Target-induced quenching for highly sensitive detection of nucleic acids based on label-free luminescent supersandwich DNA/silver nanoclusters†
Guangfeng
Wang
*,
Yanhong
Zhu
,
Ling
Chen
,
Lun
Wang
and
Xiaojun
Zhang
*
Key Laboratory of Chem-Biosensing, College of Chemistry and Materials Science, Center for Nano Science and Technology, Anhui Normal University, Wuhu 241000, Anhui Province, P.R. China. E-mail: xjzhang@mail.ahnu.edu.cn; Fax: +86-553-3869303; Tel: +86-553-3869303
First published on 8th October 2013
Abstract
Luminescent silver nanoclusters (AgNCs) were anchored by designed oligonucleotides, acting as fluorescent labels. They hybridized with specific nucleic acid targets to form a supersandwich structure resulting in the fluorescence intensity of the DNA/AgNCs decreasing linearly with respect to the concentration of the target DNA.
Since the double-stranded structure of DNA was discovered in 1953, it has advanced our understanding of the significant role that nucleic acids play in life processes.1 Nucleic acid probes are essential tools for exploring the biological processes of nucleic acid duplication, recombination, transcription, and expression. The advances in molecular biology and the chemical synthesis of nucleic acids prompted the development of nucleic acid probes designed and applied in various fields, medical diagnostics, clinical diagnosis, molecular biology, food safety, environment and biodefense applications. Recently, great efforts have been made to develop biotechnologies to improve the sensitivity, the selectivity and the ease of operation for DNA analysis including electrical, optical, or microgravimetric, amplified DNA sensors, quartz crystal microbalance, and surface plasmon resonance techniques, Raman scattering, electrochemical detection, colorimetry, photoluminescence, field effect transistors, and chemoluminescence.2 Among them, homogeneous hybridization assays with fluorescent DNA probes have been extensively used due to the inherent advantages, such as operation convenience, high sensitivity and potential compatibility.3 It comes no surprise that quantum dots (QDs) and organic dyes are attractive fluorescent labels because they exhibit high fluorescence.4 Unfortunately, even though water-soluble QDs have very strong absorption and lower photobleaching quantum yields than organic dyes as single molecular probes, when applied to biological systems both QDs and organic dyes present a number of issues, including cell toxicity, large physical size, and strong fluorescence intermittency on all time scales.5 Therefore, it is crucial to advance studies of individual biomolecule functions to create a versatile set of highly stable fluorophores with high emission rates. As promising substitutes for organic dyes and QDs, few atom metallic nanoclusters have received significant attention acting as new and biological labels due to their strong photoluminescence, good photostability, large Stokes shift and high emission rates.6 In particular, gold and silver nanoclusters (AgNCs) have already been broadly seen in the fields of chemical sensing, in vivo bioassays, and in vivo biological imaging due to their unique clearly identifiable, size-dependent and relatively air-stable fluorescence properties.7 Particularly, using DNA as scaffolds to synthesize fluorescent AgNCs in aqueous solution has attracted particular attention due to their ultrasmall size, facile synthesis, tunable fluorescence emission, high luminescence quantum yields, low toxicity, good water solubility, and good photostability and biocompatibility.8 And the structure of DNA plays an important role not only in the successful synthesis of AgNCs but also in their fluorescence properties.9 Different strand lengths, base sequences, or secondary structures of DNA would create different AgNCs with fluorescence emission bands ranging from the visible to near-IR range.10 The applications of AgNCs have expanded a great deal in the past few years including the detection of metal ions, small biomolecules, and DNA or proteins.9g,11 DNA/AgNCs represent a novel class of fluorophores that already have applications in chemical/biological detection and cellular imaging.7b,12 However, there are few reports about the study of signal amplified quenching of DNA/AgNCs. Here we demonstrated that DNA-templated AgNCs can be used to detect specific nucleic acid targets through DNA hybridization induced quenching of DNA/AgNCs.
