Open Access Article
Open Access ArticleMolecular interactions of nanomaterials and organisms: defining biomarkers for toxicity and high-throughput screening using traditional and next-generation sequencing approaches
Rebecca
Klaper
*,
Devrah
Arndt
,
Jared
Bozich
and
Gustavo
Dominguez
School of Freshwater Sciences, University of Wisconsin-Milwaukee, 600 East Greenfield Ave., Milwaukee, Wisconsin, USA. E-mail: rklaper@uwm.edu; Tel: +1 414-382-1713
First published on 5th December 2013
Abstract
The toxicity of nanomaterials depends on the basic interaction of the chemistry of the material with the molecular pathways in an organism. To design safe and sustainable nanomaterials, more detailed information on the molecular interaction and biochemical machinery that is altered in an organism upon contact with a nanomaterial is needed. There are a multitude of papers now on the toxicity of nanomaterials to various model organisms from human to ecological models, but many focus on acute high dose exposures and research on the toxicity of other chemicals has shown that the dose of a chemical can have a tremendous impact on the pathways that are affected within the organism. The most common pathways investigated in nanotoxicity experiments are related to oxidative stress, yet oxidative stress can be a temporary and natural response to an insult without a negative outcome. There are a multitude of other potential mechanisms that may be triggered in response to a toxin at sublethal exposures. Here we present a review documenting the evidence to date on the indicators of the molecular response to nanomaterials from in vitro and in vivo studies. Alternative pathways as indicated by single biomarker, global gene expression studies and next generation sequencing approaches are discussed as well as the impacts of nanomaterial type, dose, and the types of system studied. Specific mechanisms that are impacted by a nanomaterial can be used as the basis of better high-throughput methods for evaluating how nanomaterial chemistry impacts toxicity and support models to predict the toxicity of future nanomaterials.
Introduction
A major question in the field of nanoscience is whether nanomaterials will have a negative impact on human health and the environment due to their novel properties. There is some difficulty in answering this question due to the fact it is unclear as to whether those novel properties impart some unique toxicological impact that is different from other contaminants. In addition, nanoscience is somewhat in its infancy and it is anticipated that in the future nanomaterials will have more novel properties and will be more complex than what is currently being used and developed. Future materials have the potential to create unknown hazards, as they may be unlike the materials that are currently being evaluated for their impact on environmental health and safety. There is a clear need for a strategy to evaluate the impacts of not only today's nanomaterials but also those that have yet to be created.There are currently many papers on the toxicology of nanomaterials that describe acute mortality to cells and a select number of organisms. These include a variety of human, mouse, and rat cell lines as well as whole mice and rat studies and limited studies on invertebrates and other non-human model organisms. Overall, it appears from the literature that many of the current suites of available nanomaterials are not acutely toxic. Estimated exposure to nanomaterials, based on data from similar sized particulates, estimated wastewater treatment effluents, and life-cycle modeling efforts are predicted to be orders of magnitude below what is considered lethal for many nanomaterials.1 Select nanomaterials, such as metallic nanomaterials are toxic at lower doses, but this can be due to dissolution in certain aquatic environments rather than by the nanomaterial itself. Interactions with the environment can also increase toxicity, as is the case with metal oxides that are photoreactive.2,3 Realistically, the greatest impacts from nanomaterials will most likely be the result of long-term low-dose exposures. There is a significant gap regarding these types of potential impacts.
Low-dose chronic exposures often have more subtle impacts and alter different biochemical pathways in an organism than corresponding to high-dose exposures. High concentration exposures of many chemicals initiate a “global” stress response that often includes oxidative stress and pathways associated with necrosis. At lower concentrations the same chemicals trigger reactions that can show a very different pattern of molecular response, including unique gene or protein expression patterns that reflect the specific interaction of a chemical with cellular components.4,5 Slight variations in the chemical composition of a drug or pollutant have also been shown change the molecular responses and these gene expression signatures are predictive of impacts on important endpoints such as reproduction or development.6–9 These low-dose chronic exposures have become particularly important in the study of other emerging contaminants as these chemicals, like many nanomaterials, are not acutely toxic. However, research over the last two decades has shown that there is a potential for such chemicals to exert impacts on pathways associated with the reproductive system, immune system, nervous system, cancer pathways, metabolic pathways and others, even at these low doses.10,11
Long-term chronic studies do not readily lend themselves to rapid high-throughput analyses, which are desirable to quickly screen for potential impacts of exposures. Toxicity testing using molecular biomarkers that are known to be linked to negative outcomes may provide a mechanism to develop high-throughput tools that are predictive of long-term outcomes. Global gene expression patterns can be used as a sensitive tool to interrogate the interaction of the cell, tissue or whole organism to nanomaterials of differing chemistries and provide an indication of potential future impacts. Currently, there are a few individual biomarkers being explored in this capacity and they are often limited to pathways involved in oxidative stress, which is known to be a complicated biomarker. Here we review the current status of the field on the molecular impacts of nanomaterials, the limitations on the number of pathways that have been investigated to date, and the potential for next-generation sequencing platforms to provide novel information on the toxicity of nanomaterials. We discuss the potential impact of these methods on the development of high-throughput assays to evaluate the environmental health and safety of nanomaterials.
Biomarkers and molecular response
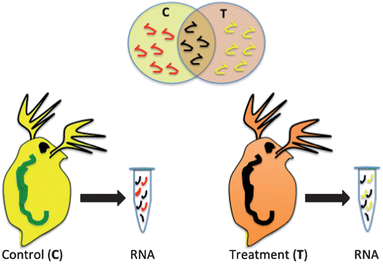
