Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
T7 bacteriophage induced changes of gold nanoparticle morphology: biopolymer capped gold nanoparticles as versatile probes for sensitive plasmonic biosensors
Palanisamy
Kannan
a,
Marcin
Los
ab,
Joanna M.
Los
b and
Joanna
Niedziolka-Jonsson
*a
aInstitute of Physical Chemistry Polish Academy of Sciences, Kasprzaka 44/52, 01-224 Warsaw, Poland. E-mail: joaniek@ichf.edu.pl; Fax: +48 22 343 33 33; Tel: +48 22 343 31 30
bDepartment of Molecular Biology, University of Gdansk, Wita Stwosza 59, 80-308 Gdansk, Poland
First published on 6th May 2014
Abstract
The morphological changes of gold nanoparticles induced by T7 virus (bacteriophage) and the determination of its femtomolar concentration by a plasmonic method are presented. Carboxymethyl chitosan capped gold nanoparticles (CMC-AuNPs) are used as plasmonic probes and are synthesized by a simple one pot wet chemical method. HR-TEM images show that the spherical structure of the CMC-AuNPs is changed into chain-like nanostructures after the addition of T7 virus due to the strong coordination of CMC-AuNPs with T7. Since T7 capsids comprise a repeating motif of capsomers built from proteins that bind to the acid groups of chitosan, the conjugation of carboxymethyl chitosan-linked AuNPs with T7 virions enables colorimetric biosensing detection. The absorbance intensity (∼610 nm) increases in the concentration range of T7 from 2 × 10−15 M to 2 × 10−13 M and the detection limit is found to be 2 × 10−15 M (2 fM). The present work demonstrates eco-friendly biopolymer stabilized AuNPs as potential nanomaterials for biosensing of viruses. Our method is very simple, low cost, selective and highly sensitive, and provides new insight into virus induced chain-like morphology of AuNPs.
Introduction
Plasmonic metallic nanoparticles are the object of intensive research as they exhibit a characteristic optical response governed by the excitation of localized surface plasmon resonance (LSPR).1,2 The undisputed distinctive optical properties of plasmonic nanostructures have allowed rapid advances in many interdisciplinary subjects, such as surface enhanced spectroscopy, solar energy conversion, integrated optical nanocircuitry, LSPR sensing, etc. Due to their specific interaction with light and surface chemical properties, gold nanoparticles (AuNPs) provide a versatile sensing platform (nanoprobes) for biological applications such as drug delivery,3 cell labelling and imaging4 or cancer diagnostics and therapeutics, DNA, lectin, protein A, and heavy metal ions.5–9Colloidal AuNPs, especially those obtained by wet chemical synthesis, are interesting because of their ease of preparation with controlled size and morphology.10–12 More importantly, the optical properties of AuNPs are dominated by their LSPR which can be optimized for specific frequencies by modifying the particle size and shape.13,14 Due to the free electrons present on their surface, uncoated AuNPs are very sensitive to environmental factors such as pH, temperature, electrolytes and solvent, having the tendency to easily aggregate when used in these media. In order to obtain stable colloids, there is a need for the addition of protective species usually presenting thiols or amine functional groups,15 in the majority of the synthesis processes. Therefore, in this work we have used carboxymethyl chitosan (CMC) as a capping agent for the synthesis of AuNPs16 for the first time in virus recognition. Chitosan is a degradable biopolymer and it has several attractive properties including excellent film forming ability, high permeability toward water, good adhesion, non-toxicity and bio-compatibility.17–19 Derivatives of chitosan have been applied for various biological applications.20–22
It has been shown that hemagglutinin is a surface protein on various viral species that can bind to sialic acid on targeted cell surfaces.23 The binding of viral proteins to sialic acid has been studied for different pathogenic viruses, such as influenza virus,23–25 human parainfluenza virus,26 human coronavirus27,28 and a specific serotype of rhinovirus.29 For instance, Niikura et al. reported the use of gold nanoparticles functionalized by sialic acid-linked lipids to detect virus-like particles. That study demonstrated the synthesis of the sialic acid-linked lipid in four complicated steps and a subsequent step to finally obtain the sialic acid-functionalized gold nanoparticles for virus-like particle detection. Recently, Lee et al.30 synthesized sialic acid capped AuNPs at 80 °C for colorimetric detection of influenza virus. It has also been known that sialic acid is expensive, and several functional modification steps or heat treatment is required for preparing AuNPs. Moreover, the development of alternative new and low cost nanomaterials is very important for both synthesis and biosensor aspects.
Bacteriophages, also called phages, are viruses which infect bacteria. They cannot develop in different e.g. human cells. One of the best studied phage groups is the so-called “T” series of phages.31 One representative of this group is the bacteriophage T7, belonging to the phage family Podoviridae, which is an excellent model for development of novel virus detection methods. The structure of this virus is quite similar to adenoviruses, which constitute medical concerns in humans, causing a wide range of diseases. The phage T7 is smaller compared to adenoviruses (55 and 90 nm respectively), but both are non-enveloped viruses. T7 does not contain spikes, which are characteristic for adenoviruses, but instead it has a tail with tail fibres. The tail of T7 is very short and does not contribute to the overall capsid mass as much as in tailed bacteriophages from other families, namely Myoviridae and Siphoviridae. Except for morphological similarities the phage T7 provides an easy and safe working environment, as it is very easy to quantify, stable in solution, non-contagious for humans, animals and plants and easy to produce. Although T7 does not constitute any health hazard, rapid detection methods capable of sensing the presence of phages may be very useful in the biotechnology industry, since phages constitute one of the greatest concerns in bacteria-based industrial processes.32
In the present work, we describe a plasmonic biosensor for T7 virus using CMC-AuNPs as plasmonic transducers. In our method we used carboxymethyl chitosan as a stabilizing agent16 for preparing AuNPs, which is a low cost material with no special modifications required, highly selective and highly sensitive for viral detection and less time consuming than the previously reported sialic acid functionalized gold nanoparticles (vide infra).30,33,34 The absorption intensity of CMC-AuNPs is decreased at 520 nm and a new absorption band is observed in the range 610–670 nm while adding T7 virions. Based on the decrease in absorption intensity at 520 nm followed by the growth of the new absorption band at ca. 610 nm, we have detected the presence of T7 phages and measured their concentration. Our experimental detection limit is found to be 2 × 10−15 M (2 fM) of T7 virus.