Since Xia et al. developed a novel “supersandwich” assay, the supersandwich DNA structure has attracted growing attention in recent years.13 Through DNA hybridization each capture probe can capture multiple signal probes on the electrode surface. In comparison with the traditional sandwich assay, such selective and sensitive supersandwich approaches can effectively amplify the signal and gain higher sensitivity. As a typical amplified structure, supersandwich related assays have played significant roles in various detection fields such as heavy metals, tumor markers, nucleic acids, etc.14 Then an interesting question stems from these results: can this supersandwich structure in DNA/AgNCs get amplified signals for detecting nucleic acids? Herein we designed a novel DNA/AgNC luminescent sensor based on the supersandwich structure through DNA hybridization. For analytical applications, the supersandwich DNA/AgNC sensor enhanced the quenching efficiency while simplifying the operation process, and is successfully used for the detection of specific nucleic acids in our work. To the best of our knowledge, the present supersandwich structure, for the first time, acts as an appropriate quencher in fluorescence detection.
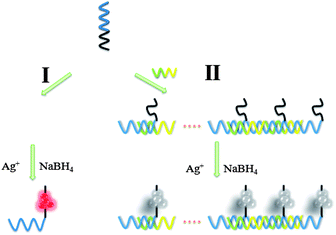
A schematic representation of the fabrication of the supersandwich DNA/AgNC luminescent sensor is shown in Scheme 1. On the basis of the work of Liu et al.,15 we chose a sequence that successfully created luminescent emitting AgNCs (S0: 5′-CCCCCCCCCCCC-3′). Previous studies have indicated that the S0 created luminescent AgNCs, with an emission maximum at 640 nm and shortened the time of producing fluorescence emission. Therefore, we believed that the S0 would be an adaptive candidate to be used for developing a AgNC-based probe for nucleic acid detection. In order to create the probe for detecting target DNA, the artificial oligonucleotide scaffold was designed (5′-CCCCCCCCCCCC-AGCTTGCAT-CGGTCAGAG-3′, termed S1) with two regions: AgNC biomineralizing region (S0) and target DNA recognizing region (-AGCTTGCAT-CGGTCAGAG-3′). S1 can produce and anchor luminescent AgNCs just like S0 with the addition of AgNO3 and reduction with NaBH4 (S1/AgNO3/NaBH4 in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17
17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 17 ratio, route I). And the recognizing region of S1 can hybridize with target DNA (5′-ATGCAAGCT-CTCTGACC-3′, termed S2) to form a supersandwich structure. In route II, hybridization between S1 and S2 can form a supersandwich structure. After the same reduction process, the emission of the supersandwich AgNC probe is significantly lower versus the case when S3 is present. The change of luminescence intensity revealed the presence of S2. The degree of fluorescence quenching is relative to the concentration of S2.
17 ratio, route I). And the recognizing region of S1 can hybridize with target DNA (5′-ATGCAAGCT-CTCTGACC-3′, termed S2) to form a supersandwich structure. In route II, hybridization between S1 and S2 can form a supersandwich structure. After the same reduction process, the emission of the supersandwich AgNC probe is significantly lower versus the case when S3 is present. The change of luminescence intensity revealed the presence of S2. The degree of fluorescence quenching is relative to the concentration of S2.
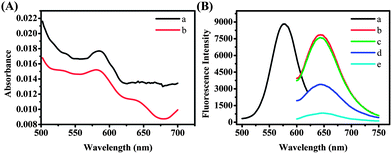
The feasibility of the experimental principle was examined under different conditions. The UV-vis absorption spectrum of the DNA/AgNCs (Fig. 1A, curve a) showed one peak at 580 nm. After the addition of the S2 target, the peak at 580 nm decreased (Fig. 1A, curve b). Then we studied the full spectral scan of the emission as a function of excitation wavelength for the S1 probe which can be found in Fig. S1, ESI.† From the results we chose 580 nm as the optimal excitation wavelength. Fig. 1B shows the excitation spectrum (curve a) and emission spectra (curves b–e) of the luminescent AgNCs obtained using S1 as the synthetic scaffold. Upon excitation at 580 nm, the formed DNA/AgNCs showed strong fluorescence intensity (Fig. 1B, curve b) that was even slightly higher than that of purified DNA/AgNCs (Fig. 1B, curve c), indicating that the small number of Ag nanoparticles (Ag NPs) in the product had little impact on the fluorescence behavior of the DNA/AgNCs. In a traditional way (ESI, Scheme S1†), S1 was hybridized with a completely complementary DNA S3 leading to the fluorescence quenching but with a low efficiency. After the addition of S3, a complementary sequence to the recognizing region caused 57% quenching of DNA/AgNCs (Fig. 1B, curve d). However, 91% quenching was observed for DNA/AgNCs in the presence of 0.9 μM S2 (Fig. 1B, curve e). This is consistent with the UV-vis spectrum results. The DNA hybridization between S1 and S2 induced the formation of a supersandwich DNA structure, resulting in the quenching of the luminescence of the DNA/AgNCs. Hybridization of the S2 target with the S1 probe was the likely cause of the significant drop in the fluorescence intensity. But the exact mechanistic details behind the drop of the fluorescence are currently being studied.
