentration range of 0 to 500 μg L−1, using the same optimized conditions for ricin. Fig. 2c shows the calibration curve with the midpoint at 13.2 μg L−1 and the detection limit at 1.4 μg L−1. The specified operating range is between 3.2 and 54.1 μg L−1. The use of anti-idiotypic antibodies is the first step for the successful combination of the two assay principles into one antibody microarray platform. It also has the advantage of using the same conditions for the immobilization, incubation and blocking steps.
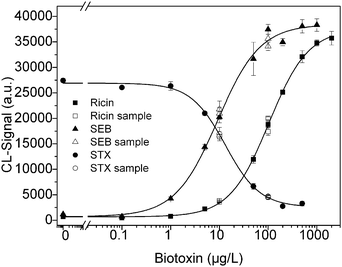
Combination of sandwich and indirect competitive assays on the same microarray
Microarrays were produced by immobilization of capture antibody anti-ricin (R109) and anti-SEB (S1001) together with the anti-idiotypic antibodies. The calibration curves for the biotoxins were measured three times independently and obtained from 0 to 500 μg L−1 with additional concentrations for SEB (1000 μg L−1) and for ricin (1000 and 2000 μg L−1). The curves are depicted in the Fig. 3.The calibration data of the multiplexed measurements are listed in Table 3. To compare the three calibration curves, the coefficient of variation (CV) was calculated from the midpoint of the three multiplex calibration curves. The variation coefficient was 13.9% for ricin, 9.3% for SEB and 28.0% for STX. The detection limit obtained for STX is similar to the LOD described by Szkola et al.29 in an indirect microarray. This indicates the successful adaptation to the competitive assay.
| Toxin | IC50 (μg L−1) | WR (20–80%) (μg L−1) | LOD (μg L−1) | CV (%) |
|---|---|---|---|---|
| Ricin | 93.7 ± 13.1 | 23.8 ± 2.7–301.9 ± 43.8 | 2.9 ± 3.1 | 13.9 |
| SEB | 8.7 ± 0.8 | 1.7 ± 0.4–48.6 ± 15.2 | 0.1 ± 0.1 | 9.3 |
| STX | 10.1 ± 2.8 | 2.6 ± 1.5–37.2 ± 3.3 | 2.3 ± 1.7 | 28.0 |
Furthermore, the LOD for ricin and SEB are as low as the available microarrays for this biotoxins, as shown in the Table 1.
Reproducibility measurements were performed using a new microarray chip per analysis and different biotoxin concentrations: ricin 500 μg L−1, SEB 100 μg L−1 and saxitoxin 10 μg L−1. The results of the four measurements indicated stable CL signals with low standard deviation: 1.8% for ricin, 4.1% for SEB and 3.5% for STX. These results prove that a parallel measurement of biotoxins with different molecular weight is possible.
For the determination of recovery rates (Table 4), the biotoxins were first calibrated simultaneously, followed by sample measurement. The samples were plotted together with the calibration curve (Fig. 3). The CL signals of the sample agree with the calibration curves. Table 3 lists the values of the recovery rates measured. An average recovery rate for ricin was 11.1 μg L−1 for the sample with a concentration of 10 μg L−1 and 100.9 μg L−1 for the sample with 100 μg L−1. Comparable good recoveries were obtained for SEB with 11.1 μg L−1 and 92.5 μg L−1 and for STX with 10.2 μg L−1 and 94.5 μg L−1, even for the concentrations out of the working range between 20 and 80%. The spiked sample of water is an example of matrix, which can assume different types in real analysis. Water and food are cited as the most probable, but other matrices can also be analyzed, such as contaminated soil. Real samples may bring some difficulties, which prevent the directly application in the MCR3. Water samples, for example, must be filtered in order to avoid blocking of the microfluidic channel and solid samples should be digested and brought to a liquid form to inject in the machine. Although the antibodies are highly specific for the toxins, cross-reactions may also be considered.
| Amount | Recovery | |||||
|---|---|---|---|---|---|---|
| Ricin | SEB | STX | ||||
| (μg L−1) | (μg L−1) | (%) | (μg L−1) | (%) | (μg L−1) | (%) |
| 10 | 10.7 | 106.6 | 10.6 | 106.5 | 9.8 | 98.6 |
| 10 | 11.5 | 115.3 | 11.6 | 116.3 | 10.6 | 105.9 |
| Average | 11.1 ± 0.6 | 110.9 ± 6.2 | 11.1 ± 0.7 | 111.4 ± 6.9 | 10.2 ± 0.5 | 102.3 ± 5.1 |
| 100 | 87.6 | 87.6 | 113.7 | 113.7 | 97.0 | 97.0 |
| 100 | 114.1 | 114.1 | 71.4 | 71.4 | 92.5 | 92.5 |
| Average | 100.9 ± 18.6 | 100.9 ± 18.6 | 92.5 ± 29.9 | 92.5 ± 29.9 | 94.5 ± 3.2 | 94.5 ± 3.2 |
Conclusions
For the first time, an antibody microarray chip was produced to detect proteotoxins and small biotoxin, combining sandwich and indirect competitive immunoassay principles. The combination of both assays on one microarray was possible by the use of anti-idiotypic antibodies, which mimic the structure of STX and is also recognized by the detection Ab. The chemiluminescence signal was amplified by using polyHRP40, providing an assay with comparable or lower detection limits than the available microarrays for the biotoxins. The described microarray platform proved to be a promising tool for biowarfare applications, not only for the mentioned toxins but for any other relevant toxin. This work opens the possibility to produce parallel detection arrays for large and small analytes in different applications, as highly required to food48–50 and water51 analysis.Acknowledgements
The authors would like to thank K. Campbell and C. Elliott from Queen's University in Ireland, for providing STX. We acknowledge the company Huntsman (Salt Lake City, USA) for offering the Jeffamine ED-2003 polyetheramine. We also kindly thank S. Wiesemann and R. Hoppe from the Institute of Hydrochemistry's workshop for producing the plastic covers.Notes and references
- H. Russmann, Gesundheitsschutz, 2003, 46, 989–996 Search PubMed.
