Assessing DNA damage from enzyme-oxidized single-walled carbon nanotubes†
Shenmin Pana,
Naimish P. Sardesaia,
Hongyun Liub,
Dandan Lia and
James F. Rusling*acd
aDepartment of Chemistry, University of Connecticut, USA. E-mail: james.rusling@uconn.edu
bDepartment of Chemistry, Beijing Normal University, China
cDepartment of Cell Biology, University of Connecticut Health Center, USA
dInstitute of Materials Science, University of Connecticut, USA
First published on 2nd August 2013
Abstract
Peroxidase enzyme digests of oxidized single-wall carbon nanotubes (SWCNTs) were shown to damage DNA in potentially genotoxic reactions for the first time using an electro-optical array with and without metabolic activation.
Carbon nanotubes (CNTs) have found applications in various areas including electronics, nanomedicine, biosensors, and drug delivery.1,2 As the likelihood of exposure increases, concerns about the safety of CNTs in humans have been raised.3 In this communication, interactions between DNA and species produced by enzyme-catalyzed oxidation of carboxylated SWCNTs (cSWCNTs) were uncovered by using genotoxicity screening arrays with electrochemiluminescent (ECL) readout and confirmed by Comet assays. Carbonaceous fragments were observed during the oxidations by transmission electron microcopy (TEM). Results of ECL arrays suggest that potentially genotoxic reactions of oxidation products of cSWCNTs with DNA occur with and without metabolic bioactivation.
Genotoxicity denotes genetic damage induced by interactions of DNA with toxic materials or their metabolites. Genotoxicity bioassays such as the micronucleus test and Comet assays revealed that pristine and surface modified CNTs may damage DNA.4,5
Toxicity of CNTs appears to be related to the administration route, source, length, morphology, surface chemistry, and clearance rate.6,7 Inhaled pristine CNTs suppress the immune response in mice, inducing pulmonary inflammation and oxidative stress.3,8 Functionalization and coating of CNTs may decrease toxicity.9 For example, cSWCNTs become more hydrophilic when coated with polyethylene glycol (PEG), and showed less accumulation in vital organs than equivalent uncoated SWCNTs.10
CNTs in the environment and in vivo are likely to be slowly oxidized. The enzyme horseradish peroxidase (HRP) oxidizes cSWCNTs when activated by H2O2, and is a reasonable model for human oxidases that could interact with CNTs.11,12 Experiments and modeling were used to explore possible mechanisms of HRP-catalyzed degradation of carbon nanotubes12 (see Table S1†). Human myeloperoxidases may also catalyze the oxidation of cSWCNTs.13 Enzyme-oxidized cSWCNTs showed decreased pulmonary inflammation in mice.13 However, these studies provide minimal information concerning chemical effects of CNTs oxidation products or their metabolites on DNA.
In this work, we employ novel molecular-based in vitro screening arrays that we have developed for assessing possible genotoxic reactivity. These electro-optical arrays produce metabolites from test chemicals and detect their subsequent damage to DNA.14 Arrays feature multiple spots combining metabolic enzymes, DNA and ECL light emitting polymer ruthenium(II)polyvinylpyridine [Ru(bpy)2PVP10]2+ (RuPVP) on a conductive pyrolytic graphite (PG) chip. In the reaction step, metabolic enzymes in these spots convert test chemicals into metabolites that can interact with DNA to induce structural changes, e.g. SN2 addition to a nucleobase or generation of reactive oxidants (ROS) that oxidize DNA. In the detection step, RuIIPVP is electrochemically oxidized to RuIIIPVP, which reacts with intact guanines in DNA to produce excited state *RuIIPVP sites that decay to give ECL light at 610 nm.15 Detection of DNA damage involves applying voltage to the chip and capturing the ECL image using a CCD camera. Compared with intact dsDNA, damaged DNA increases ECL output since the partially disrupted DNA structure provides better access of guanines to RuIIIPVP sites, thus increasing the reaction rate with guanines.14 The increase of ECL with enzyme reaction time correlates with the rate of DNA damage as confirmed by LC-MS determination of individual nucleobase adducts.14
High purity pristine SWCNTs were carboxylated using piranha solution to produce cSWCNTs.11,12 This treatment is widely used to make drug delivery and gene therapy vehicles.1,10 The resulting cSWCNTs were characterized by Raman spectroscopy (ESI, Fig. S1†). cSWCNTs were biodegraded by HRP activated by H2O2 as reported previously (S).11,12 Negative controls (C1, C2, C3) were incubated without HRP or without H2O2 or without both (Table 1). To ensure adequate HRP activity during degradation, fresh H2O2 was added daily, and fresh HRP was added every 5 days (see ESI† for details). For TEM analyses, HRP-degraded cSWCNTs were filtered with 3000 molecular weight cutoff filters and washed to remove salts. Images show tubular bundles of cSWCNTs in decreased amounts with increased degradation time (Fig. 1). Small particulate residues (red arrows) were observed after 3 days, with more residues after 10 days, but were not visible before degradation (Fig. 1A). Similar residues were observed by TEM of cSWCNT degradation products by Star et al.,11,12 and may be carbonaceous products resulting from nanotube oxidation.