In a next step, we tried to explain the quenching mechanism in this system. Then we found several observations to prove that the electron transfer may induce the quenching process. (1) The oxidation potential was used to demonstrate the mechanism. As shown in Fig. S2A (ESI†), the oxidation potential of DNA/AgNCs was measured to be 0.362 V vs. SCE (saturated KCl). The supersandwich DNA/AgNCs exhibited an oxidation peak and the potential of DNA/AgNCs changed to be 0.285 V vs. SCE (saturated KCl). On the basis of the above potential test results, a comparison of the redox state of the supersandwich DNA/AgNCs and the energy levels of this supersandwich structure (ESI, Fig. S2B†) implied that the electron transfer may happen between AgNCs and the supersandwich structure. In addition, we tested the potential of DNA/AgNCs-S3 which was measured to be 0.334 V vs. SCE (saturated KCl). We can conclude from Fig. S2C (ESI†) that electron transfer indeed may happen between AgNCs and target nucleotide acids by hybridization, while the supersandwich structure exhibited stronger ability of electron transfer. (2) We used the lifetime measurements to explore the possible quenching mechanism of the system. Fig. S2D and E (ESI†) showed the fluorescence decay dynamics of the DNA/AgNCs in the absence and presence of S2 and S3. Using a monoexponential function the fluorescence decay of the DNA/AgNCs could be fitted with a time constant of 10.41 ± 0.07 μs. While we can see that in the presence of S2, the fluorescence decay could be fitted with a time constant of 3.589 ± 0.021 μs. Nevertheless, the addition of S3 made small signal changes (ESI, Fig. 2E†). It can be concluded from the result that the S1 probe exhibited a significantly shorter fluorescence lifetime in the presence of S2, presumably as a result of electron transfer, where the electrons were transferred to the supersandwich DNA structure.
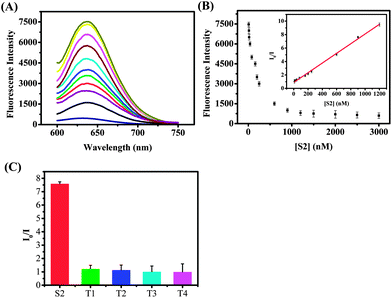
The time revolution of the fluorescence intensity of the S1 probe is shown in Fig. S3 (ESI†), and on the basis of the above-mentioned results, we tested whether we can use this designed sensor to detect specific nucleic acid targets. Fig. 2A shows the luminescence emission spectra of supersandwich DNA/AgNCs in the presence of different concentrations of S2 (10 nM to 1.2 μM). It was clearly seen that the luminescence intensity decreased with increasing S2 concentration, demonstrating that the S1 probe can be used for detecting the target nucleic acids by monitoring the generated luminescence of the DNA/AgNCs with the formation of a supersandwich DNA structure. In this publication we show two ways of mixing the S1 with DNAs (including the S2 or other mismatched DNA sequences, ESI†). One of them was performed by first mixing the S1 probe and DNAs together for 10 h and followed by adding AgNO3 and NaBH4 and monitoring the emission after 2 h. The other way first formed DNA/AgNCs, followed by addition of the DNAs. The emission spectrum (ESI, Fig. S4†) showed that no matter what concentrations of S2 we added, the luminescence of this sensor almost remained unchanged.