Experimental
Materials
Hydrogen tetrachloroaurate(III) trihydrate (HAuCl4·3H2O), chitosan, monochloroacetic acid, glucose, trisodium citrate, CTAB, poly(allylamine), poly(lysine) and sodium borohydride acid were purchased from Sigma-Aldrich. The T7 bacteriophages (4.8 × 108 PFU mL−1 = 8 × 10−13 M) were stored in phosphate buffer saline at a pH of 7.2. The M13 bacteriophages (8 × 108 PFU mL−1 = 1.3 × 10−12 M) were stored in Tris–HCl buffer at a pH of 7.4. All other chemicals used in this investigation were of analytical grade and used directly without further purification. Millipore water (18 MΩ cm) was used to prepare the solutions in the present work.Instrumentation
Absorption spectra were recorded in 1 cm quartz cuvettes at room temperature by using a Thermo Evolution 300 UV-visible spectrophotometer. High resolution transmission electron microscopy (HR-TEM) images of the AuNPs were obtained using a JEOL JEM 2010 operating at 200 kV. For TEM measurements, the sample was prepared by dropping 2 μL of a CMC-AuNP colloidal solution onto a carbon-coated copper grid. A large volume (200 mL) of CMC-AuNPs was prepared, and particles were separated by centrifugation (8000 rpm). Then the CMC-AuNPs were repeatedly washed with water and dried in a vacuum. The dried CMC-AuNP powder was used for XPS and XRD measurements. X-ray diffraction (XRD) patterns were obtained on a Rigaku D/max-IIIB diffractometer using Cu-Kα (l = 1.5406 Å) at a step scan of 0.02θ, from 30 to 80θ. The accelerating voltage and the applied current were 40 kV and 20 mA. X-ray photoelectron spectroscopy (XPS) analysis was performed on a VG ESCALAB MK II with an MgKα (1253.6 eV) achromatic X-ray source.Preparation of carboxymethyl chitosan (CMC)
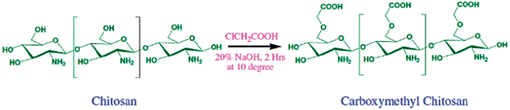
Carboxymethyl chitosan was synthesized using procedures similar to the method described by Schauer.16 Preparation of CMC was carried out by dissolving 5 g of chitosan in 20% NaOH (w/v, 100 mL) with constant stirring and then monochloroacetic acid (15 g) was added drop-wise to the reaction mixture and the reaction was continued for the next 2 h at 40 ± 2 °C with stirring. Then the reaction mixture was neutralized by using 10% acetic acid, then it was poured into an excess of 70% methanol. The produced carboxymethyl chitosan was filtered using a G2 sintered funnel and was washed with more methanol. The product was dried in a vacuum at 55 °C for 2 h to yield 6.5 g of dried carboxymethyl chitosan (Scheme 1). The degree of substitution of CMC was determined to be 0.75 according to the method described in the literature.35,36Synthesis of CMC-AuNPs
All glassware was thoroughly cleaned with freshly prepared aqua regia (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
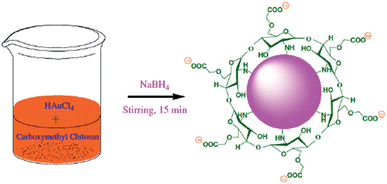
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, HCl–HNO3) and rinsed comprehensively with Millipore water prior to use. CMC-AuNPs were synthesized using a slight modification of the procedure described by Schauer16 through a wet chemical method. 0.5 mL of carboxymethyl chitosan (CMC 1%) was added to 46 mL of Millipore water in a round bottom flask with constant stirring. Then, 0.5 mL of HAuCl4·3H2O (0.03 M) was added to the stirred solution of CMC followed by slow addition of 1.5 mL of NaBH4 (0.1%). The colour of the solution immediately turns wine red (Scheme 2). Stirring was continued for another 30 min and the solution was stored in a refrigerator for further use.
1, HCl–HNO3) and rinsed comprehensively with Millipore water prior to use. CMC-AuNPs were synthesized using a slight modification of the procedure described by Schauer16 through a wet chemical method. 0.5 mL of carboxymethyl chitosan (CMC 1%) was added to 46 mL of Millipore water in a round bottom flask with constant stirring. Then, 0.5 mL of HAuCl4·3H2O (0.03 M) was added to the stirred solution of CMC followed by slow addition of 1.5 mL of NaBH4 (0.1%). The colour of the solution immediately turns wine red (Scheme 2). Stirring was continued for another 30 min and the solution was stored in a refrigerator for further use.