Biomarkers, such as mRNA transcripts, enzyme or protein expression have been proposed as health indicators for a variety of human diseases and now are being developed to indicate the health status of other model and non-model organisms in response to a xenobiotic insult.5,12,13 The premise of the technology is that proteins, and mRNA that codes for those proteins, are expressed differently in an organism that has come in contact with a xenobiotic than an organism that has not been exposed (Fig. 1). In addition, the genes or proteins that are differentially expressed give an indication of the specific pathways that are impacted in an exposed organism and potential negative impacts of the chemical on the organism.5,14 If many genes or proteins are used, differences in the global expression pattern can be used to differentiate chemicals with differing modes of action or even chemicals in the same class with similar modes of action.6,15 This is not a simple analysis and challenges include the fact that there is a time and dose-dependent impact on the expression of many genes and proteins. This adds variability in response, which makes it difficult to assign a specific gene expression state as a negative outcome. In addition, there are challenges in linking genomic changes to physiological endpoints that are meaningful to the organism or the population of organisms.4,16 However, regulatory agencies in the United States and Europe are exploring the potential of gene and protein high-throughput assays as a screen for the thousands of existing chemicals in the marketplace where we have limited toxicity information. Efforts such as the U.S. EPA ToxCast initiative are exploring the potential for in vitro cellular assays to be used to extrapolate simplified assays in conjunction with genomic and proteomic endpoints to predict whole organism impacts.9 Implicit in these efforts are the current limitations to this type of assay including the insufficient number of toxicological pathways explored and the need for considering metabolism in modeling efforts.17 There is some effort to include nanomaterials in these trials and various groups are exploring the extrapolation of select biomarkers such as oxidative stress into high-throughput nanomaterial testing as a key indicator of toxicity.18Oxidative stress as a molecular response to nanomaterial exposure
Currently, a majority of the research on molecular impacts of nanomaterials involves examining the response of oxidative stress and related pathways. Free radical generation by certain nanomaterials has been documented in select media and is thought to be the main route for oxidative stress responses.12 However, free radical generation is not always present with nanomaterials and there are toxic responses with these materials in the absence of ROS generation, demonstrating the oxidative stress response is not always representative of the entirety of the interaction of the nanomaterial and the biological entity.15,17,19 Toxicologically, it is well known that oxidative stress is highly temporal and can dissipate quickly and inflammatory mediators change in a time dependent manner.20,21 Nanomaterials may cause oxidative stress and inflammation over a 24 hour period but this effect subsides after this time period.21,22 For example, mouse lung cells upon exposure to 54 μg rutile TiO2 (in 40 μL of a bronchoalveolar liquid suspension through an intratrachael installation) expressed immune, inflammatory and metabolic pathways on the first day after initial exposure.20 Yet after three days this response subsided and calcium signaling, actin cytoskeleton, and fatty acid metabolism pathways dominated. On day 28 in this same study calcium ion and cation homeostasis pathways and pathways important in muscle regulation were significantly affected. This result indicates that rutile TiO2 might induce acute oxidative stress and lung inflammation but long-term effects relate more to smooth muscle activity. In some cases there may be a complete absence of oxidative stress response pathways associated with the interaction of nanomaterials with cells in organisms.47Oxidative stress can also be dose dependent where high levels of a toxin can cause an initial oxidative stress response that ultimately indicates a decline in cell viability, but lower doses do not instigate the same response. For example, in mice exposed to silver nanoparticles at concentrations of 100, 500 and 1000 mg kg−1, only the highest concentrations significantly induced genes involved in oxidative stress in the frontal cortex, while the lowest dose administrated minimally affected the same genes.23 Similarly, E. coli exposed to a high, medium and low concentration of TiO2 and silver nanoparticles (1, 10, 50 ppm) exhibited dose-dependent differential gene expression, with higher concentrations inducing many oxidative stress related genes (50 and 42, respectively) and the low concentration affecting only a few genes involved in stress (four and three, respectively).24
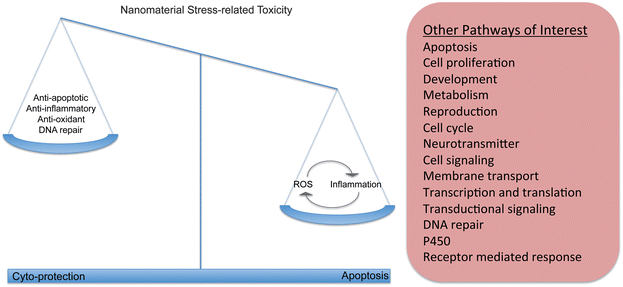
Other pathways associated with oxidative stress include inflammation, apoptosis, and general stress pathways (Fig. 2). Each of these mechanisms has been commonly studied in response to nanomaterial exposure across in vitro and in vivo study systems and also indicate that the effect of nanomaterials on these pathways is concentration and nanomaterial dependent (Table 1). Inflammation is a protective immune associated response that serves to instigate phagocytosis of a foreign object or pathogen, destroy the invading organism using ROS mechanisms. It also stimulates the secondary immune system to recognize and destroy future similar invasions. In an acute time frame inflammation is a beneficial response, however, if stimulated chronically it can destroy surrounding tissues and create disease. Cytokines such as IL-1, IL-6 and TNF are generic primary immune responses and have been measured by several studies in vertebrates in response to mainly titanium dioxide nanomaterials, carbon black and silica nanomaterials but at relatively high concentrations. Some secondary immune pathways have also been studied including IL-5 and IL-10 indicating that the reaction to nanomaterials can move beyond a simple inflammatory reaction. Exposures of mice to TiO2 indicate differential regulation of genes and proteins important in the COX-2 and MAPK/P13-k/Akt signaling pathways, apoptosis and inflammation at higher concentrations.25,26
| Pathways | Gene/protein | Model/cell line | Nanoparticle dimensions (nm); shape | Nanoparticle typea | Modeb | Dose | Durationc | Ref. |
|---|---|---|---|---|---|---|---|---|
| a Nanoparticle/NP functional group: QDs – quantum dots, DP – 4,6-diamino-2-pyrimidinethiol, SWCNT – single walled carbon nanotubes, CIT – citrate, PEI – polycationic polyethylenimine, PMAO – polyanionic polymaleic anhydride-alt-1-octadecene, PVP – polyvinyl pyrrolidone, OH – hydroxylated, THF – tetrahydrofuran. b Mode of administration: IV – intravenous, IN – intranasal, IG – intragastric, IT – intratracheal, IP – intraperitoneal, O – oral, IH – inhalation, AM – added to media. c Exposure duration: h – hour(s), d – day(s), w – week(s). | ||||||||
| Apoptosis | TLR-2 and B-actin | C28/I2, PDL, SCC-9 | 56 | Ag | AM | 10 μg mL−1 | 20 h | 31 |
| p53, Bax, parp-1 and smac/DIABLO, caspase-3,-7,-8 and -9 | A549 | Ag | AM | 43 μg mL−1 | 6 h | 32 | ||
| p53, Bcl-xl, Bcl-2, NF-κB, CDkNIA, TNFSF10, TNFSR1A and GAPDH | CET | 160 | Al2O3 | AM | 280 μg mL−1 | 12 h | 34 | |
| Bax, Bak, caspase-3, caspase-8 and GAPDH | A549, A431 | 17; spheres | Au | AM | 48.9 (A549) and 65.2 (A431) μg mL−1 | 24 h | 75 | |
| p53, Bax, caspase-3 & caspase-9, Bcl-2, GAPDH and B-actin | MCF-7 | — | Au | AM | 200 μg mL−1 | 24 h | 40 | |
| Bax, caspase-3 and -8 | HEPG2 | 5 | CdTe | AM | 10 μg mL−1 | 24 h | 39 | |
| Bcl2-like 4, HyCasp3, foxO and Hsp-70 | H. vulgaris | 3.2 | CdTe QDs | AM | 10 nM | 1–48 h | 21 | |
| Bax and p53 | CD14+ | 30 | CeO2 | AM | 5 μg mL−1 | 20 and 40 h | 37 | |
| Akt, procaspase-3, Bcl-2, Bax and caspase-3 | BGC-823 | 77; spheres | PEG-PCL-paclitaxel/tetrandrine | AM | 0.05 and 10 μM | 48 h | 38 | |
| MRP-1, LRP, Bcl2 and NFκB | Y79 | 236 | Polymer–folate–curcumin | AM | 2 μg mL−1 | 48 h | 36 | |
| Bax, caspase-3, Bcl-2 and p-p53 | HUVEC | 20; spheres | Si | AM | 200 μg mL−1 | 24 h | 44 | |
| p53, Bax, Bcl-2 and caspase-3 | HEPG2 | 14 | Si | AM | 200 μg mL−1 | 72 h | 30 | |
| Bcl-2, BclXL, Mcl-1, Bax, Bak and Bim | MM, PMB, U266 | 300; spheres | Si–snake venom | AM | 10 ng mL−1 | 12 h | 35 | |
| p53, cytochrome C, Bax, Bcl-2 and caspase-9 | HFL-I | 20 and 80 | SiO2 | AM | 100 mg mL−1 | 24 h | 46 | |
| Pdai2 and ada | Mice (CD-1 ICR) | 6 | TiO2 | IN | 10 mg kg−1 | 90 d | 26 | |
| Birc5, Crap2 and Tfrc | Mice | 6 | TiO2 | IG | 10 mg kg−1 | 90 d | 29 | |
| BNIP3 | BEAS-2B | 14–25 | ZnO | AM | 5 μg mL−1 | 24 h | 28 | |
| Cell functions | Adh5 | Zebrafish (wild type WIK eggs) | 10 | Ag | AM | 5 μg L−1 | 24 and 48 h | 81 |
| FliG, fliN, fliM, CheW, cheB, cheY, cheZ, cheR, cheA, and motB | E. coli | — | Au-DP | AM | 10 mg L−1 | 4 h | 61 | |