A possible explanation could be that after 10 hours the DNA/AgNCs displayed very weak luminescence, and this result is consistent with the time evolution of the fluorescence intensity of the S1 probe (ESI, Fig. S3†). The relationship between the fluorescence intensity and the concentrations of S2 is shown in Fig. 2B. A good linear dependence of the I0/I intensity versus S2 target (I0 being the value without addition of S2) was obtained. The linear correlation was from 10 nM to 1200 nM and the linear regression equation was I0/I = 0.89243 + 0.0072C (R2 = 0.9978). As can be seen in Fig. 2C, the S2 target (900 nM) has the largest effect on the I0/I ratio (a 7.5 times drop in the fluorescence intensity), while other DNA strands (each 9 μM) with one (T1), two (T2), three (T3) mismatched nucleotides and non-target DNA (T4) only had a limited effect on the fluorescence intensity of the S1 probe. We can conclude from Fig. 2C that the detection approach showed a high selectivity toward the target DNA, allowing even one mismatched nucleotide to be distinguished.
In conclusion, we have demonstrated the proof of principle that the quenching of luminescent supersandwich DNA/AgNCs can be used for the sensitive and selective detection of target DNA. Even though the presented results indicate that high specificity toward detecting specific nucleic acid target sequences is possible, we should also investigate a larger set of relevant DNA probes and procedure optimization in further studies. Compared with other luminescence or fluorescence assays, this supersandwich DNA/AgNC sensor shows some unique advantages: (1) the sensor does not need any complex labeling processes, and the DNA/AgNCs serve as the fluorescent probes and the target DNA induced the formation of a supersandwich structure acting like the quencher. (2) The whole experiment is very simple and does not require separation and multi-step procedures. (3) The background fluorescence is very low, which is beneficial for improving the sensitivity of the sensor. (4) With only changing the sensing sequence it can be used for detecting various targets. These promising studies indicate that this sensor based on supersandwich DNA/AgNCs offers distinct features with respect to target specificity, and we are exploring these perspectives for developing these supersandwich structured DNA/AgNCs into a new technology for target DNA or protein detection.
Acknowledgements
This work was financially supported by the projects (21073001, 21005001 and 21371007) from the National Natural Science Foundation of China, Anhui Provincial Natural Science Foundation (1208085QB28), and the Natural Science Foundation of Anhui (KJ2012A139).Notes and references
- (a) J. D. Watson and F. H. Crick, Nature, 1953, 171, 737 CrossRef CAS; (b) J. D. Watson and F. H. Crick, Nature, 1953, 171, 964 CrossRef CAS; (c) K. M. Wang, Z. W. Tang, C. Y. J. Yang, Y. M. Kim, X. H. Fang, W. Li, Y. R. Wu, C. D. Medley, Z. H. Cao, J. Li, P. Colon, H. Li and W. H. Tan, Angew. Chem., Int. Ed., 2009, 48, 856 CrossRef CAS PubMed.