Production and purification of bacteriophage T7
40 mL of LB media (Roth) in a 250 mL conical flask was inoculated from a single colony of Escherichia coli MG1655. After overnight incubation at 37 °C with shaking (150 rpm), the culture of E. coli MG1655 was used to inoculate in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 ratio 2 5 L conical flasks containing 2 L of LB each. Shake flasks were incubated at 37 °C with shaking (150 rpm) and the growth was monitored. When OD600 of bacterial culture reached 0.2 approximately 106 bacteriophage T7 per flask were added. After lysis occurred (approx. 3–4 h after phage addition), lysate was centrifuged in a Beckman Avanti J20 using a JA10 rotor at RCF = 6000g for 20 minutes. The supernatant was collected, the NaCl concentration was adjusted to 1 M, and PEG8000 was added to a final concentration of 10%, and the solution was left in a cold room with stirring overnight. The next day phage lysate was centrifuged in the Beckman Avanti J20 using the JA10 rotor at RCF = 10
100 ratio 2 5 L conical flasks containing 2 L of LB each. Shake flasks were incubated at 37 °C with shaking (150 rpm) and the growth was monitored. When OD600 of bacterial culture reached 0.2 approximately 106 bacteriophage T7 per flask were added. After lysis occurred (approx. 3–4 h after phage addition), lysate was centrifuged in a Beckman Avanti J20 using a JA10 rotor at RCF = 6000g for 20 minutes. The supernatant was collected, the NaCl concentration was adjusted to 1 M, and PEG8000 was added to a final concentration of 10%, and the solution was left in a cold room with stirring overnight. The next day phage lysate was centrifuged in the Beckman Avanti J20 using the JA10 rotor at RCF = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 25 minutes, the supernatant was discarded and a pellet containing the precipitated phages was resuspended in a solution containing 1 M NaCl, 10 mM MgSO4 and 10 mM Tris–HCl pH 7.4. The suspension was mixed with an equal volume of chloroform in order to extract the remaining PEG8000 and centrifuged in the Beckman Avanti J20 using a JS13.1 rotor at 5000 rpm for 20 minutes. Supernatants containing phages were then purified using CsCl gradient phage purification according to the method described by Sambrook et al.37 Phages during CsCl gradient purification were concentrated and formed a clearly visible band, which was collected. Concentrated phages then were dialysed against PBS pH 7.2.
000g for 25 minutes, the supernatant was discarded and a pellet containing the precipitated phages was resuspended in a solution containing 1 M NaCl, 10 mM MgSO4 and 10 mM Tris–HCl pH 7.4. The suspension was mixed with an equal volume of chloroform in order to extract the remaining PEG8000 and centrifuged in the Beckman Avanti J20 using a JS13.1 rotor at 5000 rpm for 20 minutes. Supernatants containing phages were then purified using CsCl gradient phage purification according to the method described by Sambrook et al.37 Phages during CsCl gradient purification were concentrated and formed a clearly visible band, which was collected. Concentrated phages then were dialysed against PBS pH 7.2.
Results and discussion
Spectral characterization and stability of the CMC-AuNPs
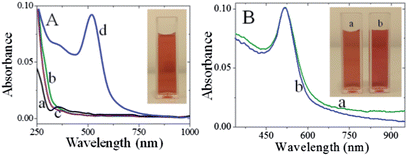
When metal nanoparticles are exposed to light, the oscillating electromagnetic field of the light induces collective coherent oscillation of the free electrons (conduction band electrons) of the metal nanoparticles.10 This electron oscillation around the particle surface causes charge separation with respect to the ionic lattice, forming dipole oscillation along the direction of the electric field of the light. The amplitude of the oscillation reaches maximum at a specific frequency, called the surface plasmon resonance (SPR) (vide supra).10,38,39 The SPR induces strong absorption of the incident light and thus can be measured using a UV-visible spectrophotometer. Fig. 1 shows the absorption spectra recorded for CMC, HAuCl4 and a mixture of CMC and HAuCl4 in the presence and absence of NaBH4.CMC shows an absorption maximum at 350 nm (curve a), whereas HAuCl4 in water exhibits a broad absorption maximum at 290 nm (curve b). Addition of HAuCl4 solution to an aqueous solution of CMC causes the absorption band characteristics of CMC to decrease while the absorption intensity at 290 nm was 20 nm blue shifted (curve c). The observed changes in intensity at 290 nm were ascribed to the complex formation between CMC and HAuCl4. When 1.5 mL of 0.01% NaBH4 was slowly added to a mixture of CMC and HAuCl4 solution, the pale-yellow solution becomes wine red immediately (inset of Fig. 1A) and shows a new absorption band at 520 nm (curve d). This new band corresponds to the SPR band of the AuNPs, confirming the successful formation of AuNPs. Additionally, the presence of the band at 350 nm proves their modification with CMC. The stability of the AuNPs was usually checked from the changes in their absorption characteristics such as shift in the absorption maximum and decrease in the absorbance. In comparison to freshly prepared CMC-AuNPs, no change in the absorption characteristics of one month aged CMC-AuNPs was observed (Fig. 1B). Further, the wine red colour of the solution also remains the same (inset of Fig. 1B). These results indicate that the synthesized CMC-AuNPs were highly stable. For highly reproducible virus detection it is imperative to accurately control the parameters and results of the synthesis.