| CXC, CXCL9 and 1/CCL2 | Mice (BALB/c) | 14 and 95; spheres | Carbon Black (CB) | IN | 125 μg | 4 w | 71 | |
| c-Jun, ERK 1/2 and p38 | HEPG2 | 5 | CdTe | AM | 10 μg mL−1 | 24 h | 39 | |
| c-Jun | SMMC-7721 | 20–30; hexahedrals | CeO2 | AM | 50 μg mL−1 | 24 h | 55 | |
| p-c-Jun and c-Jun | HUVEC | 20; spheres | Si | AM | 25–200 μg mL−1 | 24 h | 44 | |
| Sri-74, srv-7, srx-22 and srx-69 | C. elegans | 73, nanotubes | SWCNT | AM | 500 mg L−1 | 48 h | 59 | |
| CCL11, GM-CSF, IFN-g, CXCL1, CCL2, CCL3, CCL4, FGF-basic, MCSF and VEGF | Mice (C57BL/6) | — | TiO2–rutile | IT | 18 54 and 162 μg | 1, 3 and 28 d | 20 | |
| Inflammation immune | IL-6, IL-8 and NFκB | IMR-90, U251 | 6–20 | Ag | AM | 400 μg mL−1 | 48 h | 78 |
| IL-1b and TNF-alpha | Rats (liver) | 10 and 50; spheres and hexagons | Au | IP | 22 μg kg−1 | 1 d | 22 | |
| IL-6 and TNF-alpha | Rats (liver) | 50 nm hexagons | Au | IP | 22 μg kg−1 | 1 d | 22 | |
| IL-6 and TNF-alpha | Rats (kidney) | 50 nm hexagons | Au | IP | 22 μg kg−1 | 1 d | 22 | |
| 1 alpha/CCL3, Il-1-beta and TNF-alfa | Mice (BALB/c) | 14 and 95; spheres | CB | IN | 125 μg | 4 w | 71 | |
| Il1rn, Il1b, Il18pb, Il18, Ifi47, Igtp, Irf1, Irf8, RT1-A2, RT1CE12, RT1-M6-2, RT1-CE1 and RT1-M3-1 | Rats | 20; spheres | Cd doped Si | IT | 1 mg | 7 d | 62 | |
| IL-1b, TNF-a and CCL5 | A549, THP-1 | 20 | CdSe QDs-PEG-COOH | AM | 2 nM | 24 h | 73 | |
| p-ERK, ERK, p-JNK, JNK, p-p53 and NF-κB | HUVEC | 20; spheres | Si | AM | 200 μg mL−1 | 24 h | 44 | |
| COX-2 MAPKs and P-13/Akt, JNK and ERK | Mice (CD-1 ICR) | 5.5 | TiO2 | IG | 10 mg kg−1 | 15, 30, 45, 60, 75, 90 d | 25 | |
| Def-b4, H2-Oa, Chi313, Alox5ap, and IL1b | Mice (CD-1 ICR) | 6 | TiO2 | IN | 10 mg kg−1 | 90 d | 26 | |
| Bcl6, Cfi and Cfd | Mice | 6 | TiO2 | IG | 10 mg kg−1 | 90 d | 29 | |
| Akr1c18 | Mice (CD-1 ICR) | 6 | TiO2 | IG | 10 mg kg−1 | 90 d | 63 | |
| Ccl3, Ccl6, clec5a, cxcl1, Il1r2 | Mice (C57BL/6) | — | TiO2–rutile | IA | 54 μg | 1 d | 20 | |
| C3, Ccl2, Ccl3, Ccl4, Ccl7, Ccl8, Ccl9, Ccl9, Ccl12, Ccl17, Ccl20, Ccl22, Ccr1, Ccr2, Ccr3, Ccr4, Ccr5, clec5a, Cxcl1, Cxcl9, Cxcl10, Cxcr2, Cxcr3, Ifng, Il10ra, 1l1a, Il1b, Ilr2, Il4, Itgam, spp1, TgFb1, Timp1, Tnf, Tnfrsf1b | Mice (C57BL/6) | — | TiO2–rutile | IA | 162 μg | 1 d | 20 | |
| C3, Ccl2, Ccl3, Ccl4, Ccl9, Ccr5, Ccr8, clec5a, Cxcl5, Oas1f, spp1, Timp1, Vnn1 | Mice (C57BL/6) | — | TiO2–rutile | IA | 162 μg | 28 d | 20 | |
| Cell cycle/proliferation | Cyclin B, cyclin E, CDC2 and CDK5R21 | IMR-90, U251 | 6–20 | Ag | AM | 400 μg mL−1 | 48 h | 78 |
| Pim3, Cdkn1b, Ptprf, Fkbp1a, GClm, Adamts1, Ddx24, Usp7, Uba5, Rchy1 | Rats | 20; spheres | Cd doped Si | IT | 1 mg | 30 d | 62 | |
| Hymyc1 | H. vulgaris | 3.2 | CdTe Qds | AM | 10 nM | 1–48 h | 21 | |
| Akt-1, cbp-1, daf-2, daf-12, daf-21, mek-2 and mpk-1 | C. elegans | 73; nanotubes | SWCNT | AM | 500 mg L−1 | 48 h | 59 | |
| Metabolism | atpD and atpA | E. coli | — | Au-DP | AM | 10 mg L−1 | 4 h | 61 |
| fruA, fruK, nuoK, nuoG, nuoF, frdD, frdC and frdB | S. typhimurium (TA100) | 71 | C60 | AM | 2 mg L−1 | 24 h | 57 | |
| CYP1A and POR | MSC | <50 | CB | AM | 50 and 100 μg mL−1 | 24 h | 27 | |
| rnt, thiS, cysW, yciW, cysI, ilvG, cysN, and pyrB | E. coli | 6–40 | CeO2 | AM | 100 mg L−1 | 1 h | 56 | |
| GULP1, SLC30A8, NEGR1, SEC16B, MTCH2, MAF, MC4R, TMEM195, INSIG2, NAMPT, MTMR9, PFKP, KCTD15, LPL and GNPDA2 | Adipocytes | — | FeO | AM | — | 30 h | 66 | |
| Cytochrome C | HFL-I | 20 and 80 | SiO2 | AM | 100 mg mL−1 | 24 h | 46 | |
| Cdkn1a and Cdkn1c | Mice (CD-1 ICR) | 6 | TiO2 | IN | 10 mg kg−1 | 90 d | 26 | |
| Cyp171a | Mice (CD-1 ICR) | 6 | TiO2 | IG | 10 mg kg−1 | 90 d | 63 | |
| Ch25h, Fabp3, Aldob | Mice (C57BL/6) | — | TiO2–rutile | IA | 54 μg | 28 d | 20 | |
| Membrane transport | ar, tsr and tap | E. coli | — | Au-DP | AM | 10 mg L−1 | 4 h | 61 |
| Slc25a30, Rgn, Tmlhe, Sec62, Cldn16, Ktn1 | Rats | 20; spheres | Cd doped Si | IT | 1 mg | 30 d | 62 | |
| cmr, yajr, and emrE, and dnaK | E. coli (k12) | 10 | TiO2 | AM | 10 mg L−1 | 2 h | 24 | |
| DNA repair | recN, uvrA, ybfE, yebG, ssb, sbmc and nfo | E. coli (k12) | 60 | Ag | AM | 10 mg L−1 | 2 h | 24 |
| XRCC1, XRCC3, FEN1, RED51C and RPA1 | IMR-90, U251 | 6–20 | Ag | AM | 400 μg mL−1 | 48 h | 78 | |
| RecA and lexA | E. coli (k12) | 10 | TiO2 | AM | 10 mg L−1 | 2 h | 24 | |
| Oxidative stress/general stress response | sodA, sodC, and katE | E. coli (k12) | 60 | Ag | AM | 10 mg L−1 | 2 h | 24 |
| Sod-3 | C. elegans | <100; spheres | Ag | AM | 0.1 mg L−1 | 24 h | 41 | |
| pkm2a, pkmb2 and etv5a | Zebrafish (WT/WIK eggs) | 10 | Ag | AM | 5 μg L−1 | 24 and 48 h | 81 | |
| AhpC | E. coli | — | Au-DP | AM | 10 mg L−1 | 4 h | 61 | |
| CAT, GST | D. magna | 100 | C60-Hx, C60-OH and TiO2 | AM | 7.5 (C60-Hx) 100 (C60-OH) and 7.5 mg L−1 (TiO2) | 24 h | 17 | |
| GSTM3 and GSR | MSC | <50 | CB | AM | 50 and 100 μg mL−1 | 24 h | 27 | |
| GSH, SOD, CAT, GST, and Nrf2 | HEPG2 | 5 | CdTe | AM | 10 μg mL−1 | 24 h | 39 | |
| ERK 1/2, p38, GAPDH, MDA, SOD, GSH and CAT | SMMC-7721 | 20–30; hexahedrals | CeO2 | AM | 50 μg mL−1 | 24 h | 55 | |
| Sod-3, Hsp-70, cyp-35A2, dnaK and katG | C. elegans | 73, nanotubes | SWCNT | AM | 500 mg L−1 | 48 h | 59 | |
| Axud1, Cyp4a12a, Cyp4a12b, Cyp4a14 and Cyp2d9 | Mice | 6 | TiO2 | IG | 10 mg kg−1 | 90 d | 29 | |
| Cryab and Alkbh7 | Mice (CD-1 ICR) | 6 | TiO2 | IN | 10 mg kg−1 | 90 d | 26 | |
| PRDX3, PRNP and TXNRD1 | BEAS-2B | 14–25 | ZnO | AM | 5 μg mL−1 | 24 h | 28 | |
| Reproduction | CrEcR | C. riparius (larvae) | — | Ag | AM | 0.2 mg L−1 | 1–72 h | 45 |
| chc-1, col-51, col-183, flna1, lit and par-3 | C. elegans | 73, nanotubes | SWCNT | AM | 500 mg L−1 | 48 h | 59 | |
At lower exposure doses, which are most likely the more realistic environmental exposure scenario for many organisms, nanomaterials illicit changes in a wider range of pathways (Table 1). For example mice exposed to three concentrations of TiO2 NPs (18, 54 and 162 μg per mouse), responded differently depending upon concentration. At 28 days, exposure of mice to 18 μg (the lowest dose) TiO2 induced changes in muscle contraction and striated tissue development, whereas after the same 28 day duration, exposure to 162 μg (the highest dose) was dominated by the inflammatory response.20
Generalized responses to xenobiotics are also implicated in the nanotoxicity literature. Metabolizing enzymes such as CYP1A, involved in xenobiotic metabolism has been shown to be triggered by carbon black nanoparticles in cell cultures.27 Some of these genes are also related to antioxidant enzymes that protect the cells from oxidative stress from contaminants, including PRDX3 and BNIP3 which have been found to be expressed in cell lines in response to ZnO nanomaterials28 or oxidize compounds such as CYP2d9 in response to TiO2 nanomaterial exposure.29
Apoptosis is a normal process important in balancing cellular structure associated with growth and development within an organism. However apoptosis can also be associated with cellular damage and death due to injury. This can be due to oxidative or free radical damage, or it can be an independent signal of an inflammatory response that induces ROS generation by the primary immune cells of the organism. The apoptosis process also instigates cellular alterations such as shrinkage, DNA degradation, and cell surface alterations that trigger phagocytosis by other cells to remove foreign substances. In cell cultures a variety of nanomaterials have been shown to directly increase genes associated with apoptosis such as caspases and cytochrome C.30–40 The suppression of certain gene families that in turn suppress cell death can also trigger apoptosis and immune response and in studies of titanium dioxide25,26,29 these genes including Birc5 and Crap2 were suppressed after 90 day exposures which increased apoptosis in whole organism models. However, in contrast to in vitro cell line studies, whole organism studies to date do not suggest significant expression of pathways associated with apoptosis associated with nanomaterial exposures.