- (a) W. Tan, K. Wang and T. J. Drake, Curr. Opin. Chem. Biol., 2004, 8, 547 CrossRef CAS PubMed; (b) C. Fan, K. W. Plaxco and A. J. Heeger, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 9134 CrossRef CAS PubMed; (c) Y. Lu and J. Liu, Curr. Opin. Biotechnol., 2006, 17, 580 CrossRef CAS PubMed; (d) N. K. Navani and Y. Li, Curr. Opin. Chem. Biol., 2006, 10, 272 CrossRef CAS PubMed; (e) F. Lucarelli, S. Tombelli, M. Minunni, G. Marrazza and M. Mascini, Anal. Chim. Acta, 2008, 609, 139 CrossRef CAS PubMed; (f) Y. Weizmann, F. Patolsky and I. Willner, Analyst, 2001, 126, 1502 RSC; (g) F. Wang, J. Elbaz, C. Teller and I. Willner, Angew. Chem., Int. Ed., 2011, 50, 295 CrossRef CAS PubMed; (h) S. J. Ye, H. X. Li and W. Cao, Biosens. Bioelectron., 2011, 26, 2215 CrossRef CAS PubMed; (i) Y. C. Li, Z. Wang, L. M. L. Ou and H. Z. Yu, Anal. Chem., 2007, 79, 426 CrossRef CAS PubMed; (j) M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013 CrossRef CAS; (k) W. C. W. Chan and S. M. Nie, Science, 1998, 281, 2016 CrossRef CAS; (l) Y. C. Cao, R. C. Jin, J. M. Nam, C. S. Thaxton and C. A. Mirkin, J. Am. Chem. Soc., 2003, 125, 14676 CrossRef CAS PubMed; (m) F. Patolsky, R. Gill, Y. Weizmann, T. Mokari, U. Banin and I. Willner, J. Am. Chem. Soc., 2003, 125, 13918 CrossRef CAS PubMed; (n) D. S. Grubisha, R. J. Lipert, H. Y. Park, J. Driskell and M. D. Porter, Anal. Chem., 2003, 75, 5936 CrossRef CAS PubMed; (o) Y. Xiao, X. Lou, T. Uzawa, K. J. I. Plakos, K. W. Plaxco and H. T. Soh, J. Am. Chem. Soc., 2009, 131, 15311 CrossRef CAS PubMed; (p) F. Jelen, A. B. Olejniczak, A. Kourilova, Z. J. Lesnikowski and E. Palecek, Anal. Chem., 2009, 81, 840 CrossRef CAS PubMed; (q) C. P. Chen, A. Ganguly, C. H. Wang, C. W. Hsu, S. Chattopadhyay, Y. K. Hsu, Y. C. Chang, K. H. Chen and L. C. Chen, Anal. Chem., 2009, 81, 36 CrossRef CAS PubMed; (r) N. C. Tansil, H. Xie, F. Xie and Z. Gao, Anal. Chem., 2005, 77, 126 CrossRef CAS PubMed; (s) R. Elghanian, J. J. Storhoff, R. C. Mucic, R. L. Letsinger and C. A. Mirkin, Science, 1997, 277, 1078 CrossRef CAS; (t) P. Alivisatos, Nat. Biotechnol., 2003, 22, 47 CrossRef PubMed; (u) E. Katz and I. Willner, Angew. Chem., Int. Ed., 2004, 43, 6042 CrossRef CAS PubMed.
- (a) C. Y. Yang, C. D. Medley and W. H. Tan, Curr. Pharm. Biotechnol., 2005, 6, 445 CrossRef CAS; (b) W. R. Algar and U. J. Krull, Anal. Chem., 2009, 81, 4113 CrossRef CAS PubMed; (c) I. Willner and E. Katz, Angew. Chem., Int. Ed., 2004, 43, 6042 CrossRef PubMed.
- (a) P. Alivisatos, Nat. Biotechnol., 2004, 22, 47 CrossRef CAS PubMed; (b) F. Chen and D. Gerion, Nano Lett., 2004, 4, 1827 CrossRef CAS.
- (a) W. C. W. Chan and S. M. Nie, Science, 1998, 281, 2016 CrossRef CAS; (b) C. Y. Zhang, H. Ma, S. M. Nie, Y. Ding, L. Jin and D. Y. Chen, Analyst, 2000, 125, 1029 RSC; (c) M. Bruchez, M. Moronne, P. Gin, S. Weiss and A. P. Alivisatos, Science, 1998, 281, 2013 CrossRef CAS; (d) M. Kuno, D. P. Fromm, H. F. Hamann, A. Gallagher and D. J. Nesbitt, J. Chem. Phys., 2000, 112, 3117 CrossRef CAS; (e) A. M. Derfus, W. C. W. Chan and S. N. Bhatia, Nano Lett., 2004, 4, 11 CrossRef CAS; (f) S. F. Lee and M. A. Osborne, J. Am. Chem. Soc., 2007, 129, 8936 CrossRef CAS PubMed.