HR-TEM study
The size and morphology of the synthesized CMC-AuNPs were examined by HR-TEM. Fig. 2 shows the HR-TEM images of CMC-AuNPs taken at different magnifications. The HR-TEM images show that the CMC-AuNPs were spherical in shape with a size of ∼6 nm and have a narrow size distribution (Fig. 2A and B). The high magnification HR-TEM image of a CMC-AuNP originating from a single crystal shows the lattice images (Fig. 2C). Fig. 2D shows the selected area diffraction (SAED) pattern of CMC-AuNPs. The SAED pattern of CMC-AuNPs illustrates the predominant Au(111) crystalline nature of AuNPs.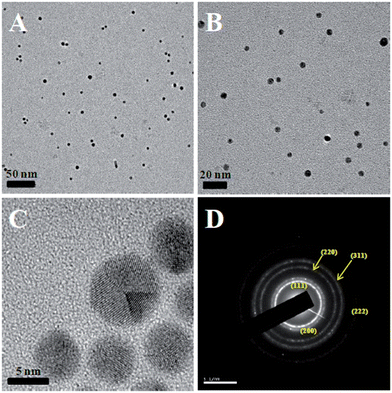 | ||
| Fig. 2 HR-TEM images of different magnifications (A–C) and selected area diffraction (D) of CMC-AuNPs. | ||
Determination of the concentration of AuNPs and surface coverage of CMC ligands
We have estimated the concentration of AuNPs from the number of atoms per particle “N” using the following equation.40| N = πρD3NA/6M | (1) |
| C = NT/NVNA | (2) |
Characterization of AuNPs by XRD and XPS
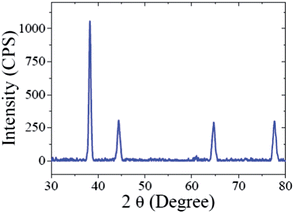
The crystalline nature of the CMC-AuNPs was confirmed by the XRD analysis. Fig. 3 shows the diffraction features appearing at 38.17°, 44.43°, 64.52°, and 77.48° corresponding to (111), (200), (220), and (311) respectively.15 The peak corresponding to the (111) plane is more intense than the other planes. The ratio between the intensity of the (200) and (111) diffraction peaks was much lower (0.5), suggesting that the (111) plane is a predominant orientation. The width of the (111) peak was employed to calculate the average crystalline size of the CMC-AuNPs using the Scherrer equation.42 The calculated average size of the AuNPs is ∼6.5 nm, which closely matches with the particle size obtained from HR-TEM.Further, the CMC-AuNPs were characterized by XPS. Fig. 4 shows the XPS spectra obtained for CMC-AuNPs. The survey spectrum of the CMC-AuNPs (Fig. 4A) indicates the presence of Au, C and N species in the AuNPs. The XPS spectrum of Au 4f shows two peaks at 85.75 eV for 4f7/2 and 89.45 eV for 4f5/2 (Fig. 4B). The position and binding energy difference between the two peaks (3.7 eV) exactly match with the value reported for Au0.43 The absence of a peak at 84.9 eV corresponding to Au(III) indicates that the gold atoms are present as Au0.43 The C 1s spectrum shows a binding energy at 285.41 eV (Fig. 4C), suggesting that CMC remains on the surface of AuNPs. The N 1s band is observed at 399.20 eV, which corresponds to the nitrogen atom of CMC molecules attached to Au nanoparticles.44 Moreover, the binding energy at 401 eV (Fig. 4D) was absent in our XPS N 1s spectrum, which was assigned to a free amine group (–NH2).44 The obtained XPS results for CMC-AuNPs clearly confirmed the presence of CMC molecules on the surface of AuNPs and the absence of Au ions in the colloidal nanoparticle solution.
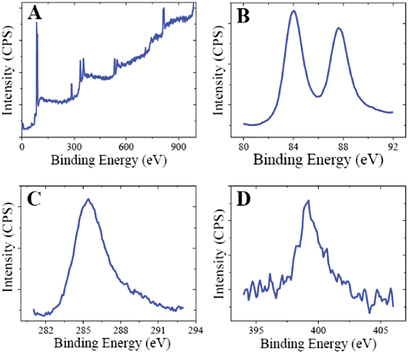 | ||
| Fig. 4 XPS spectra of CMC-AuNPs: (A) survey spectrum and enlarged XPS spectra of CMC-AuNPs in the (B) Au 4f, (C) C 1s and (D) N 1s regions. | ||
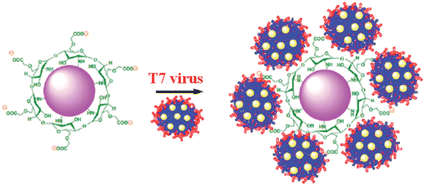
T7 virus in CMC-AuNPs
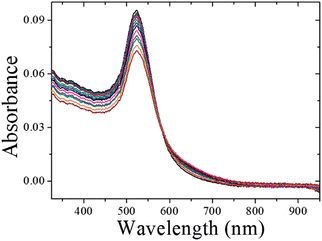
The schematic representation of using CMC-AuNPs for virus detection is shown in Scheme 3.It is known that the carboxylic acid group of CMC plays a critical role in the binding30,33 of the viral capsid protein on the viral particles to CMC on the surface of cells. The CMC layer on the AuNPs' surface prevents the aggregation of the AuNPs in solution and the carboxylic acid group will face outward after CMC-AuNPs are formed. In the presence of virus, the AuNPs are attached to the viral capsid protein on the viral particle surface, resulting in a shorter distance between each AuNP and chain-like nanostructures. It is expected that interaction among AuNPs would lead to a change in the absorption spectra, thus signalling the presence of viral particles. The progress of the T7 virus biosensor was followed by UV-visible spectral measurements. CMC-AuNPs (2 mL) and T7 virus (5 μL of 4.8 × 108 PFU mL−1 = 4.0 × 10−15 M) were mixed and introduced into the quartz cell, and spectra were recorded at regular intervals. The colour of the CMC-AuNP solution changed from wine red to purple within ∼45 min of reaction. Fig. 5 shows the time-dependent spectral response obtained during the detection of T7 virus on the AuNPs. The spectra recorded in the early stage (up to 3 to 15 min) show a slight change in the absorbance around 575 nm.