 | ||
| Fig. 1 TEM micrographs confirming enzyme-catalyzed degradation of cSWCNTs. Samples were prepared using 3000 molecular weight cutoff filters with washing to remove salt. | ||
Alternatively, degraded cSWCNTs were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min and the supernatant was discarded to remove salts. Fig. S3† shows TEM images suggesting tubular bundles of cSWCNTs at decreased amounts with increased degradation time. This result suggests that some long cSWCNTs are shortened, and smaller particles possibly remain dispersed during centrifugation.
000g for 10 min and the supernatant was discarded to remove salts. Fig. S3† shows TEM images suggesting tubular bundles of cSWCNTs at decreased amounts with increased degradation time. This result suggests that some long cSWCNTs are shortened, and smaller particles possibly remain dispersed during centrifugation.
A major fraction of chemicals that cause toxicity undergo bioactivation in the liver to generate active metabolites that damage DNA.14 Human liver microsomes (HLM) were included in the ECL array as a source of cytochrome (cyt) P450s, the enzyme family responsible for most metabolic oxidations.16 An NADPH regeneration system in the buffer activates the cyt P450s. Cytosolic bioconjugation enzymes were also included for a more complete metabolic representation.
After HRP oxidation, dispersions S, C1, C2 and C3 (Table 1) were acidified by 0.1 M HCl and extracted 3× with ethyl acetate. After removing solvent, the residue was reconstituted in 50 μL acetonitrile, and 4 μL were added to incubation buffer containing NADPH (see ESI†) to activate cyt P450 enzymes on the arrays. These solutions were spotted onto ECL arrays and incubated.
Fig. 2 presents reconstructed, colorized ECL images of a representative set of array spots containing HLM and the cytosol exposed to extracts of degraded cSWCNTs. Styrene and benzo[a]pyrene (B[a]P) were used as positive controls. Increased ECL was found after exposure to extracts for DNA/RuPVP/enzyme spots with and without NADPH. This suggests DNA damage both with and without metabolic activation. Metabolized extracts (+NADPH) produced more ECL light than extracts with no metabolic activation (−NADPH). Extracts from negative controls (C1, C2 and C3) did not induce a significant ECL increase, suggesting no DNA damage.
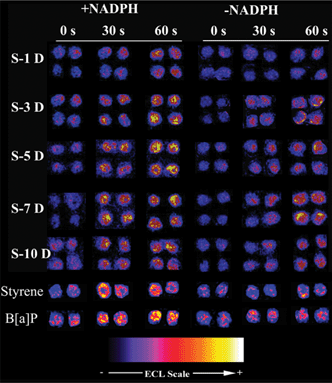 | ||
| Fig. 2 Reconstructed ECL array images of DNA/RuPVP/enzyme spots exposed to extracts of cSWCNTs degraded by HRP for 0, 30 s and 60 s of incubation on the chip. S-xD denotes extracts from solution S after x days (D) of degradation. Enzyme sources in the spots were human microsomes + cytosol. +NADPH indicates an NADPH regeneration system was included and −NADPH indicates it was not included in the assay. | ||
Array results from cSWCNT extracts were presented as % ECL increase over 0 s ECL (Fig. 3) along with styrene and B[a]P. Error bars reflect reproducibility within ±10% for ≥8 replicates. Styrene is moderately genotoxic,17 and is metabolized by HLM to styrene oxide that attacks DNA at N7 of guanine. B[a]P is a more potent genotoxic agent and produces a more reactive metabolite, B[a]P diol epoxide (BPDE) that adducts guanines and adenines in DNA.18,19 ECL intensity increased ∼10% for 1 mM styrene and ∼50% for 25 μM B[a]P after 1 min enzyme reaction (Fig. 3). The ECL increases of spots exposed to C1 (black bars) were all within 5%, indicating negligible DNA damage. ECL after incubation with S with or without metabolic activation increased 10 to 50%. This suggests that some products in the extract react with DNA directly without bioactivation, and that bioactivation generates additional DNA damage. Metabolized extracts induced ∼2 fold more ECL (red bars) than those with no bioactivation (blue bars). % ECL increase reached a peak at ∼7 days.