- (a) T. Vosch, Y. Antoku, J. C. Hsiang, C. I. Richards, J. I. Gonzalez and R. M. Dickson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12616 CrossRef CAS PubMed; (b) J. Zheng and R. M. Dickson, J. Am. Chem. Soc., 2002, 124, 13982 CrossRef CAS PubMed; (c) T. Huang and R. W. Murray, J. Phys. Chem. B, 2001, 105, 12498 CrossRef CAS; (d) Z. Shen, H. Duan and H. Frey, Adv. Mater., 2007, 19, 349 CrossRef CAS; (e) L. Shang and S. J. Dong, Chem. Commun., 2008, 1088 RSC; (f) W. Lesniak, A. U. Bielinska, K. Sun, K. W. Janczak, X. Shi, J. R. Baker and L. P. Balogh, Nano Lett., 2005, 5, 2123 CrossRef CAS PubMed; (g) J. Zheng, C. W. Zhang and R. M. Dickson, Phys. Rev. Lett., 2004, 93, 077402 CrossRef; (h) J. Zheng, P. R. Nicovich and R. M. Dickson, Annu. Rev. Phys. Chem., 2007, 58, 409 CrossRef CAS PubMed; (i) J. Yu, S. A. Patel and R. M. Dickson, Angew. Chem., Int. Ed., 2007, 46, 2028 CrossRef CAS PubMed; (j) I. Díez, M. Pusa, S. Kulmala, H. Jiang, A. Walther, A. S. Goldmann, A. H. E. Müller, O. Ikkala and R. H. A. Ras, Angew. Chem., Int. Ed., 2009, 48, 2122 CrossRef PubMed; (k) J. Zhang, S. Xu and E. Kumacheva, Adv. Mater., 2005, 17, 2336 CrossRef CAS; (l) L. Maretti, P. S. Billone, Y. Liu and J. C. Scaiano, J. Am. Chem. Soc., 2009, 131, 13972 CrossRef CAS PubMed; (m) L. A. Peyser, A. E. Vinson, A. P. Bartko and R. M. Dickson, Science, 2001, 291, 103 CrossRef CAS PubMed.
- (a) J. Zheng, P. R. Nicowich and R. M. Dickson, Annu. Rev. Phys. Chem., 2007, 58, 409 CrossRef CAS PubMed; (b) H.-C. Yeh, J. Sharma, J. Han, J. S. Martinez and J. H. Werner, Nano Lett., 2010, 10, 3106 CrossRef CAS PubMed; (c) Y. Bao, H. C. Yeh, C. Zhong, S. A. Ivanov, J. K. Sharma, M. L. Neidig, D. M. Vu, A. P. Shreve, R. B. Dyer, J. H. Werner and J. S. Martinez, J. Phys. Chem. C, 2010, 114, 15879 CrossRef CAS; (d) Y. Bao, C. Zhong, D. M. Vu, J. P. Temirov, R. B. Dyer and J. S. Martinez, J. Phys. Chem. C, 2007, 111, 12194 CrossRef CAS.
- (a) B. Sengupta, C. M. Ritchie, J. G. Buckman, K. R. Johnsen, P. M. Goodwin and J. T. Petty, J. Phys. Chem. C, 2008, 112, 18776 CrossRef CAS; (b) W. W. Guo, J. P. Yuan and E. K. Wang, Chem. Commun., 2009, 3395 RSC; (c) J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207 CrossRef CAS PubMed; (d) J. H. Yu, S. Choi and R. M. Dickson, Angew. Chem., Int. Ed., 2009, 48, 318 CrossRef CAS PubMed; (e) C. M. Ritchie, K. R. Johnsen, J. R. Kiser, Y. Antoku, R. M. Dickson and J. T. Petty, J. Phys. Chem. C, 2007, 111, 175 CrossRef CAS PubMed; (f) C. I. Richards, S. Choi, J. C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y. L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038 CrossRef CAS PubMed; (g) E. G. Gwinn, P. O'Neill, A. J. Guerrero, D. Bouwmeester and D. K. Fygenson, Adv. Mater., 2008, 20, 279 CrossRef CAS; (h) C. I. Richards, J. C. Hsiang, D. Senapati, S. Patel, J. H. Yu, T. Vosch and R. M. Dickson, J. Am. Chem. Soc., 2009, 131, 4619 CrossRef CAS PubMed.