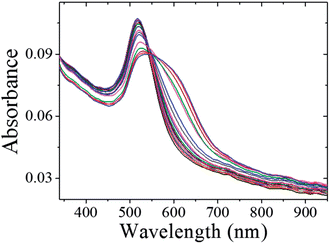 | ||
| Fig. 5 UV-vis spectra CMC-AuNPs with 2.0 × 10−15 M of T7 phages. UV-vis absorption spectra measured at 0, 1, 3, 5, 7, 9, 11, 13, 15, 20, 25, 30, 35, 40, 45 and 50 min after the addition of T7 phages. | ||
Then, the absorbance at ∼575 nm increases with respect to time while a new shoulder around 575–675 nm (longitudinal peak) appeared after 20 min on the longer wavelength side. With time, the shoulder shifts to longer wavelength and stabilizes at ∼625 nm after the completion of the reaction (50 min). These time-dependent features can be ascribed either to the aggregation of spherical nanoparticles or to the formation of anisotropic nanostructures. Because the absorbance of both bands at 520 and ∼625 nm remains at the end of the reaction, it is concluded that the observed feature is not due to the aggregation of nanoparticles. The intense band observed at 520 nm is ascribed to the existence of the transverse component of the SP absorption whereas the shoulder observed at ∼625 nm is assigned to the longitudinal component of SP absorption. The SP absorption of the coinage metal nanoparticles, in general, is more sensitive to the particle shape and surrounding medium than to the size.45,46 The presence of two well-separated absorption bands is a characteristic feature of anisotropic nanoparticles. Such features have been observed for nanostructured Au particles such as nanorods and nanodots.47,48 The position and intensity of the longitudinal band depend on the size, surface morphology, and aspect ratio. For instance, in the case of nanorods the longitudinal absorption band is dominant.48 However, branched nanocrystals have a single broad band, and nanocrystals having bipod and tripod structures have a less intense longitudinal band and a dominant transverse band.49,50 In our study, we hypothesized that the observed visible color change and appearance of a new peak in a higher wavelength region are due to the structural changes of CMC-AuNPs. This was confirmed by HR-TEM. It has been well established that when the spherical structure of AuNPs was changed, a new absorption band would appear at higher wavelength.51Fig. 6 shows the HR-TEM images of CMC-AuNPs recorded at different magnifications after the addition of T7 virus (4.0 × 10−15 M). In contrast to the image of CMC-AuNPs (Fig. 2), the HR-TEM images recorded at different magnifications in the presence of T7 virus show that the spherical morphology of the CMC-AuNPs was changed into a chain-like structure (Fig. 6; images: A–D).
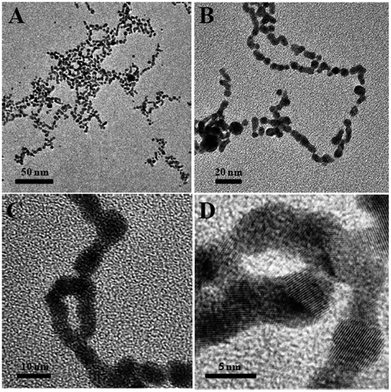 | ||
| Fig. 6 HR-TEM images of different magnifications of CMC-AuNPs after the addition of 4.0 × 10−15 M T7 virus. | ||
The changes in the morphology of CMC-AuNPs can be explained as follows. While adding T7 virus into CMC-AuNPs, the monodispersed spherical AuNPs come closer due to the decrease in electrostatic repulsion and they coordinate with the T7 virus. This leads to the formation of a chain-like structure. Since T7 virus comprises a repeating viral capsid protein that binds to the acid group of CMC, the conjugation of CMC-linked Au particles with T7 virus enables the chain-like arrangement of Au particles. This structure produced a red shift in the absorption spectrum due to plasmon coupling between adjacent Au particles, leading to the construction of an optical virus detection system. Moreover, the transverse band is more intense than the longitudinal band. The longitudinal band appears as a shoulder-like peak, indicating that the T7 phages induced the anisotropic chain-like shape of the CMC-AuNPs. These spectral features are very similar to those observed for Au nanowires, and monopod and bipod nanocrystals.50 Our system depends not on the simple linking of T7 virus and Au particles, but on the formation of ordered Au chain-like nanostructures. The results obtained from the UV-visible and HR-TEM analyses are also in good agreement with Mie theory.52 According to Mie theory, when the distance between the two nanoparticles becomes smaller than the sum of their radii, the UV-visible spectra show a broad band in a higher wavelength region.51,52 The changes in the absorption characteristics of AuNPs after the addition of T7 virus may be used to determine the concentration of T7 virus. Notably, the spectral changes were observed in the femtomolar concentration range of T7 virus. This indicates that our CMC-AuNPs were highly suitable to determine trace level concentrations of T7 virus.
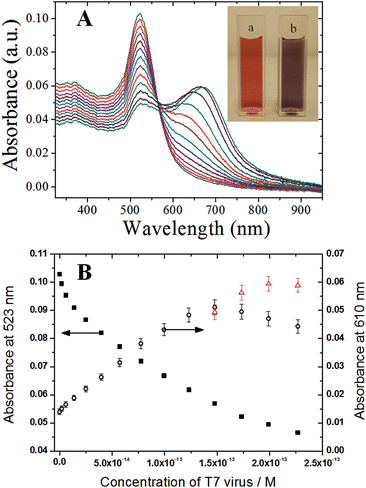
The dynamic range and the sensitivity of the present colorimetric viral sensor were quantitatively determined by the examination of the absorption change and peak wavelength shifts of the colorimetric viral sensor upon the affinity of T7 virus on CMC-AuNPs ranging from 2.0 × 10−15 M to 2.3 × 10−13 M (Fig. 7). Calibration was done at 610 nm, following the new band formation. Spectra were recorded 20 min after T7 addition. At higher concentrations the shoulder turns into a new peak. Therefore additional points were added for the last four concentrations obtained at the peak maxima at 626 nm, 647 nm, 664 nm and 670 nm respectively (red triangles). The decrease of the absorption band at 523 nm includes both the effect of the structural changes of the CMC-AuNPs and dilution of the sample by addition of solution. To exclude the dilution effect of the CMC-AuNPs and its contribution to the virus recognition the experiment was performed with adding only PBS buffer. In this case no changes in the spectrum were recorded at 610 nm.