![ECL increase over the 0 s level at different cSWCNT biodegradation times for incubation on the RuPVP/DNA/HLM chip of 1 min. Red solid bars are array spots incubated with the S extract in the presence of NADPH, blue solid bars denote ECL array spots incubated with the S extract in the absence of NADPH, and black bars denote ECL array spots incubated with the C1 extract in the presence of NADPH. Lighter red and blue bars represent the positive controls incubated with either 1 mM styrene or 25 μM B[a]P in the presence of NADPH. Error bars represent the standard deviations of responses from 8 array spots exposed to cSWCNT extracts obtained from two individual degradation experiments. (See ESI for experiment details.)](/image/article/2013/TX/c3tx50022e/c3tx50022e-f3.gif) | ||
| Fig. 3 ECL increase over the 0 s level at different cSWCNT biodegradation times for incubation on the RuPVP/DNA/HLM chip of 1 min. Red solid bars are array spots incubated with the S extract in the presence of NADPH, blue solid bars denote ECL array spots incubated with the S extract in the absence of NADPH, and black bars denote ECL array spots incubated with the C1 extract in the presence of NADPH. Lighter red and blue bars represent the positive controls incubated with either 1 mM styrene or 25 μM B[a]P in the presence of NADPH. Error bars represent the standard deviations of responses from 8 array spots exposed to cSWCNT extracts obtained from two individual degradation experiments. (See ESI† for experiment details.) | ||
Comet assays validated the ECL array results. In Comet assays, damaged DNA in cells produces a “tail” during electrophoresis. The product of tail length and fractional amount of DNA in the tail is related to the extent of DNA damage.20,21 Results in Fig. S4 in ESI† show that cells incubated with S-3D, S-5D and S-7D extracts presented more obvious tailing from the cell nucleus compared with those incubated with control extracts (C1-5D, C2-5D and C3-5D) or buffer. Quantitative results presented in Fig. S5 ESI† obtained by averaging tail moments of 10 cells clearly showed that maximum DNA damage occurred from S-7D, followed by S-5D and S-3D. These results were similar to those from ECL arrays (Fig. 3).
The results above clearly indicate reactivity of oxidative degradation products of carbon nanotubes with DNA that may lead to possible genotoxicity in living systems. Significantly, extracts from HRP-degraded cSWCNTs induce DNA damage with or without metabolic activation (Fig. 2 and 3, Fig. S4 and S5†).
There are several studies with which our findings can be meaningfully compared. Using HRP treatment of cSWCNTs, Star et al. observed carbonaceous intermediates in TEM as we did (Fig. 1 and Fig. S3†), and increased carbon dioxide evolution over 10 days.12 They speculated that oxidized polyaromatic hydrocarbons form during degradation based on mass spectrometry results.12 However, degradation of cSWCNTs was not complete in our study even after a month, although active HRP was maintained in the degradation solution. Our degradation solutions were extremely complex mixtures in which it was not possible to identify significant amounts of individual molecules using LC-MS or GC-MS. Clearly, some low MW compounds could be volatilized during sample handling, may be present below our detection limits, or are poorly resolved from the complex mixture in LC- or GC-MS. Our observations are consistent with a report that cSWCNTs are degraded slowly and incompletely.22 Furthermore, degradation starts from defect areas of cSWCNTs12 and may depend on the density of defects originally present.23 It is possible that our starting cSWCNTs had less defects compared with the CNTs used by Star, and thus underwent slower degradation. However, all studies show that oxidative degradation of cSWCNTs by peroxidases occurs.