- (a) J. Zheng, P. R. Nicovich and R. M. Dickson, Annu. Rev. Phys. Chem., 2007, 58, 409 CrossRef CAS PubMed; (b) B. Adhikari and A. Banerjee, Chem. Mater., 2010, 22, 4364 CrossRef CAS; (c) M. H. Muhammed, S. Ramesh, S. Sinha, S. Pal and T. Pradeep, Nano Res., 2008, 1, 333 CrossRef CAS; (d) M. Muhammed, P. Verma, S. Pal, R. Kumar, S. Paul, R. Omkumar and T. Pradeep, Chem.–Eur. J., 2009, 15, 10110 CrossRef CAS PubMed; (e) C. A. Lin, T. Y. Yang, C. H. Lee, S. H. Huang, R. A. Sperling, M. Zanella, J. K. Li, J. L. Shen, H. H. Wang, H. I. Yeh, W. J. Parak and W. H. Chang, ACS Nano, 2009, 3, 395 CrossRef CAS PubMed; (f) T. U. B. Rao and T. Pradeep, Angew. Chem., Int. Ed., 2010, 49, 3925 CrossRef CAS PubMed; (g) T. U. B. Rao, B. Nataraju and T. Pradeep, J. Am. Chem. Soc., 2010, 132, 16304 CrossRef CAS PubMed.
- J. Zheng and R. M. Dickson, J. Am. Chem. Soc., 2002, 124, 13982 CrossRef CAS PubMed.
- (a) J. Zheng, J. T. Petty and R. M. Dickson, J. Am. Chem. Soc., 2003, 125, 7780 CrossRef CAS PubMed; (b) J. Zheng, C. Zhang and R. M. Dickson, Phys. Rev. Lett., 2004, 93, 077402 CrossRef; (c) Y. P. Bao, C. Zhong, D. M. Vu, J. P. Temirov, R. B. Dyer and J. S. Martinez, J. Phys. Chem. C, 2007, 111, 12194 CrossRef CAS; (d) Y. C. Jao, M. K. Chen and S. Y. Lin, Chem. Commun., 2010, 46, 2626 RSC; (e) J. G. Zhang, S. Q. Xu and E. Kumacheva, Adv. Mater., 2005, 17, 2336 CrossRef CAS; (f) Z. Shen, H. W. Duan and H. Frey, Adv. Mater., 2007, 19, 349 CrossRef CAS; (g) L. Shang and S. J. Dong, Chem. Commun., 2008, 1088 RSC.
- (a) S. W. Yang and T. Vosch, Anal. Chem., 2011, 83, 6935 CrossRef CAS PubMed; (b) G. Y. Lan, W. Y. Chen and H. T. Chang, Biosens. Bioelectron., 2011, 26, 2431 CrossRef CAS PubMed; (c) J. Sharma, H. C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280 RSC; (d) J. Sharma, H. C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2011, 47, 2294 RSC; (e) J. Li, X. Zhong, H. Zhang, X. C. Le and J. Zhu, Anal. Chem., 2012, 84, 5170 CrossRef CAS PubMed; (f) Z. Zhou, Y. Du and S. Dong, Biosens. Bioelectron., 2011, 28, 33 CrossRef CAS PubMed.
- F. Xia, R. J. White, X. L. Zuo, A. Patterson, Y. Xiao, D. Kang, X. Gong, K. W. Plaxco and A. J. Heeger, J. Am. Chem. Soc., 2010, 132, 14346 CrossRef CAS PubMed.
- (a) T. Yuan, Z. Y. Liu, L. Z. Hu, L. Zhang and G. B. Xu, Chem. Commun., 2011, 47, 11951–11953 RSC; (b) G. F. Wang, H. Huang, B. J. Wang, X. J. Zhang and L. Wang, Chem. Commun., 2012, 48, 720–722 RSC; (c) G. F. Wang, X. P. He, B. J. Wang, X. J. Zhang and L. Wang, Analyst, 2012, 137, 2036 RSC; (d) G. F. Wang, X. P. He, L. Wang and X. J. Zhang, Biosens. Bioelectron., 2013, 42, 337–341 CrossRef CAS PubMed.
- X. Q. Liu, F. Wang, A. Niazov-Elkan, W. W. Guo and I. Willner, Nano Lett., 2013, 13, 309 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, EIS, control experiments and gel-electrophoresis characterization. See DOI: 10.1039/c3an01702h |
| This journal is © The Royal Society of Chemistry 2014 |