- R. Franz, Defense against toxin weapons, Medical Aspects of Chemical and Biological Warfare, Textbook of Military Medicine, Washington DC, 1997 Search PubMed.
- S. Worbs, K. Köhler, D. Pauly, M. A. Avondet, M. Schaer, M. B. Dorner and B. G. Dorner, Toxins, 2011, 3, 1332–1372 CrossRef CAS PubMed.
- W. Burrows and S. Renner, Environ. Health Perspect., 1999, 107, 975–984 CrossRef CAS.
- B. Katzin, Proteins: Struct., Funct., Bioinf., 1991, 10, 251–259 CrossRef CAS PubMed.
- CDC-10.20. 2013, http://www.bt.cdc.gov/agent/agentlist-category.asp.
- WWCA-10.20.2013, http://www.opcw.org/chemicalweapons-convention/download-the-cwc.
- W. Catterall, Neuron, 2000, 26, 13–25 CrossRef CAS.
- E. Schantz and E. Johnson, Microbiol. Rev., 1992, 56, 80–99 CAS.
- H. Tanino, T. Nakata, T. Kaneko and Y. Kishi, J. Am. Chem. Soc., 1977, 99, 2818–2819 CrossRef CAS.
- E. Schantz, Ann. N. Y. Acad. Sci., 1960, 90, 843–855 CrossRef CAS PubMed.
- H. D. Raj and M. S. Bergdoll, J. Bacteriol., 1969, 98, 833–834 CAS.
- A. Brosnahan and P. Schlievert, FEBS J., 2011, 278, 4649–4667 CrossRef CAS PubMed.
- N. Balaban and A. Rasooly, Int. J. Food Microbiol., 2000, 61, 1–10 CrossRef CAS.
- A. Lakoff and S. J. Collier, Biosecurity interventions: Global health and security in question, Columbia University Press, New York, 2012 Search PubMed.
- J. Gooding, Anal. Chim. Acta, 2006, 559, 137–284 CrossRef CAS PubMed.
- H. Bates and H. Rapoport, J. Agric. Food Chem., 1975, 23, 237–239 CrossRef CAS.
- R. Manger, L. Leja, S. Lee, J. Hungerford and M. Wekell, Anal. Biochem., 1993, 214, 190–194 CrossRef CAS PubMed.
- J. Jellett, L. Marks, J. Stewart, M. Dorey, W. Watson-Wright and J. Lawrence, Toxicon, 1992, 30, 1143–1156 CrossRef CAS.
- S. R. Kalb and J. R. Barr, Anal. Chem., 2009, 81, 2037–2042 CrossRef CAS PubMed.
- S. Kull, D. Pauly, B. Störmann, S. Kirchner, M. Stämmler, M. B. Dorner, P. Lasch, D. Naumann and B. G. Dorner, Anal. Chem., 2010, 82, 2916–2924 CrossRef CAS PubMed.
- T. McGrath, C. Elliott and T. Fodey, Anal. Bioanal. Chem., 2012, 403, 75–92 CrossRef CAS PubMed.
- E. Ezan, E. Duriez, F. Fenaille and F. Becher, Functional Assays for Ricin Detection, in Detection of Biological Agents for the Prevention of Bioterrorism, ed. J. Banoub, Springer Netherlands, Dordrecht, 2011, pp. 131–147 Search PubMed.
- W. Lian, D. H. Wu, D. V. Lim and S. G. Jin, Anal. Biochem., 2010, 401, 271–279 CrossRef CAS PubMed.
- O. G. Weingart, O. G. Weingart, H. Gao, F. Crevoisier, F. Heitger, M. A. Avondet and H. Sigrist, Sensors, 2012, 12, 2324–2339 CrossRef CAS PubMed.
- E. J. Cho, J. R. Collett, A. E. Szafranska, A. E. Szafranska and A. D. Ellington, Anal. Chim. Acta, 2006, 564, 82–90 CrossRef CAS PubMed.