In addition to these studies, other studies show the same response in a variety of organisms and cell lines, with higher concentrations of nanoparticles inducing genes associated with general stress, oxidative stress, and apoptosis and lower concentrations affecting other molecular pathways such as groups of genes in the major facilitator superfamily, such as drug resistance or detoxification genes and other cell signaling genes and transcription factors.24,41,42 Variations in dose will differentially alter gene expression with low and high concentrations displaying a unique molecular fingerprint.21,43,44 However, there are some exceptions of genomic overlapping, where higher concentrations trigger different genes but the same pathways of genes or affect the same genes.27,41,45,46
These studies demonstrate the need for testing nanomaterials at lower exposure concentrations so the more specific interactions of nanomaterials and cells that occur at environmentally relevant concentrations can be elucidated. This shows that there are other non-oxidative stress mechanisms present regarding nanoparticle toxicity. Testing these materials at low, environmentally relevant concentrations will better enable the development of biomarkers for assessing nanomaterial toxicity.
Modeling the impacts of nanomaterials and the need to investigate other molecular mechanisms
One of the goals of nanotoxicology research is to inform the design of safe and sustainable materials to minimize potential impacts to human health and the environment. If the properties that can make a given nanomaterial harmful can be predicted during the design process then production can shift to create a less harmful version. To make these predictions feasible, data is needed to model the interactions of these chemicals with organisms. Examining acute high dose interactions of nanomaterials with biological entities provides limited information as to the interaction of nanomaterials on a molecular level as is seen in the previous discussion. These extreme exposures overwhelm cellular system and ultimately lead to tissue necrosis and failure. Pesticide studies, which include some of the most well studied toxins, have provided a wealth of information regarding the potential for modeling responses of chemicals. They have shown that examining acute exaggerated endpoints limit the ability to predict the impacts of new toxins through modeling such as quantitative structure activity relationships (QSAR). These models consistently overestimate toxicity when data is based on necrosis.48,49 For example Reuschenback and collaborators50 found that using traditional ECOSAR data, 69% of 1000 industrial chemicals tested fell into the correct category. Mode-of-action based QSARs may more accurately predict effects, yet there is less information to feed these types of models given traditional testing strategies.51 Ultimately the diversity of training compounds for a model and their corresponding molecular toxicology information may be the limiting factor in creating models that may consistently and accurately predicts effects.52,53 Therefore identifying a greater number of pathways involved in response to nanomaterials will provide more robust modeling and predictive power in determining the impacts of nanomaterials.Other factors that limit current models include the measurement of a limited set of endpoints and time points as well as large dose exposures. Effects are also often time and chemical dependent therefore using an exaggerated response over a short time period introduces bias into modeling.54 In addition, low-dose effects are often hormetic in nature, where low-dose exposures and high-dose exposures have opposite effects, low can be stimulatory to many pathways and high doses cause a toxic response. As a result models using higher concentrations will inaccurately predict effects at doses that are most environmentally realistic. Developing assays that provide unbiased parameters and give information regarding different mechanisms of action will ultimately provide better models. As has been seen with other emerging contaminants such as endocrine disruptors, alternative pathways of effect that occur over chronic low-dose exposures and potentially over generations may in the end be the ultimate concern. There is a need to test nanomaterials at multiple concentrations and time points to help develop biomarkers to assess nanomaterial toxicity. More relevant and useful information for modeling is garnered from low-dose exposure studies that examine multiple endpoints and pathways of impact. Sublethal concentrations elicit more specific biological pathway responses.
Evidence for alternative mechanisms of nanomaterial impact
The interaction of nanoparticles with other pathways that do not directly involve apoptosis, oxidation or inflammation is slowly being explored and reported in the literature but is still underrepresented compared to other pathways and mechanisms of action or effect. A simple survey of articles in Pubmed using nanomaterials, toxicity and specific pathways as key words shows a significant bias in the literature towards oxidative stress and inflammation (Table 2). Other pathways impacted by nanomaterial exposure include to a smaller extent include those involved in reproduction,45,55 metabolism,56–58 cell cycle and cell proliferation,21,59 membrane transport,24,30,60–62 cellular motility,61 steroidogenic pathways,63 and others.| Processes | # Results |
|---|---|
| Cell function | 2470 |
| Oxidative stress | 694 |
| Apoptosis | 574 |
| Cell proliferation | 490 |
| Inflammation | 474 |
| Biological development | 449 |
| Lipid metabolism | 327 |
| Immune | 240 |
| Reproductive | 191 |
| Cell cycle | 184 |
| Neurotransmitter | 181 |
| Cell signaling | 173 |
| Membrane transport | 139 |
| Translation | 121 |
| Transductional signaling | 91 |
| Transcription | 89 |
| Glucose metabolism | 45 |
| DNA repair | 40 |
| P450 | 11 |
| Receptor mediated response | 11 |
| Ion homeostasis | 9 |
| Muscle regulation | 5 |
The types of nanomaterials that have been tested across different toxicity studies differ tremendously and the diversity of nanomaterials studied in any one study is very narrow. This makes it difficult to draw conclusions regarding which nanomaterial may cause the greatest impact, what size and shape may be most toxic, and the pathways that may provide the most information. For example, individual studies have shown separately that pathways involved in cell cycle and cell proliferation have been shown to be impacted by TiO2,26,29 SiO2,64 CdSi,62 CuNP,47 and Ag nanomaterials65 at various concentrations and exposure conditions. Cell signaling pathways, which are heavily involved in disease and impact development and immunity as well as other functions have been shown to be impacted by only TiO2 (ref. 20 and 29) and CdSi nanomaterials.62 Pathways associated with metabolism transcription, translation and some metabolism pathways have been significantly less studied. The lack of information on these other pathways speaks not to the lack of impact on these pathways but to the general lack of data and consistency of evaluating multiple pathways of effect.