Small standard deviations over several replicates at different concentrations of T7 virus were obtained (RSD = 2–5%, n = 3–7), which shows that the response of the colorimetric viral sensor is highly consistent during the recognition process. The reproducible properties of the CMC-AuNPs allowed for the establishment of an accurate calibration curve. As a result, a dynamic range of 2.0 × 10−15 M to 1.5 × 10−13 M was obtained. In addition, based on experimental data, a limit of detection (LOD; defined as the smallest quantity of T7 that can be distinguished from its absence) of 2.0 × 10−15 M (2 fM) was achieved, which is 1–2 orders of magnitude better than the sialic acid based viral particle biosensor system, recently reported by Niikura et al.33 and Lee et al.30
Comparison of the CMC-AuNPs vs. other functionalized AuNPs and selectivity towards T7 virus detection
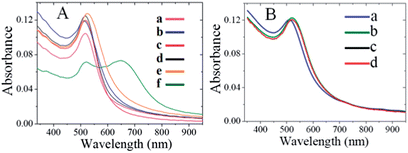
Plasmonic detection of T7 virus (1.7 × 10−13 M) was performed in the presence of citrate-AuNPs, CTAB-AuNPs, PAA-AuNPs, PL-AuNPs and glucose-AuNPs under optimized conditions and the results are presented in Fig. 8.The absorption spectra of citrate-AuNPs, CTAB-AuNPs, PAA-AuNPs, and PL-AuNPs do not change after spiking with T7 phages into the respective AuNP solution (curves a–d), However, while adding 1.7 × 10−13 M T7 virus into the glucose stabilized AuNP solution (curve e), the absorption was broadened in a small amount compared to the other AuNPs, this impact may be due to the structure similarity effect i.e., the presence of a monosaccharide unit in glucose-AuNPs like CMC-AuNPs. Interestingly, while adding the same amount of T7 viruses into CMC-AuNP solution, a distinguished new absorption around 650 nm was clearly observed (curve f). To check the selectivity of the CMC-AuNPs, a very high concentration of 1 × 10−6 M of BSA was added to the fresh suspension (Fig. 8B). No change in the colour of the solution was observed and only a slight (4 nm) red-shift was noticed, probably due to the change in the closest CMC-AuNP environment. This proves that this system is selective towards viruses and not proteins.
To further ascertain the selectivity of the present plasmonic biosensor, we have tested M13 bacteriophage as an example. UV-visible spectra of CMC-AuNPs in different concentrations of M13 virus are shown in Fig. 9, indicating that the CMC-Au nanoparticles afford a suitable control for the selectivity toward other family viruses (filamentous M13 type virus). The addition of M13 virus to a solution of CMC-Au nanoparticles did not cause a plasmon shift (Fig. 9). Moreover, no red shift of the CMC-Au nanoparticles was observed in the presence of M13 virus. Moreover, the presence of acid functionality in chitosan plays a key role in specific interactions toward the viruses. These results confirmed that the CMC-AuNPs showed high affinity toward T7 viruses. The difference in binding capabilities toward T7 and M13 may arise from the different properties of the virions of both virus types which arise from their different adsorption strategies. T7, similar to many types of viruses including adenoviruses, during adsorption is subject to two different interactions – repulsing electrostatic force of the negatively charged cell surface against the negatively charged head of the bacteriophage, and attraction of the positively charged tail with the adsorption apparatus. In the case of adenoviruses the positive charge is localized in spikes, which protrude from the capsid. M13 does not adsorb onto cell surfaces. Its receptor is sex pilus, which protrudes far away from bacterial cells, where electrostatic interactions may not be strong enough to repulse the phage and prevent binding. Thus the highly specialized adsorption modules, which are found in many viruses, may be the key factor responsible for the observed effect. The evidence supporting such a hypothesis is the lack of detection of the M13 phage, which has an adsorption apparatus that comprises only 5 small proteins and the lack of detection of BSA, which is a protein well known for its good binding to different surfaces, and moreover, a transportation protein capable of binding different molecules. It is known that chitosan and its carboxylic derivative form complexes with transition and heavy metal ions.53 We tested our CMC-AuNPs in the presence of a relatively high concentration of Fe3+ (1 mg L−1 equal to the total Fe concentration in human blood). A noticeable response was seen, on the level comparable to a high concentration of M13. We also performed HR-TEM measurements for glucose-AuNPs before and after the addition of T7 phages, shown in Fig. 10. The HR-TEM images show that the glucose-AuNPs were closely spherical in shape with a size of ∼8 nm and have a narrow size distribution (Fig. 10A). In contrast to the image of glucose-AuNPs (Fig. 10A), the HR-TEM images were recorded after spiking of 1.7 × 10−13 M.