- M. Huebner, K. Wutz, A. Szkola, R. Niessner and M. Seidel, Anal. Sci., 2013, 29, 461–466 CrossRef CAS.
- J. Cooper, N. Yazvenko, K. Peyvan, K. Maurer, C. R. Taitt, W. Lyon and D. L. Danley, PLoS One, 2010, 5, e9781 Search PubMed.
- A. Szkola, K. Campbell, C. T. Elliott, R. Niessner and M. Seidel, Anal. Chim. Acta, 2013, 787, 211–218 CrossRef CAS PubMed.
- S. E. McNamee, C. T. Elliot, P. Dalahnt and K. Campbell, Environ. Sci. Pollut. Res., 2013, 20, 6794–6807 CrossRef CAS PubMed.
- D. Pauly, S. Kirchner, B. Stoermann, T. Schreiber, S. Kaulfuss, R. Schade, R. Zbinden, M. Avondet, M. B. Dorner and B. Dorner, Analyst, 2009, 134, 2028–2039 RSC.
- M. Seidel and R. Niessner, Anal. Bioanal. Chem., 2008, 391, 1521–1544 CrossRef CAS PubMed.
- S. Oswald, X. Y. Z. Karsunke, R. Dietrich, E. Märtlbauer, R. Niessner and D. Knopp, Anal. Bioanal. Chem., 2013, 405, 6405–6415 CrossRef CAS PubMed.
- F. S. Ligler, C. R. Taitt, L. C. Shriver-Lake, K. E. Sapsford, Y. Shubin and J. P. Golden, Anal. Bioanal. Chem., 2003, 377, 469–477 CrossRef CAS PubMed.
- Y. Fang, A. G. Frutos and J. Lahiri, Langmuir, 2003, 19, 1500–1505 CrossRef CAS.
- M. Hartmann, M. Schrenk, A. Doettinger, S. Nagel, J. Roeraade, T. O. Joos and M. F. Templin, Clin. Chem., 2008, 54, 956–963 CAS.
- S. L. Lim, H. Ichinose, T. Shinoda and H. Ueda, Anal. Chem., 2007, 79, 6193–6200 CrossRef CAS PubMed.
- V. Parro, G. de Diego-Castilla, J. A. Rodriguez-Manfredi, L. A.Rivas, Y. Blanco-Lopez, E. Sebastian, J. Romeral, C. Compostizo, P. L. Herrero, A. Garcia-Marin, M. Moreno-Paz, M. Garcia-Villadangos, P. Cruz-Gil, V. Peinado, J. Martin-Soler, J. Perez-Mercader and J. Gomez-Elvira, Astrobiology, 2011, 11, 15–28 CrossRef CAS PubMed.
- P. Fernández-Calvo, L. A. Rivas, C. Naeke, M. García-Villadangos, J. Gómez-Elvira and V. Parro, Planet Space Sci., 2006, 54, 1612–1621 CrossRef PubMed.
- A. Akkoyun, V. Kohen and U. Bilitewski, Sens. Actuators, B, 2000, 70, 12–18 CrossRef CAS.
- Y. Pan, S. C. Yuhasz and L. M. Amzel, FASEB J., 1995, 9, 43–49 CAS.
- D. Pauly, S. Worbs, S. Kirchner, O. Shatohina, M. B. Dorner and B. G. Dorner, PLoS One, 2012, 7, e35360 CAS.
- A. Wolter, R. Niessner and M. Seidel, Anal. Chem., 2007, 79, 4529–4537 CrossRef CAS PubMed.
- V. Langer, R. Niessner and M. Seidel, Anal. Bioanal. Chem., 2011, 399, 1041–1050 CrossRef CAS PubMed.
- K. Kloth, R. Niessner and M. Seidel, Biosens. Bioelectron., 2009, 24, 2106–2112 CrossRef CAS PubMed.
- D. Li, Y. Ying, J. Wu, R. Niessner and D. Knopp, Microchim. Acta, 2013, 180, 711–717 CrossRef CAS.
- D. Pauly, M. Dorner, X. Zhang, A. Hlinak, B. Dorner and R. Schade, Poult. Sci., 2009, 88, 281–290 CrossRef CAS PubMed.
- P. D. Patel, Trends Anal. Chem., 2002, 21, 96–115 CrossRef CAS.
- E. C. Alocilja and S. M. Radke, Biosens. Bioelectron., 2003, 18, 841–846 CrossRef CAS.
- L. D. Mello and L. T. Kubota, Food Chem., 2002, 77, 237–256 CrossRef CAS.
- S. D. Richardson and T. A. Ternes, Anal. Chem., 2005, 77, 3807–3838 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ESI S1 Experimental details; S2 comparison between horseradish peroxidase–streptavidin (SA–HRP) and poly(horseradish peroxidase)–streptavidin (SA–PolyHRP40); S3 influence of the sequential and parallel addition of reactants; S4 influence of the flow rate and interaction time on the CL signal. See DOI: 10.1039/c4an00345d |
| This journal is © The Royal Society of Chemistry 2014 |