There are singular studies, most often cell-based assays, which have investigated impacts on other less commonly studied pathways. For example, in one study the response of adipocytes to superparamagnetic iron oxide nanoparticles included differential regulation of genes associated with lipid and glucose metabolism.66 Expression of genes associated with the transduction signaling of TGF-beta pathway was altered after exposure of Fe3O4 nanoparticles in HeLa cells indicating interference with that pathway.67 Other studies have indicated endoplasmic reticulum stress response,68 transduction signaling of Epidermal Growth Factor (EGF) receptor,69 and hypoxia associated responses to exposure to nanomaterials.70
Information obtained from molecular indicators: how nanomaterial characteristics may influence toxicity
Molecular indicators, such as gene or protein expression, have provided some useful information regarding the impact of specific nanomaterial properties on their toxicity. For example, molecular studies have shown that nanomaterial size has an impact on the general molecular response of a cell, tissue or organism to exposure. Depending on the assay and endpoint considered, studies suggest that smaller particles are able to cross cell barriers and induce a greater response or in some cases larger nanomaterials have a greater impact. For example, 14 nm carbon black (CB) nanoparticles cause an induction in proinflammatory cytokines, chemokines and monokines in the olfactory bulb of mice where 95 nm CB particle do not.71 In addition, human lung fibroblasts exposed to 20 nm SiO2 nanoparticles induced p53, and Bax expression, inhibited Bcl-2 production and activated caspase-9 where 80 nm nanoparticles did not.46 In contrast, 50 nm GNPs induce a larger immunotoxic response in liver cytokines and induce immune related IL-6 and TNF-a expression than the smaller 10 nm sized GNPs in the liver and kidney of injected rats.22 Similarly 500 nm silica nanoparticles elicited a greater response in murine macrophages than 10 nm nanoparticles.64 Silica particles that are 500 nm impact pathways in macrophage cells related to cell cycle progression, DNA transcription, inflammatory response, apoptosis, signal transduction and cell differentiation, which are not differentially expressed in 10 nm particle exposure.64 Yet overall the processes represented by these pathways are not enriched in one particle size or another and the authors hypothesize that any differences may be due to the either the level of disturbance to the cell membrane differing with surface area or the ability of smaller particles to enter the cell.64Surface chemistry may play a large role in the interaction of a nanomaterial with an organism or cell. Some studies suggest that the charge of the surface of the nanomaterial is a big determinant of impact and that positively charged surfaces have caused a greater biological response. For example Yang and co-workers72 found that Azotobacter vinelandii (a nitrogen fixing bacteria commonly found in wastewater treatment facilities) exposed to quantum dots (QD) coated with cationic polyethylenimine (PEI) exhibited the up-regulation of gene cad R, a gene associated with metal contamination, more so than QD coated with anionic polymaleic anhydride-alt-1-octadecene (PMAO). Additionally, the up-regulation of several types of nitrogenases (nif D, nif H, anf D, anf K, vnf D and vnf H) were observed in the QD-PEI (but not QD-PMAO) exposed bacteria, indicating that nitrogen fixation is stimulated upon exposure to the positively charged QD-PEI particles.72 In another study QD COOH-pQDs were more toxic and instigated the expression of scavenger receptor (SRA) endocytic pathway and the downstream NF-kB signaling cascades which are involved in innate and adaptive immunity, inflammation, and stress responses, in comparison to NH2-PEG-pQDs and HO-PEG-pQD which had an overexpression of p38 AP-1 cell signaling cascades instead.73 In a whole organism study, Daphnia magna exposed to polyvinyl pyrrolidone (PVP) and citrate (CIT) capped silver nanoparticles reported differences in gene expression related to stress response upon exposure to the differently coated silver nanoparticles. PVP coated particles, although less toxic, induced stress genes metallothionein (MT) and DNA repair gene (REV1) significantly, while CIT coated particles did not.74 Collectively, these results show that the various surface coatings and functional groups of a variety of nanoparticles can affect the expression of genes involved in numerous pathways.
Molecular data has also indicated that other factors such as the interactions of nanomaterials with cellular media and endogenous biomolecules, differences in the tissue or cell line, and location of accumulation of nanomaterial can impact the molecular interaction with a nanomaterial. The interactions of nanoparticles with chemical compounds present in cell culture media and their subsequent impacts on toxicity tests and molecular response have been minimally explored. However several studies have shown that cellular media and molecules can coat nanomaterial and change their properties and their interactions with cells. For example, cytotoxicity decreases for citrate coated Au-NPs due to, in the presence of fetal bovine serum (FBS) medium.75 The media components were shown to alter the citrate-coated gold crystals as Au crystals, naked and citrate-coated, deposited in FBS exhibited a frequency decrease that was higher than the Au crystals, naked or functionalized, incubated in RPMI without FBS present. Due to the importance of the interactions of nanoparticles with biological systems,76,77 there is a significant concern that there are only a handful of human cell studies that mention this interaction with free molecules present in the supplemented FBS media.
Differences among cell lines used for toxicity studies lead to differences regarding the pathways instigated by exposure to nanomaterials. Cancer cells lines such as U251 cells78 and MCF7 (ref. 79) are more susceptible to nanoparticle exposure than normal cells. The animal where the cell line originated is also important. For example murine macrophages were shown to be more sensitive than human cell lines when exposed to BSA-stabilized silica nanoparticles80 and silver-doped silica nanoparticles induce more toxicity in human hepatoma cell line (Huh7) than fibroblast-like fathead minnow (FHM, Pimephales promelas) cells.68 Cell lines from different tissues of the same organism also respond differently to nanoparticle treatment. Toll-like receptor 2 (TLR-2) gene expression, a gene responsible for cell membrane receptors that recognize foreign substances, was expressed to a greater extent in human chondrocytes (C28/I2) and periodontal ligament (PDL) cells in response to Ag-NPs exposure than in squamous cell carcinoma from the tongue (SCC-9).31 Because of these differences among cell types one might expect that the deposition location in an organism can also impact the molecular response. If nanomaterials induce toxicity to vital organs, including the lung, brain, liver, kidney, spleen, and ovary, each of these tissues may be specific in their response due to their differing functionality. All of these factors point towards the need to explore a diversity of cells, tissues and organisms to fully understand these molecular interactions.
Microarrays and next-generation gene expression technologies to identify new mechanisms and biomarkers of effect
Global gene expression patterns generated using microarrays or more recently, next-generation sequencing technologies, hold promise for providing a more diverse profile of the impacts of nanomaterials on various biological systems. Using the pattern of expression of thousands of genes at once provides a systems overview of the reaction of an organism or cell. In addition, these data provide a method to differentiate among nanomaterials as to their impact across a range of potential. Microarrays probe a pre-prescribed set of tens of thousands of genes at once. This technology is now relatively cheap with analysis costs of approximately $200 USD per sample, allowing for multiple comparisons across treatments or individuals within an experiment. Alternatively, using next-generation sequencing, direct sequencing representative of RNA expressed provides millions of data points per sample to compare responses across exposures. Next-generation sequencing analyses probes a multitude of genes and pathways at once with no preconceived idea of the genes that may be relevant and therefore the limitation of the predesigned array is removed. Semiquantification of transcripts and gene discovery can be done simultaneously. However, there is also a more significant expense associated with this type of analysis and quantification of gene transcripts need to be confirmed using quantitative PCR or similar quantitative analyses.There have been several studies to date that have investigated the biological response of cells or organisms to nanomaterials using global gene expression patterns from microarrays that have provided some key insights to the molecular response to exposure.20,26,29,47,62,63 Similar to other xenobiotics these studies have shown that when comparing a nanomaterial exposed organism or cell to a control there can be nanomaterial specific gene regulation42,56 and the gene expression patterns can be used to separate the effects of the nanomaterial from the other components in a suspension such as any metal ions that may be emitted from a metal nanomaterial.47 Toxicity of metal nanomaterials has been investigated more than other nanomaterial types and such experiments have identified that common pathways of impact include oxidative stress as well as inflammation and apoptosis. Genomics tools have been used as a tool to separate the impacts of metal ions from the dissolution of metal nanomaterials in solution from the nanomaterial itself to determine the underlying mechanism of toxicity of nanomaterials. For example similar genomic profiles are expressed in silver nanomaterial or silver ion exposures suggesting that both types of exposures induce toxicity by similar mechanisms.81,82 However, other studies show that nanomaterial toxicity acts by mechanisms that are unique to the nanomaterial and not a consequence of metal ion dissolution. Some novel genetic pathways associated with metal nanomaterials versus their metal ion counterparts in these studies include apoptosis, cell proliferation and differentiation, and cancer progression for copper nanoparticles;47 energy metabolism for copper nanoparticles;83 ribosome activity and elongation factors for TiO2 nanoparticles47 and metal detoxification, metabolic processes, and radical scavenging for silver nanoparticles.84 Variations in the responses found in these studies might be a result of the type of exposure (terrestrial, aqueous, or inhalation/instillation) and the type of particle, as different nanomaterials behave differently in various types of media.