Enzymatic degradation of cSWCNTs could yield products including oxidized polycyclic aromatic hydrocarbons (PAH) like phenols or phenolic acids. They have been speculated as degradation products from the oxidation of CNTs by HRP or myeloperoxidase.12,13 In another study of carboxylated SWCNT degradation by H2O2 alone, fluorescent products were observed23a that may be related to PAHs. We found no fluorescence in our extracts. If oxidized PAHs are produced during our experiments, we suspect that they undergo further oxidation to yield smaller molecules.
There are many reports demonstrating that cSWCNTs cross cell membranes and even interact with nuclei.6,10,24 However, intermediates generated during degradation in vivo can be a genotoxicity concern. The ECL array and Comet assay results strongly suggest that some products of oxidized SWCNTs can damage DNA directly or after bioactivation. An important issue is whether cSWCNT oxidative degradation occurs in humans. A major challenge in drug delivery vehicles, for example, is to deliver the drug on a cSWCNT carrier and have the cSWCNT excreted by the body shortly afterwards, so that it has less time to degrade or embed in organs. A few studies suggest that this might be achieved by better dispersing the cSWCNTs by wrapping with PEG.10,25 Our work suggests that additional studies of toxicity of cSWCNTs need to focus on whether and to what extent DNA damage can occur in vivo, and also on detection of primary nucleobase adducts.
In summary, extracts from 5–10 day degradation of cSWCNTs by peroxide-activated HRP analyzed by an ECL array involving metabolic activation revealed damaged DNA to a similar extent as 25 μM B[a]P, a potent genotoxic agent. About half of the DNA damage did not require metabolic activation in the array as B[a]P does. Comet assays supported direct damage to cell DNA from cSWCNT degradation products. Degradation of the nanotubes by HRP oxidation was slow, and DNA damage peaked at 7 days. This result suggests that rapid clearance (e.g. <1 day) might protect humans from this kind of DNA damage resulting from carbon nanotube products, such as drug delivery vehicles.
The authors gratefully acknowledge financial support from the National Institute of Environmental Health Sciences (NIEHS), NIH (grant no. ES03154) and Natural Science Foundation of China (NSFC 21105004). The authors thank Dr Xiuling Lu, School of Pharmacy, University of Connecticut, for the gift of the A549 cells.
Notes and references
- (a) S. N. Kim, J. F. Rusling and F. Papadimitrakopoulos, Adv. Mater., 2007, 19, 3214–3228 CrossRef CAS; (b) M. F. L. deVolder, S. H. Tawfick, R. H. Baughman and A. J. Hart, Science, 2013, 339, 535–539 CrossRef CAS.
- H. C. Wu, X. Chang, L. Liu, F. Zhao and Y. Zhao, J. Mater. Chem., 2010, 20, 1036–1052 RSC.
- (a) C. W. Lam, J. T. James, R. McCluskey and R. L. Hunter, Toxicol. Sci., 2004, 77, 126–134 CrossRef CAS; (b) C. W. Lam, J. T. James, R. McCluskey, S. Arepalli and R. L. Hunter, Crit. Rev. Toxicol., 2006, 36, 189–217 CrossRef CAS.
- H. K. Lindberg, G. C. Falck, S. Suhonen, M. Vippola, E. Vanhala, J. Catalan, K. Savolainen and H. Norppa, Toxicol. Lett., 2009, 186, 166–173 CrossRef CAS.
- E. Petersen and B. Nelson, Anal. Bioanal. Chem., 2010, 398, 613–650 CrossRef CAS.
- K. Kostarelos, Nat. Biotechnol., 2008, 26, 774–776 CrossRef CAS.
- S. Beg, M. Rizwan, A. M. Sheikh, M. S. Hasnain, K. Anwer and K. Kohli, J. Pharm. Pharmacol., 2011, 63, 141–163 CrossRef CAS.
- L. A. Mitchell, F. T. Lauer, S. W. Burchiel and J. D. McDonald, Nat. Nanotechnol., 2009, 4, 451–456 CrossRef CAS.