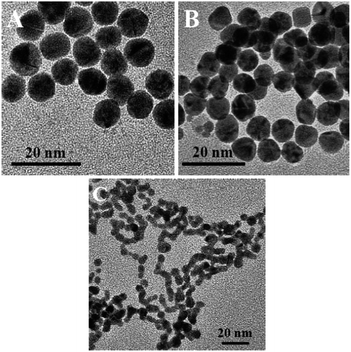 | ||
| Fig. 10 HR-TEM images of glucose-AuNPs before and after the addition of 1.7 × 10−13 M T7 virus. (C) CMC-AuNPs after the addition of 1.7 × 10−13 M T7 phages. | ||
T7 phages showed a small amount of collapsed spherical morphology of the glucose-AuNPs (Fig. 10B), observed due to the structure similarity effect (vide supra). Interestingly, in the HR-TEM images of CMC-AuNPs recorded after the addition of 1.7 × 10−13 M T7 phages, a clear chain-like AuNP morphology was observed (Fig. 10C). Therefore the above results confirm that CMC-AuNPs were potential nanomaterials for the plasmonic T7 viral biosensor. It is worth comparing the analytical performance and advantage of our studies with those of available plasmonic virus biosensors. Niikura33et al. have reported the use of gold nanoparticles functionalized by sialic acid-linked lipids to detect virus-like particles. That study included the synthesis of the sialic acid-linked lipid in four complicated steps and a subsequent step to finally obtain the sialic acid-functionalized gold nanoparticles for virus-like particle detection. A recent paper reported (Lee et al.)30 the one-pot synthesis of sialic acid–AuNPs for demonstration of colorimetric detection of actual virus. The above procedures were tedious to prepare the nanoparticles, expensive, and time consuming and gave low sensitivity. Our study demonstrates that carboxymethyl chitosan is an alternative and low cost nanomaterial for preparation of AuNPs and the resulting material shows higher sensitivity to the virus detection. Moreover, carboxymethyl chitosan possesses a monosaccharide backbone unit, which is similar to the isomeric structure of sialic acid. Thus, the present plasmonic biosensor was very simple to fabricate, highly stable, sensitive, selective, and reproducible. Our findings conclude that CMC-AuNPs can be developed as efficient and versatile high performance plasmonic nanomaterials for plasmonic T7 virus biosensor application.
Conclusions
We demonstrated the morphological changes of gold nanoparticles induced by T7 virus and the determination of femtomolar concentrations of T7 virus by a simple plasmonic method. The HR-TEM images show that the spherical structure of CMC-AuNPs is changed into a chain-like nanostructure after the addition of T7 virus due to the strong coordination (affinity) of CMC-AuNPs with T7 virus. Since T7 virions comprise a repeating viral capsid protein that binds to the acid group of chitosan, the conjugation of carboxymethyl chitosan-linked AuNPs with T7 viral particles enables the colorimetric biosensing detection. The absorbance intensity was measured vs. the concentration range of T7 phages from 2.0 × 10−15 M to 2.3 × 10−13 M and the detection limit was found to be 2.0 × 10−15 M (2 fM). The present work demonstrates simple biopolymer stabilized AuNPs as potential nanomaterials for biosensing of T7 viruses. Our method is very simple, low cost, and highly sensitive and selective, and provides new insight into the virus induced chain-like morphology of AuNPs.Acknowledgements
This work was financed by the Foundation for Polish Science within the FOCUS Program no. F3/2010/P/2013. Palanisamy Kannan thanks the European Union FP7, under the grant REGPOT-CT-2011-285949-NOBLESSE for financial support.References
- J. N. Anker, W. Paige Hall, O. Lyandres, N. C. Shah, J. Zhao and R. P. V. Duyne, Nat. Mater., 2008, 7, 442–453 CrossRef CAS PubMed.
- J. A. Schuller, E. S. Barnard, W. Cai, Y. Chul Jun, J. S. White and M. L. Brongersma, Nat. Mater., 2010, 9, 193–204 CrossRef CAS PubMed.
- G. F. Paciotti, D. G. I. Kingston and L. Tamarkin, Drug Dev. Res., 2006, 67, 47–54 CrossRef CAS.
- R. A. Farrer, F. L. Butterfield, V. W. Chen and J. T. Fourkas, Nano Lett., 2005, 5, 1139–1142 CrossRef CAS PubMed.
- C. A. Mirkin, R. L. Letsinger, R. C. Mucic and J. J. Storhoff, Nature, 1996, 382, 607–609 CrossRef CAS PubMed.
- F. X. Gu, R. Karnik, A. Z. Wang, F. Alexis, E. Levy-Nissenbaum, S. Hong, R. S. Langer and O. C. Farokhzad, Nano Today, 2007, 2, 14–21 CrossRef.
- H. Otsuka, Y. Akiyama, Y. Nagasaki and K. Kataoka, J. Am. Chem. Soc., 2001, 123, 8226–8230 CrossRef CAS PubMed.
- N. T. K. Thanh and Z. Rosenzweig, Anal. Chem., 2002, 74, 1624–1628 CrossRef CAS.
- Y. Kim, R. C. Johnson and J. T. Hupp, Nano Lett., 2001, 1, 165–167 CrossRef CAS.
- M.-C. Daniel and D. Astruc, Chem. Rev., 2003, 104, 293–346 CrossRef PubMed.
- A. J. Mieszawska, W. J. M. Mulder, Z. A. Fayad and D. P. Cormode, Mol. Pharmaceutics, 2013, 10, 831–847 CrossRef CAS PubMed.
- C.-H. Kuo, T.-F. Chiang, L.-J. Chen and M. H. Huang, Langmuir, 2004, 20, 7820–7824 CrossRef CAS PubMed.
- P. Jain, X. Huang, I. El-Sayed and M. El-Sayed, Plasmonics, 2007, 2, 107–118 CrossRef CAS.
- J. Zhang and C. Noguez, Plasmonics, 2008, 3, 127–150 CrossRef CAS.
- P. Kannan and S. Abraham John, Nanotechnology, 2008, 19, 085602 CrossRef PubMed.
- M. J. Laudenslager, J. D. Schiffman and C. L. Schauer, Biomacromolecules, 2008, 9, 2682–2685 CrossRef CAS PubMed.
- M. N. V. Ravi Kumar, React. Funct. Polym., 2000, 46, 1–27 CrossRef.
- C. K. S. Pillai, W. Paul and C. P. Sharma, Prog. Polym. Sci., 2009, 34, 641–678 CrossRef CAS PubMed.
- M. Dash, F. Chiellini, R. M. Ottenbrite and E. Chiellini, Prog. Polym. Sci., 2011, 36, 981–1014 CrossRef CAS PubMed.