Next generation sequencing technologies also hold promise for distinguishing the impacts of nanomaterials of differing properties. For example, our lab has investigated the differences in toxicity and associated molecular pathways instigated in response to fullerene and carbon nanotubes exposures with varying surface chemistries in several organisms. To examine global gene expression, RNA was extracted, cDNA created and sequenced using Roche 454 sequencer from the aquatic toxicology and genomic model Daphnia magna (Fig. 3) that had been exposed to 50 ppm fullerene nanomaterials for 48 hours. This concentration has been shown to be at the LC25 for this species in our previous work but was also shown not to instigate oxidative stress17 (for toxicity and nanomaterial characterization information see Klaper et al. 2009, and Arndt et al. 2013).17,85 As a comparison, daphnids exposed to control water and 50 ppm of fullerols were also sequenced (C60 (OH)24). Sequences were screened for quality control and approximately 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fragments were sequenced for a pool of three replicates of 20 adult daphnids for each treatment. Sequences were compared to each other and overlapping sequences (with greater than 95% overlap) from all treatments were assembled into one set of contigs, or longer gene sequence fragments, and then annotation by comparing these longer sequences to public databases. Fragments that only appeared once or had no overlap with other sequences were excluded from analysis. Associated pathway information for each gene fragment was also identified. Approximately 30
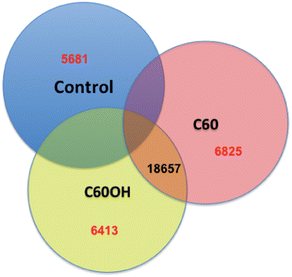
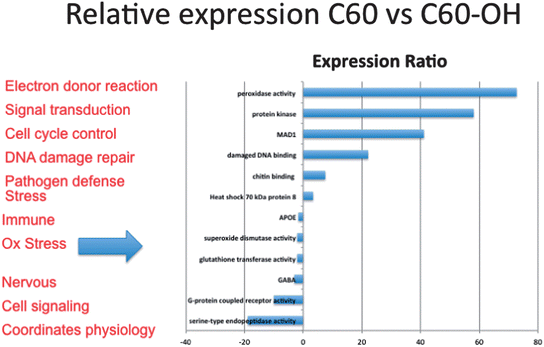
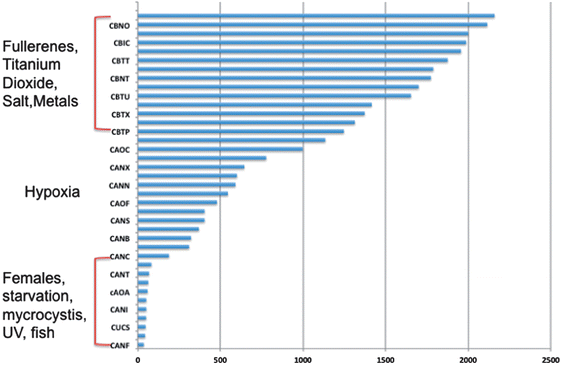
000 fragments were sequenced for a pool of three replicates of 20 adult daphnids for each treatment. Sequences were compared to each other and overlapping sequences (with greater than 95% overlap) from all treatments were assembled into one set of contigs, or longer gene sequence fragments, and then annotation by comparing these longer sequences to public databases. Fragments that only appeared once or had no overlap with other sequences were excluded from analysis. Associated pathway information for each gene fragment was also identified. Approximately 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 contigs per treatment remained after this cleanup and were used in an RNA-Seq analysis, which compares the number of times a given sequence is represented between treatments. Sequences that were overrepresented in one treatment versus another were also analyzed with respect to pathways represented. Some of the key findings of this analysis include a significant overlap in the sequences represented across treatments with a large number of unique sequences represented in each treatment (Fig. 4). In addition, when the sequences are analyzed for their associated molecular pathways it is clear that although overlaps exist the difference in the surface chemistry of these two particles causes not only a difference in the toxicity but at sublethal concentrations it causes a difference in the molecular pathways that are expressed (Fig. 5). Genomic analyses can separate not only the various impacts of xenobiotic exposures but reaction to natural stressors compared to xenobiotic exposures as well. For this same experiment we compared the expression of genes in the nanomaterial exposures above to Daphnia pulex exposed to several different xenobiotic exposures and other stressor exposures in experiments related to our lab and the lab of others conducted through the Daphnia Genome Consortium and sequenced by the Joint Genome Institute. These included titanium dioxide and fullerene exposures as well as exposures to heavy metal exposures, hypoxia, temperature stress, exposure to pheromones from predators, and different life stages. When the Daphnia magna exposed to fullerenes and fullerols are compared to the Daphnia pulex exposures it is clear that the D. magna fullerene and fullerol nanomaterials related expression clusters with the D. pulex titanium dioxide and fullerene nanomaterial exposures as well as the other chemical exposures. They are less similar to hypoxia related stress or the more biological stressors of starvation and predator presence (Fig. 6). This indicates that the genomic response to nanomaterials is more similar to other chemical exposures than more natural stressors. The types of genes expressed upon nanomaterial exposure could provide an indicator of the differences in the molecular pathways instigated by nanomaterials versus other stressors.
000 contigs per treatment remained after this cleanup and were used in an RNA-Seq analysis, which compares the number of times a given sequence is represented between treatments. Sequences that were overrepresented in one treatment versus another were also analyzed with respect to pathways represented. Some of the key findings of this analysis include a significant overlap in the sequences represented across treatments with a large number of unique sequences represented in each treatment (Fig. 4). In addition, when the sequences are analyzed for their associated molecular pathways it is clear that although overlaps exist the difference in the surface chemistry of these two particles causes not only a difference in the toxicity but at sublethal concentrations it causes a difference in the molecular pathways that are expressed (Fig. 5). Genomic analyses can separate not only the various impacts of xenobiotic exposures but reaction to natural stressors compared to xenobiotic exposures as well. For this same experiment we compared the expression of genes in the nanomaterial exposures above to Daphnia pulex exposed to several different xenobiotic exposures and other stressor exposures in experiments related to our lab and the lab of others conducted through the Daphnia Genome Consortium and sequenced by the Joint Genome Institute. These included titanium dioxide and fullerene exposures as well as exposures to heavy metal exposures, hypoxia, temperature stress, exposure to pheromones from predators, and different life stages. When the Daphnia magna exposed to fullerenes and fullerols are compared to the Daphnia pulex exposures it is clear that the D. magna fullerene and fullerol nanomaterials related expression clusters with the D. pulex titanium dioxide and fullerene nanomaterial exposures as well as the other chemical exposures. They are less similar to hypoxia related stress or the more biological stressors of starvation and predator presence (Fig. 6). This indicates that the genomic response to nanomaterials is more similar to other chemical exposures than more natural stressors. The types of genes expressed upon nanomaterial exposure could provide an indicator of the differences in the molecular pathways instigated by nanomaterials versus other stressors.
 | ||
| Fig. 3 Daphnia magna is a model organism for toxicity and the interaction of the genome with the environment. | ||
Genomic data may not provide all answers to the impacts of nanomaterials on organisms. There is some possibility that some of the pathways expressed may be similar across nanomaterials as there are a set of key responses that cells and tissues use to respond to xenobiotics. However, as shown previously the data available through in vitro cellular assays indicates the complete expression pattern can be used to distinguish among stressors. The current issue is in deciphering the endpoints related with the global gene expression response. In addition, it should be noted that the data generated in genomic experiments is complicated by the volume of sequences generated and in organisms where the genome has not been sequenced and annotated full interpretation of the data will be incomplete. However, overall the molecular response described by examining the co-expression of these multiple biochemical pathways in a systems approach illustrates that the response is more complicated than oxidative stress and inflammation. It also provides a more complete idea of the response of an organism to nanomaterial exposure.
Concluding remarks
Molecular biomarkers provide key data regarding the way in which nanomaterials interact with cells, tissues and organisms. Data indicate that pathways expressed in response to nanomaterials differ among different types of nanomaterials, either due to the size, core chemistry or surface chemistry of the nanomaterials. In addition, the molecular response to nanomaterials can differ depending on the dose and exposure time. Collectively, the sum of the responses across experiments indicates that although many studies focus on oxidative stress to predict toxicity, organisms respond with more than an oxidative stress response. Given the impacts of sublethal chronic exposures that will most likely occur, including a diversity of pathways will be necessary to ultimately determine how nanomaterials may impact organisms in more realistic exposure scenarios. Investigating a diversity of pathways can supplement modeling efforts in predicting the potential effect of new nanomaterials as they are developed. Global gene expression profiling will become an even more useful tool as more high-throughput approaches are explored in evaluating the environmental health and safety aspects of nanomaterials.Acknowledgements
This work was supported by a grant to R. Klaper from the National Science Foundation (CBET 933720). Daphnia pulex sequences obtained as part of the Daphnia Genome Consortium and Joint Genome Institute. This work was also supported by a grant to R. Klaper from the Bradley Catalyst Foundation.References
- F. Gottschalk and B. Nowack, J. Environ. Monit., 2011, 13, 1145–1155 RSC.
- L. n. Brunet, D. Y. Lyon, E. M. Hotze, P. J. J. Alvarez and M. R. Wiesner, Environ. Sci. Technol., 2009, 43, 4355–4360 CrossRef CAS.
- H. Ma, A. Brennan and S. A. Diamond, Environ. Toxicol. Chem., 1999, 31, 2099–2107 CrossRef PubMed.