- Z. Liu, X. Sun, N. Nakayama-Ratchford and H. Dai, ACS Nano, 2007, 1, 50–56 CrossRef CAS.
- A. A. Bhirde, S. Patel, A. A. Sousa, V. Patel, A. A. Molinolo, Y. Ji, R. D. Leapman, J. S. Gutkind and J. F. Rusling, Nanomedicine, 2010, 5, 1535–1546 CrossRef CAS.
- B. L. Allen, P. D. Kichambare, P. Gou, I. I. Vlasova, A. A. Kapralov, N. Konduru, V. E. Kagan and A. Star, Nano Lett., 2008, 8, 3899–3903 CrossRef CAS.
- B. L. Allen, G. P. Kotchey, Y. Chen, N. V. K. Yanamala, J. Klein-Seetharaman, V. E. Kagan and A. Star, J. Am. Chem. Soc., 2009, 131, 17194–17205 CrossRef CAS.
- V. E. Kagan, N. V. Konduru, W. Feng, B. L. Allen, J. Conroy, Y. Volkov, I. I. Vlasova, N. A. Belikova, N. Yanamala, A. Kapralov, Y. Y. Tyurina, J. Shi, E. R. Kisin, A. R. Murray, J. Franks, D. Stolz, P. Gou, J. Klein-Seetharaman, B. Fadeel, A. Star and A. A. Shvedova, Nat. Nanotechnol., 2010, 5, 354–359 CrossRef CAS.
- (a) J. F. Rusling, E. G. Hvastkovs and J. B. Schenkman, in Drug Metabolism Handbook, ed. A. Nassar, P. F. Hollenburg, and J. Scatina, J. Wiley, N.J., 2009, pp. 307–340 Search PubMed; (b) E. G. Hvastkovs, J. B. Schenkman and J. F. Rusling, Annu. Rev. Anal. Chem., 2012, 5, 79–105 CrossRef CAS.
- L. Dennany, R. J. Forster and J. F. Rusling, J. Am. Chem. Soc., 2003, 125, 5213–5218 CrossRef CAS.
- P. R. Ortiz de Montellano, Cytochrome P450, Kluwer/Plenum, New York, 3rd edn, 2005 Search PubMed.
- K. Hemminki and P. Vodicka, Toxicol. Lett., 1995, 77, 153–161 CrossRef CAS.
- PAHs and Related Compounds, ed. A. H. Neilson, Springer, Berlin, 1998 Search PubMed.
- T. Shimada, E. M. J. Gillam, Y. Oda, F. Tsumura, T. R. Sutter, F. P. Guengerich and K. Inoue, Chem. Res. Toxicol., 1999, 12, 623–629 CrossRef CAS.
- (a) P. L. Olive, J. P. Banáth and R. E. Durand, Radiat. Res., 1990, 122, 86–94 CrossRef CAS; (b) P. L. Olive, J. P. Banáth and R. E. Durand, J. Natl. Cancer Inst., 1990, 82, 779–783 CrossRef CAS.
- S. Brendler-Schwaab, A. Hartmann, S. Pfuhler and G. Speit, Mutagenesis, 2005, 20, 245–254 CrossRef CAS.
- J. Russier, C. Menard-Moyon, E. Venturelli, E. Gravel, G. Marcolongo, M. Meneghetti, E. Doris and A. Bianco, Nanoscale, 2011, 3, 893–896 RSC.
- (a) X. Liu, R. H. Hurt and A. B. Kane, Carbon, 2010, 48, 1961–1969 CrossRef CAS; (b) I. I. Vlasova, A. V. Sokolov, A. V. Chekanov, V. A. Kostevich and V. B. Vasil'ev, Bioorg. Khim., 2011, 37, 510–521 CAS.
- S. Patel, A. A. Bhirde, J. F. Rusling, X. Chen, J. S. Gutkind and V. Patel, Pharmaceutics, 2011, 3, 34–52 CrossRef CAS.
- Z. Liu, C. Davis, W. Cai, L. He, X. Chen and H. Dai, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 1410–1415 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c3tx50022e |
| This journal is © The Royal Society of Chemistry 2013 |