- S. C. Boca, M. Potara, F. Toderas, O. Stephan, P. L. Baldeck and S. Astilean, Mater. Sci. Eng., C, 2011, 31, 184–189 CrossRef CAS PubMed.
- M. Stefan, V. Melnig, D. Pricop, A. Neagu, M. Mihasan, L. Tartau and L. Hritcu, Mater. Sci. Eng., C, 2013, 33, 550–556 CrossRef CAS PubMed.
- S. C. Boca, M. Potara, A.-M. Gabudean, A. Juhem, P. L. Baldeck and S. Astilean, Cancer Lett., 2011, 311, 131–140 CrossRef CAS PubMed.
- N. K. Sauter, J. E. Hanson, G. D. Glick, J. H. Brown, R. L. Crowther, S. J. Park, J. J. Skehel and D. C. Wiley, Biochemistry, 1992, 31, 9609–9621 CrossRef CAS.
- J. N. Varghese, P. M. Colman, A. van Donkelaar, T. J. Blick, A. Sahasrabudhe and J. L. McKimm-Breschkin, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 11808–11812 CrossRef CAS.
- W. Weis, J. H. Brown, S. Cusack, J. C. Paulson, J. J. Skehel and D. C. Wiley, Nature, 1988, 333, 426–431 CrossRef CAS PubMed.
- T. Suzuki, A. Portner, R. Ann Scroggs, M. Uchikawa, N. Koyama, K. Matsuo, Y. Suzuki and T. Takimoto, J. Virol., 2001, 75, 4604–4613 CrossRef CAS PubMed.
- R. Vlasak, W. Luytjes, W. Spaan and P. Palese, Proc. Natl. Acad. Sci. U. S. A., 1988, 85, 4526–4529 CrossRef CAS.
- F. Künkel and G. Herrler, Virology, 1993, 195, 195–202 CrossRef PubMed.
- C. R. Uncapher, C. M. Dewitt and R. J. Colonno, Virology, 1991, 180, 814–817 CrossRef CAS.
- C. Lee, M. A. Gaston, A. A. Weiss and P. Zhang, Biosens. Bioelectron., 2013, 42, 236–241 CrossRef CAS PubMed.
- S. T. Abedon, Genetics, 2000, 155, 481–486 CAS.
- M. Los, in T Microbial Metabolic Engineering, 2012, vol. 834, pp. 305–315 Search PubMed.
- K. Niikura, K. Nagakawa, N. Ohtake, T. Suzuki, Y. Matsuo, H. Sawa and K. Ijiro, Bioconjugate Chem., 2009, 20, 1848–1852 CrossRef CAS PubMed.
- I. Papp, C. Sieben, K. Ludwig, M. Roskamp, C. Böttcher, S. Schlecht, A. Herrmann and R. Haag, Small, 2010, 6, 2900–2906 CrossRef CAS PubMed.
- Y. G. Wu, W. L. Chan and Y. S. Szeto, J. Appl. Polym. Sci., 2003, 90, 2500–2502 CrossRef CAS.
- R. Farag and R. Mohamed, Molecules, 2012, 18, 190–203 CrossRef PubMed.
- Molecular cloning: A laboratory manual, ed. J. Sambrook, E. F. Fritsch and T. Maniatis, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 1989 Search PubMed.
- K.-S. Lee and M. A. El-Sayed, J. Phys. Chem. B, 2006, 110, 19220–19225 CrossRef CAS PubMed.
- X. Huang and M. A. El-Sayed, J. Adv. Res., 2010, 1, 13–28 CrossRef PubMed.
- R. T. Tom, V. Suryanarayanan, P. G. Reddy, S. Baskaran and T. Pradeep, Langmuir, 2004, 20, 1909–1914 CrossRef CAS.
- http://www.cristalvillage.org/NPs%20calc.htm .
- B. D. Cullity, Elements of X-ray Diffraction, Addison-Wesley, Menlo Park, CA, 2nd edn, 1978 Search PubMed.
- B. Vincent Christ, Hand Books of Monochromatic XPS Spectra, XPS international LLC, 2006, vol. II Search PubMed.
- M. M. Kemp, A. Kumar, S. Mousa, T.-J. Park, P. Ajayan, N. Kubotera, S. A. Mousa and R. J. Linhardt, Biomacromolecules, 2009, 10, 589–595 CrossRef CAS PubMed.
- P. Mulvaney, Langmuir, 1996, 12, 788–800 CrossRef CAS.
- I. O. Sosa, C. Noguez and R. G. Barrera, J. Phys. Chem. B, 2003, 107, 6269–6275 CrossRef CAS.
- S. Link and M. A. El-Sayed, J. Phys. Chem. B, 1999, 103, 8410–8426 CrossRef CAS.
- S. Link, M. B. Mohamed and M. A. El-Sayed, J. Phys. Chem. B, 1999, 103, 3073–3077 CrossRef CAS.
- C.-H. Kuo and M. H. Huang, Langmuir, 2005, 21, 2012–2016 CrossRef CAS PubMed.
- S. Chen, Z. L. Wang, J. Ballato, S. H. Foulger and D. L. Carroll, J. Am. Chem. Soc., 2003, 125, 16186–16187 CrossRef CAS PubMed.
- Y.-R. Kim, R. K. Mahajan, J. S. Kim and H. Kim, ACS Appl. Mater. Interfaces, 2009, 2, 292–295 Search PubMed.
- G. Mie, Ann. Phys., 1908, 330, 377–445 CrossRef.
- H. A. McIlwee, C. L. Schauer, V. G. Praig, R. Boukherroub and S. Szunerits, Analyst, 2008, 133, 673–677 RSC.
| This journal is © The Royal Society of Chemistry 2014 |