- S. Bhattacharya, Q. Zhang, P. L. Carmichael, K. Boekelheide and M. E. Andersen, PLoS One, 2011, 6, e20887 CAS.
- M. E. Andersen and D. Krewski, Toxicol. Sci., 2009, 107, 324–330 CrossRef CAS PubMed.
- H. K. Hamadeh, P. R. Bushel, S. Jayadev, K. Martin, O. DiSorbo, S. Sieber, L. Bennett, R. Tennant, R. Stoll, J. C. Barrett, K. Blanchard, R. S. Paules and C. A. Afshari, Toxicol. Sci., 2002, 67, 219–231 CrossRef CAS PubMed.
- A. Brouwer, M. P. Longnecker, L. S. Birnbaum, J. Cogliano, P. Kostyniak, J. Moore, S. Schantz and G. Winneke, Environ. Health Perspect., 1999, 107, 639–649 CAS.
- F. S. Vom Saal and C. Hughes, Environ. Health Perspect., 2005, 113, 926–933 CrossRef CAS.
- R. P. Amin, H. K. Hamadeh, P. R. Bushel, L. Bennett, C. A. Afshari and R. S. Paules, Toxicology, 2002, 181–182, 555–563 CrossRef CAS.
- S. Ansar Ahmed, Toxicology, 2000, 150, 191–206 CrossRef CAS.
- A. Pal, K. Y.-H. Gin, A. Y.-C. Lin and M. Reinhard, Sci. Total Environ., 2010, 408, 6062–6069 CrossRef CAS PubMed.
- H. Yang, C. Liu, D. Yang, H. Zhang and Z. Xi, J. Appl. Toxicol., 2009, 29, 69–78 CrossRef CAS PubMed.
- P. Van Hummelen and J. Sasaki, Mutat. Res., Rev. Mutat. Res., 2010, 705, 165–171 CrossRef CAS PubMed.
- V. I. Lushchak, Aquat. Toxicol., 2011, 101, 13–30 CrossRef CAS PubMed.
- O. Choi and Z. Hu, Environ. Sci. Technol., 2008, 42, 4583–4588 CrossRef CAS.
- A. K. Goetz, B. P. Singh, M. Battalora, J. M. Breier, J. P. Bailey, A. C. Chukwudebe and E. R. Janus, Regul. Toxicol. Pharmacol., 2011, 61, 141–153 CrossRef CAS PubMed.
- R. Klaper, J. Crago, J. Barr, D. Arndt, K. Setyowati and J. Chen, Environ. Pollut., 2009, 157, 1152–1156 CrossRef CAS PubMed.
- A. Nel, T. Xia, H. Meng, X. Wang, S. Lin, Z. Ji and H. Zhang, Acc. Chem. Res., 2012, 46, 607–621 CrossRef PubMed.
- I. Fenoglio, M. Tomatis, D. Lison, J. Muller, A. Fonseca, J. B. Nagy and B. Fubini, Free Radical Biol. Med., 2006, 40, 1227–1233 CrossRef CAS PubMed.
- M. Husain, A. T. Saber, C. Guo, N. R. Jacobsen, K. A. Jensen, C. L. Yauk, A. Williams, U. Vogel, H. Wallin and S. Halappanavar, Toxicol. Appl. Pharmacol., 2013, 269, 250–262 CrossRef CAS PubMed.
- A. Ambrosone, L. Mattera, V. Marchesano, A. Quarta, A. S. Susha, A. Tino, A. L. Rogach and C. Tortiglione, Biomaterials, 2012, 33, 1991–2000 CrossRef CAS PubMed.
- H. A. Khan, M. A. Abdelhalim, A. S. Alhomida and M. S. Al-Ayed, BioMed Res. Int., 2013, 2013, 590730 Search PubMed.
- M. F. Rahman, J. Wang, T. A. Patterson, U. T. Saini, B. L. Robinson, G. D. Newport, R. C. Murdock, J. J. Schlager, S. M. Hussain and S. F. Ali, Toxicol. Lett., 2009, 187, 15–21 CrossRef CAS PubMed.
- N. Gou, A. Onnis-Hayden and A. Z. Gu, Environ. Sci. Technol., 2010, 44, 5964–5970 CrossRef CAS PubMed.
- X. Sang, B. Li, Y. Ze, J. Hong, X. Ze, S. Gui, Q. Sun, H. Liu, X. Zhao, L. Sheng, D. Liu, X. Yu, L. Wang and F. Hong, J. Agric. Food Chem., 2013, 61, 5590–5599 CrossRef CAS PubMed.
- B. Li, Y. Ze, Q. Sun, T. Zhang, X. Sang, Y. Cui, X. Wang, S. Gui, D. Tan, M. Zhu, X. Zhao, L. Sheng, L. Wang, F. Hong and M. Tang, PLoS One, 2013, 8, 1–11 Search PubMed.
- A. A. Alshatwi, V. S. Periasamy, P. Subash-Babu, M. A. Alsaif, A. A. Alwarthan and K. A. Lei, Environ. Toxicol. Pharmacol., 2013, 36, 215–222 CrossRef CAS PubMed.
- C.-C. Huang, R. S. Aronstam, D.-R. Chen and Y.-W. Huang, Toxicol. in Vitro, 2010, 24, 45–55 CrossRef CAS PubMed.
- S. Gui, X. Sang, L. Zheng, Y. Ze, X. Zhao, L. Sheng, Q. Sun, Z. Cheng, J. Cheng, R. Hu, L. Wang, F. Hong and M. Tang, Part. Fibre Toxicol., 2013, 10, 4 CrossRef CAS PubMed.
- J. Ahmad, M. Ahamed, M. J. Akhtar, S. A. Alrokayan, M. A. Siddiqui, J. Musarrat and A. A. Al-Khedhairy, Toxicol. Appl. Pharmacol., 2012, 259, 160–168 CrossRef CAS PubMed.
- A.-S. Kim, C.-H. Chae, J. Kim, J.-Y. Choi, S.-G. Kim and G. Băciucmb.cedilt, Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology, 2012, 113, 789–798 CrossRef PubMed.
- R. Govender, A. Phulukdaree, R. Gengan, K. Anand and A. Chuturgoon, J. Nanobiotechnol., 2013, 11, 5 CrossRef CAS PubMed.
- M. Ahamed, M. Karns, M. Goodson, J. Rowe, S. M. Hussain, J. J. Schlater and Y. Hong, Toxicol. Appl. Pharmacol., 2008, 233, 404–410 CrossRef CAS PubMed.
- A. A. Alshatwi, P. Vaiyapuri Subbarayan, E. Ramesh, A. A. Al-Hazzani, M. A. Alsaif and A. A. Alwarthan, J. Biochem. Mol. Toxicol., 2012, 26, 469–476 CrossRef CAS PubMed.
- G. Badr, M. K. Al-Sadoon, M. A. Abdel-Maksoud, D. M. Rabah and A. M. El-Toni, PLoS One, 2012, 7, e51661 CAS.
- M. Das and S. K. Sahoo, PLoS One, 2012, 7, e32920 CAS.
- S. Hussain, F. Al-Nsour, A. B. Rice, J. Marshburn, B. Yingling, Z. Ji, J. I. Zink, N. J. Walker and S. Garantziotis, ACS Nano, 2012, 6, 5820–5829 CrossRef CAS PubMed.
- X. Li, X. Lu, H. Xu, Z. Zhu, H. Yin, X. Qian, R. Li, X. Jiang and B. Liu, Mol. Pharmaceutics, 2011, 9, 222–229 Search PubMed.
- K. C. Nguyen, W. G. Willmore and A. F. Tayabali, Toxicology, 2013, 306, 114–123 CrossRef CAS PubMed.
- M. E. Selim and A. A. Hendi, Asian Pac. J. Cancer Prev., 2012, 13, 1617–1620 CrossRef.
- J.-y. Roh, S. J. Sim, J. Yi, K. Park, K. H. Chung, D.-y. Ryu and J. Choi, Environ. Sci. Technol., 2009, 43, 3933–3940 CrossRef CAS.
- S. I. L. Gomes, S. C. Novais, J. J. Scott-Fordsmand, W. De Coen, A. M. V. M. Soares and M. J. B. Amorim, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2012, 155, 219–227 CrossRef CAS PubMed.
- H. C. Poynton, J. M. Lazorchak, C. A. Impellitteri, M. E. Smith, K. Rogers, M. Patra, K. A. Hammer, H. J. Allen and C. D. Vulpe, Environ. Sci. Technol., 2010, 45, 762–768 CrossRef PubMed.
- X. Liu and J. Sun, Biomaterials, 2010, 31, 8198–8209 CrossRef CAS PubMed.
- P. M. G. Nair and J. Choi, Environ. Toxicol. Pharmacol., 2012, 33, 98–106 CrossRef CAS PubMed.
- Z. Xu, L. Chou and J. Sun, J. Appl. Toxicol., 2012, 32, 358–364 CrossRef CAS PubMed.
- R. J. Griffitt, K. Hyndman, N. D. Denslow and D. S. Barber, Toxicol. Sci., 2009, 107, 404–415 CrossRef CAS PubMed.
- D. R. J. Moore, R. L. Breton and D. B. MacDonald, Environ. Toxicol. Chem., 2003, 22, 1799–1809 CrossRef CAS.
- H. Sanderson and M. Thomsen, Bull. Environ. Contam. Toxicol., 2007, 79, 331–335 CrossRef CAS PubMed.
- P. Reuschenbach, M. Silvani, M. Dammann, D. Warnecke and T. Knacker, Chemosphere, 2008, 71, 1986–1995 CrossRef CAS PubMed.
- S. P. Bradbury, C. L. Russom, G. T. Ankley, T. W. Schultz and J. D. Walker, Environ. Toxicol. Chem., 2003, 22, 1789–1798 CrossRef CAS.
- R. P. Sheridan, B. P. Feuston, V. N. Maiorov and S. K. Kearsley, J. Chem. Inf. Comput. Sci., 2004, 44, 1912–1928 CrossRef CAS PubMed.
- I. V. Tetko, I. Sushko, A. K. Pandey, H. Zhu, A. Tropsha, E. Papa, T. Öberg, R. Todeschini, D. Fourches and A. Varnek, J. Chem. Inf. Model., 2008, 48, 1733–1746 CrossRef CAS PubMed.
- T. Jager and S. L. M. Kooijman, Ecotoxicology, 2009, 18, 187–196 CrossRef CAS PubMed.
- G. Cheng, W. Guo, L. Han, E. Chen, L. Kong, L. Wang, W. Ai, N. Song, H. Li and H. Chen, Toxicol. in Vitro, 2013, 27, 1082–1088 CrossRef CAS PubMed.
- D. A. Pelletier, A. K. Suresh, G. A. Holton, C. K. McKeown, W. Wang, B. Gu, N. P. Mortensen, D. P. Allison, D. C. Joy, M. R. Allison, S. D. Brown, T. J. Phelps and M. J. Doktycz, Appl. Environ. Microbiol., 2010, 76, 7981–7989 CrossRef CAS PubMed.
- D. E. Hancock, K. J. Indest, K. A. Gust and A. J. Kennedy, Environ. Toxicol. Chem., 2012, 31, 1438–1444 CrossRef CAS PubMed.
- C. Lu, D. J. Stewart, J. J. Lee, L. Ji, R. Ramesh, G. Jayachandran, M. I. Nunez, I. I. Wistuba, J. J. Erasmus, M. E. Hicks, E. A. Grimm, J. M. Reuben, V. Baladandayuthapani, N. S. Templeton, J. D. McMannis and J. A. Roth, PLoS One, 2012, 7, e34833 CAS.
- P.-H. Chen, K.-M. Hsiao and C.-C. Chou, Biomaterials, 2013, 34, 5661–5669 CrossRef CAS PubMed.
- Y. Yang, H. Zhu, V. L. Colvin and P. J. Alvarez, Environ. Sci. Technol., 2011, 45, 4988–4994 CrossRef CAS PubMed.
- Y. Cui, Y. Zhao, Y. Tian, W. Zhang, X. Lü and X. Jiang, Biomaterials, 2012, 33, 2327–2333 CrossRef CAS PubMed.
- T. Coccini, E. Roda, M. Fabbri, M. G. Sacco, L. Gribaldo and L. Manzo, J. Nanopart. Res., 2012, 14, 1–10 CrossRef.
- G. Gao, Y. Ze, B. Li, X. Zhao, T. Zhang, L. Sheng, R. Hu, S. Gui, X. Sang, Q. Sun, J. Cheng, Z. Cheng, L. Wang, M. Tang and F. Hong, J. Hazard. Mater., 2012, 243, 19–27 CrossRef CAS PubMed.
- K. M. Waters, L. M. Masiello, R. C. Zangar, B. J. Tarasevich, N. J. Karin, R. D. Quesenberry, S. Bandyopadhyay, J. G. Teeguarden, J. G. Pounds and B. D. Thrall, Toxicol. Sci., 2009, 107, 553–569 CrossRef CAS PubMed.
- L. K. Braydich-Stolle, B. Lucas, A. Schrand, R. C. Murdock, T. Lee, J. J. Schlager, S. M. Hussain and M. C. Hofmann, Toxicol. Sci., 2010, 116, 577–589 CrossRef CAS PubMed.
- S. Sharifi, S. Daghighi, M. M. Motazacker, B. Badlou, B. Sanjabi, A. Akbarkhanzadeh, A. T. Rowshani, S. Laurent, M. P. Peppelenbosch and F. Rezaee, Sci. Rep., 2013, 3, 2173 CAS.
- J. A. Khan, T. K. Mandal, T. K. Das, Y. Singh, B. Pillai and S. Maiti, Mol. BioSyst., 2011, 7, 1481–1486 RSC.
- V. Christen and K. Fent, Chemosphere, 2012, 87, 423–434 CrossRef CAS PubMed.
- J. Rauch, W. Kolch and M. Mahmoudi, Sci. Rep., 2012, 2, 868 Search PubMed.
- W. Busch, D. Kuhnel, K. Schirmer and S. Scholz, BMC Genomics, 2010, 11, 65 CrossRef PubMed.
- S. Tin Tin Win, S. Yamamoto, S. Ahmed, M. Kakeyama, T. Kobayashi and H. Fujimaki, Toxicol. Lett., 2006, 163, 153–160 CrossRef PubMed.
- Y. Yang, J. Wang, H. Zhu, V. L. Colvin and P. J. Alvarez, Environ. Sci. Technol., 2012, 46, 3433–3441 CrossRef CAS PubMed.
- Y. Zhang, H. Pan, P. Zhang, N. Gao, Y. Lin, Z. Luo, P. Li, C. Wang, L. Liu, D. Pang, L. Cai and Y. Ma, Nanoscale, 2013, 5, 5919–5929 RSC.
- H. C. Poynton, J. M. Lazorchak, C. A. Impellitteri, B. J. Blalock, K. Rogers, H. J. Allen, A. Loguinov, J. L. Heckman and S. Govindasmawy, Environ. Sci. Technol., 2012, 46, 6288–6296 CrossRef CAS PubMed.
- S. Y. Choi, S. Jeong, S. H. Jang, J. Park, J. H. Park, K. S. Ock, S. Y. Lee and S.-W. Joo, Toxicol. in Vitro, 2012, 26, 229–237 CrossRef CAS PubMed.
- M. Lundqvist, J. Stigler, G. Elia, I. Lynch, T. Cedervall and K. A. Dawson, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 14265–14270 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- P. AshaRani, S. Sethu, H. Lim, G. Balaji, S. Valiyaveettil and M. P. Hande, Genome Integr., 2012, 3, 2 CrossRef CAS PubMed.
- A. Choi, S. Brown, M. Szyf and D. Maysinger, J. Mol. Med., 2008, 86, 291–302 CrossRef CAS PubMed.
- R. Foldbjerg, J. Wang, C. Beer, K. Thorsen, D. S. Sutherland and H. Autrup, Chem.-Biol. Interact., 2013, 204, 28–38 CrossRef CAS PubMed.
- R. van Aerle, A. Lange, A. Moorhouse, K. Paszkiewicz, K. Ball, B. D. Johnston, E. de-Bastos, T. Booth, C. R. Tyler and E. M. Santos, Environ. Sci. Technol., 2013, 47, 8005–8014 CrossRef CAS PubMed.
- S. I. L. Gomes, A. M. V. M. Soares, J. J. Scott-Fordsmand and M. J. B. Amorim, J. Hazard. Mater., 2013, 254–255, 336–344 CrossRef CAS PubMed.
- S. I. L. Gomes, S. C. Novais, J. J. Scott-Fordsmand, W. De Coen, A. M. V. M. Soares and M. J. B. Amorim, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2012, 155, 219–217 CrossRef CAS PubMed.
- Y. J. Chae, C. H. Pham, J. Lee, E. Bae, J. Yi and M. B. Gu, Aquat. Toxicol., 2009, 94, 320–327 CrossRef CAS PubMed.
- D. A. Arndt, M. Moua, J. Chen and R. D. Klaper, Environ. Sci. Technol., 2013, 47, 9444–9452 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2014 |